Abstract
We report the characterization of a novel factor, Nob1p (Yor056c), which is essential for the synthesis of 40S ribosome subunits. Genetic depletion of Nob1p strongly inhibits the processing of the 20S pre-rRNA to the mature 18S rRNA, leading to the accumulation of high levels of the 20S pre-rRNA together with novel degradation intermediates. 20S processing occurs within a pre-40S particle after its export from the nucleus to the cytoplasm. Consistent with a direct role in this cleavage, Nob1p was shown to be associated with the pre-40S particle and to be present in both the nucleus and the cytoplasm. This suggests that Nob1p accompanies the pre-40S ribosomes during nuclear export. Pre-40S export is not, however, inhibited by depletion of Nob1p.
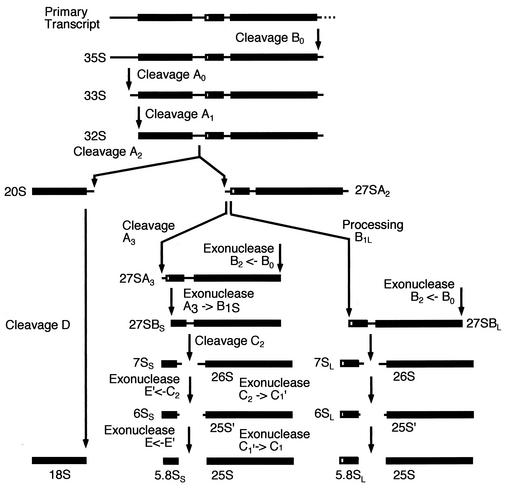
Eukaryotic ribosome synthesis takes place largely within the nucleolus, but the late steps in both pre-40S and pre-60S maturation occur after export of the subunits to the cytoplasm. In Saccharomyces cerevisiae, this cytoplasmic maturation includes an endonuclease cleavage at site D, the 3′ end of the 18S rRNA that generates the mature rRNA from the 20S pre-rRNA (Fig. 1) (27, 37). Around 150 proteins and 80 small nucleolar RNAs are known to be required for the posttranscriptional steps in yeast ribosome synthesis. Despite the identification of this large number of factors, there are a great many gaps in our knowledge. Among these are the identities of the endonucleases that perform several of the pre-rRNA cleavages including that at the 3′ end of the mature 18S rRNA (Fig. 1). The pathway of ribosome subunit synthesis in yeast was initially studied by sucrose gradient centrifugation (16, 36, 38), leading to the identification of a 90S particle that contains the 35S pre-rRNA, a 66S particle that contains the 27S pre- rRNAs, and a 43S particle containing the 20S pre-rRNA. Recent analyses have shown that the precursors to the 60S particle are substantially more complex than was initially proposed, with multiple pre-60S particles containing different complements of the precursors to the 25S and 5.8S rRNAs and different protein compositions (6, 13, 28, 31; reviewed in references 7 and 41). Equivalent analyses of the pre-40S ribosomes have not yet been reported, but a high-throughput proteomic analysis of yeast protein interactions identified three complexes that potentially represented such particles (8; reviewed in reference 7), one of which was associated with tandem affinity purification (TAP)-tagged Nob1p (Yor056c).
FIG. 1.
Pre-rRNA processing pathway. In wild-type cells, the 35S pre-rRNA is cleaved at site A0, producing the 33S pre-rRNA. This molecule is rapidly cleaved at site A1 to produce the 32S pre-rRNA, which is cleaved at site A2 releasing the 20S and 27SA2 pre-rRNAs. The 20S pre-rRNA is exported to the cytoplasm, where it is dimethylated by Dim1p and then cleaved at site D, by an unidentified enzyme, to generate the mature 18S rRNA. 27SA2 is processed via two alternative pathways. It is either cut at site A3 to generate 27SA3, which is then trimmed to site B1S, producing 27SBS. Alternatively, it can be processed to 27SBL by an as yet unknown mechanism. 27SBS and 27SBL are matured to the 5.8S and 25S following identical pathways. Cleavage at site C2 generates the 7S and 26S pre-rRNAs. The 7S pre-rRNA is digested 3′ to 5′ to 6S pre-rRNA and then to the mature 5.8S rRNA. The 26S pre-rRNA is digested 5′ to 3′ to the 25S pre-rRNA and then to the mature 25S rRNA. For a review on pre-rRNA processing and the known processing enzymes, see reference (40).
Each of these complexes contained Tsr1p, which is required for 20S-to-18S processing (9), and one also contained the dimethylase Dim1p, which modifies the 20S pre-rRNA, probably in the cytoplasm, and is additionally required for nucleolar pre-rRNA processing (4, 19, 22). Rrp10p (Rio1p) is also required for 20S-to-18S processing (39) but was not identified in these putative pre-40S particles.
Nob1p (for “Nin one binding protein”) was initially reported to interact in a two-hybrid screen with Nin1p/Rpn12p, a subunit of the 19S regulatory particle of the yeast 26S proteosome (34), and is required for proteosome biogenesis (35). Immunoprecipitation of Nob1-TAP did not detect the interaction with Nin1p (8), but Nob1p was copurified with each of the putative pre-40S complexes. Sequence analyses indicated that Nob1p is an evolutionarily conserved protein with a predicted RNA-binding domain at the C terminus and an N-terminal PIN domain (24), a motif proposed to be associated with RNase activity (5). These features made suggested that it might be the long-sought 18S rRNA 3′ endonuclease, and its role in ribosome synthesis was therefore assessed. Here we report that Nob1p is indeed a component of pre-40S ribosomal particles and is strictly required for cleavage of the 20S pre-rRNA to generate the mature 18S rRNA.
MATERIALS AND METHODS
Strains and microbiological techniques.
Standard techniques were employed for growth and handling of yeast. The yeast strains used in this work were: BMA64 (MATa ade2-1 his3-11,15 leu2-3,112 trp1-Δ ura3-1), YAF34 (MATa ade2-1 his3-11,15 leu2-3,112 trp1-Δ ura3-1 KAN::GAL::HA-NOB1), BMA38 (MATa his3-Δ200 leu2-3,112 ura3-1 trp1-Δ ade2-1), YAF36 (MATa his3-Δ200 leu2-3,112 ura3-1 trp1-Δ ade2-1 NOB1-TAP::TRP1). Strain YAF34 was created from strain BMA64 by use of a one-step PCR strategy as previously described (20, 23). TAP tagging of Nob1p was performed as described previously (29).
RNA extraction, Northern hybridization, and primer extension.
For depletion of the Nob1p protein, cells were harvested at intervals following a shift from RSG medium (2% galactose, 2% sucrose, 2% raffinose), or YPGal medium containing 2% galactose, to YPD medium containing 2% glucose. Otherwise strains were grown in YPD medium. RNA was extracted as described previously (17). Northern hybridizations and primer extensions were as described previously (17). Standard 1.2% agarose-formaldehyde and 6% acrylamide-urea gels were used to analyze the high- and low-molecular-weight RNA species, respectively.
Oligonucleotides.
For RNA hybridization and primer extension, the following oligonucleotides were used: 001 (5′-CCAGTTACGAAAATTCTTG), 002 (5′-GCTCTTTGCTCTTGCC), 003 (5′-TGTTACCTCTGGGCCC), 004 (CGGTTTTAATTGTCCTA), 005 (5′-ATGAAAACTCCACAGTG), 006 (5′-GGCCAGCAATTTCAAGTTA), 007 (5′-CTCCGCTTATTGATATGC), 008 (5′-CATGGCTTAATCTTTGAGAC), 013 (5′-GCGTTGTTCATCGATGC), 020 (5′-TGAGAAGGAAATGACGCT), 029 (TAATGATCCTTCCGCA), 033 (5′-CGCTGCTCACCAATGG), 041 (5′-CTACTCGGTCAGGCTC), anti-NOB1 (AACGCCCTTACATGTGCGGTTTGGTTTTCGGTCAT), and ITS1 (5′-TGGACTC5CCATCTCTTGAC5TCTTGCCCAG5AAAAGCTC5CATGCTC5T).
Sucrose gradient analysis and affinity purification.
Sucrose gradient centrifugation was performed as described previously (3, 33). RNA was extracted from each fraction and resolved on a standard 1.2% agarose-formaldehyde gel. Mature rRNAs and pre-rRNA species were detected by ethidium bromide staining and Northern hybridization, respectively. Sedimentation of proteins was assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and TAP-tagged Nob1p was detected by Western immunoblotting with peroxidase-conjugated rabbit immunoglobulin G (Sigma). Affinity purification of TAP-tagged Nob1p and analysis of copurified RNAs were performed as previously described (7).
Pulse-chase labeling.
Metabolic labeling of RNA was performed as described previously (6). The strains GAL::HA-nob1 and BMA64 were transformed with a plasmid containing the URA3 gene, pregrown in galactose medium lacking uracil, and transferred to glucose minimal medium for 8 h. Cells with an optical density at 600 nm of 0.3 units were labeled with [5,6-3H]uracil for 1 min; this was followed by a chase with excess unlabeled uracil for 0, 1, 2.5, 5, 10, and 20 min. Standard 1.2% agarose-formaldehyde and 6% polyacrylamide-urea gels were used to analyze the high- and low-molecular-weight RNA species, respectively.
Fluorescence microscopy.
Indirect immunofluorescence was performed with the Nob1-TAP strain as previously described (11). Plasmid pUN100 DsRed-Nop1 was introduced into yeast cells by transformation and selected on SD-URA medium (containing 0.67% yeast nitrogen base and 2% glucose and lacking uracil). Individual transformants were grown in selective medium, fixed by incubation in 4% (vol/vol) formaldehyde for 30 min at 25°C, and spheroplasted. The TAP fusion was detected with a rabbit anti-protein A antibody (Sigma) and a secondary goat anti-rabbit antibody coupled to fluorescein isothiocyanate (Sigma) at 1:100 and 1:200 dilutions, respectively. Fluorescent in situ hybridization was performed with the GAL::nob1 strain as previously described (2) with some modifications. The probe for the ITS1 5′ RNA was a 50-bp oligonucleotide fluorescently labeled with Cy3. Cells were grown to an optical density of 0.5 to 1.0 unit at 600 nm and fixed with 2% (vol/vol) formaldehyde-5% (vol/vol) acetic acid for 10 min at 25°C. Samples were hybridized with 100 ng of Cy3-labeled RNA oligonucleotide for 16 h at 37°C. To stain nuclear DNA, 4′,6-diamidino-2-phenylindole (DAPI) was included in the mounting medium (Vectashield; Vector Laboratories). Cells were viewed on a Zeiss Axioscop fluorescence microscope, and pictures were obtained with a Xillix Microimager charge-coupled device camera.
RESULTS
Nob1p is a conserved protein with putative nuclease and RNA-binding domains.
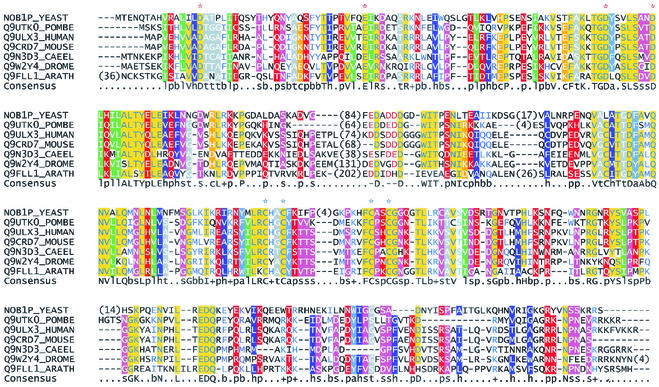
Sequence analyses using BLASTP (1) revealed that Nob1p has close relatives in higher eukaryotes. Each of these aligns with Nob1p over the entire length (Fig. 2), and they are all confidently predicted to be orthologues. Subsequent searches using PSI-BLAST (1) identified archaeal proteins similar to the N-terminal half of Nob1p. Each of these proteins shares with Nob1p a region with a compact predicted structure, termed a PIN domain. This domain was originally proposed to have a function in signaling (24), but a more recent analysis has proposed that the PIN domain is associated with a phosphodiesterase activity (5). This was based on the predicted structure and the complete conservation of several acidic residues (denoted by red asterisks in Fig. 2) between the PIN domain and the 5′ exonuclease domain of DNA polymerase I. This degrades double-stranded nucleic acids exonucleolytically but also has a single-stranded DNA “flap” endonuclease activity. PIN domains were identified in several proteins involved in RNA degradation, including the eukaryotic 3′ → 5′ exonuclease Dis3p/Rrp44p, the worm mRNA turnover factor Smg5p, and the putative yeast turnover factor Nmd4p (5). In archaeal proteins, the PIN domain is immediately followed by a short zinc ribbon module, whereas in eukaryotes, these two regions are separated by a domain of variable length. Metal-binding zinc ribbons have four conserved cysteines or histidines (marked by blue asterisks in Fig. 2) and are commonly found in ribosomal proteins (25). The C-terminal part of Nob1p is missing in related archaeal proteins but is present and well conserved in the eukaryotic orthologues. There was, however, no convincing sequence similarity between this region of the Nob1p family and known protein domains. We therefore attempted to identify related domains by fold recognition. The results obtained from the 3D-PSSM server (15; http://www.sbg.bio.ic.ac.uk/∼3dpssm/) indicate that this part of Nob1p is similar to ribosomal protein L2/L8. The primary sequence homology is not statistically significant, but its relevance is supported by a good match between predicted and known secondary-structure elements of these proteins and raises the possibility that this domain has a function in RNA binding.
FIG. 2.
Multiple sequence alignment of Nob1p and related proteins. The full sequences of Nob1p and its putative eukaryotic orthologues were aligned using T-Coffee and corrected manually where required. Sequences are identified by the SwissProt number and species abbreviation. Numbers within the sequences indicate residues omitted due to unreliable alignments in these regions. Letters and symbols on the consensus line are as follows: s, small residues; t, tiny; b, big; h, hydrophobic; a, aromatic; l, aliphatic; p, polar; c, charged; −, negatively charged; +, positively charged. Individual residues with more than 80% identity in the entire alignment are shown as capital letters on the consensus line. Conserved acidic (Asp/Glu) residues in the PIN domain are denoted by red asterisks above the alignment. Conserved cysteines marked by blue asterisks are part of the zinc ribbon motif. The figure was prepared using CHROMA (10).
Nob1p associates with pre-40S ribosomal particles.
A systematic high-throughput proteomic analysis (8) identified Nob1p (Yor056c) in complexes that also included several proteins previously implicated in 40S biogenesis, including Enp1p, Tsr1p, Rrp12p, Dim1p, and Rio2p, a homologue of Rrp10p (Rio1p) (9, 12, 21, 30; M. Oeffinger and D. Tollervey, unpublished data). Nob1p was also coprecipitated with overexpressed, tagged Tsr1p (14). These observations suggested that Nob1p might be a component of a pre-40S ribosomal particle.
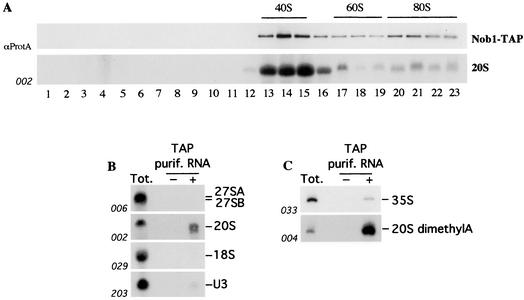
To monitor the distribution and associations of Nob1p, a strain expressing a C-terminal TAP tag (29) fusion with Nob1p under the control of its own promoter was constructed using of a one step-PCR strategy (20). The fusion construct supported wild-type growth, indicating that it was fully functional (data not shown). To assess the association of Nob1p with pre-ribosomal particles, a sucrose gradient analysis was performed with a lysate from the Nob1-TAP strain. The sedimentation of Nob1-TAP was determined by Western blotting with antibodies that recognize the protein A moiety of the TAP tag and was compared to that of the rRNA species and pre-rRNAs (Fig. 3). Comparison to the positions of the mature rRNAs detected by ethidium staining (data not shown) indicated that Nob1-TAP is enriched in the 40S region of the gradient, with a weaker peak around 80S to 90S (Fig. 3A). Northern hybridization showed that the peak of Nob1-TAP coincides with the peak of the 20S pre-rRNA (Fig. 3A). Consistent with the presence of Nob1p in purified preribosomal complexes, these analyses indicate that Nob1p is a component of the pre-40S particles.
FIG. 3.
Nob1p is associated with pre-40S ribosomal particles. (A) The upper panel shows sedimentation of TAP-tagged Nob1p on a 10 to 50% sucrose gradient. The levels of the Nob1-TAP protein were determined by immunoblot analysis. The lower panel shows Northern analysis of the levels of the 20S pre-rRNA. Positions of 40S and 60S ribosomal subunits and 80S ribosomes are indicated, as determined by ethidium staining of the RNA recovered from each fraction (data not shown). (B and C) Northern analysis (B) and primer extension analyses (C) of rRNAs and pre-rRNAs coprecipitated with Nob1-TAP. RNA was extracted from whole cells (Tot.) and affinity-purified fractions from tagged (+ lane) and nontagged isogenic wild-type strain (− lane). The Northern blot membrane was consecutively hybridized with the probes indicated (see Fig. 6A for the locations of the probes used). The primer extension stop detected with oligonucleotide 004 is due to the cytoplasmic dimethylation of the 20S pre-rRNA at two consecutive A residues near the 3′ end of the 18S rRNA.
RNA species that coprecipitated with Nob1-TAP were analyzed by Northern hybridization (Fig. 3B) and primer extension (Fig. 3C). The 20S pre-rRNA was strongly precipitated with Nob1p, consistent with association with pre-40S particles. Primer extension showed that 20S pre-rRNA associated with Nob1p had undergone adenine dimethylation (Fig. 3C). This modification is performed by Dim1p at two sites, m26A1779 and m26A1780, near the 3′ end of the 18S rRNA (19) and is reported to occur in the cytoplasm (4), suggesting that Nob1p is exported to the cytoplasm as a component of the pre-40S ribosomes. In agreement with the sedimentation data, weaker precipitation was seen for the 35S pre-rRNA and the U3 snoRNA, both of which are components of the 90S pre-ribosomes. No clear precipitation was seen for the mature 18S rRNA or the 27SA and 27SB pre-rRNA components of the pre-60S particles.
Construction of a conditional NOB1 allele.
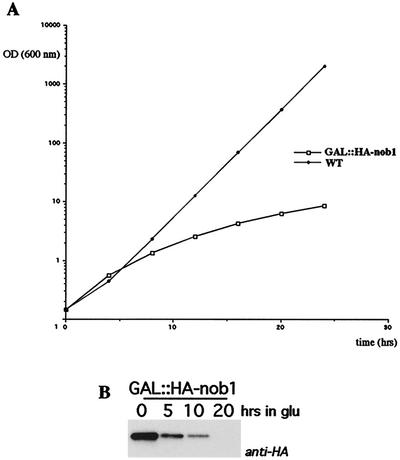
The NOB1 gene is essential for viability (34), and a conditional allele was therefore constructed. A one-step PCR technique was used to create a chromosomal N-terminally HA-tagged Nob1p fusion construct under the control of the repressible GAL1 promoter, as previously described (20, 23). The chromosomal GAL::HA-nob1 construct is the only source of Nob1p in this strain. In galactose liquid media, growth of the GAL::HA-nob1 strain was identical to that of the isogenic wild-type strain (data not shown), showing the fusion protein to be functional. Following transfer to glucose liquid medium, the growth rates of both strains were initially the same, but growth of the GAL::HA-nob1 strain decreased progressively, commencing 6 h after transfer (Fig. 4A). By 20 h after transfer, growth was severely reduced. Western blot analysis showed that the decrease in growth rate on glucose medium was concomitant with depletion of HA-Nob1p (Fig. 4B).
FIG. 4.
Depletion of Nob1p inhibits growth. (A) Growth rates of the GAL::HA-nob1 (squares) and wild-type (diamonds) strains following a shift from galactose to glucose medium. (B) Western blot analysis of Nob1p depletion. Whole-cell extracts were prepared from samples harvested at the indicated times. Equal amount of proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide), and the HA tag on HA-Nob1p was detected by Western blotting.
Nob1p is required for processing of the 20S pre-rRNA.
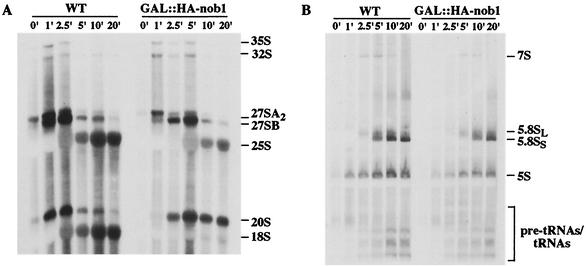
The association of Nob1p with pre-40S particles suggested its involvement in subunit maturation. Pre-rRNA processing was initially assessed by pulse-chase labeling in vivo with [5,6-3H]uracil 8 h after transfer to glucose medium (Fig. 5). Processing of the 35S pre-rRNA was mildly delayed in the Nob1p-depleted strain, with a concomitant delay in the appearance of the 32S, 27SA, 27SB, and 7S pre-rRNAs and the mature 25S and 5.8S rRNAs. A much more dramatic defect was seen for the 20S pre-rRNA, which was apparently stable over the chase time course, with very little conversion to the mature 18S rRNA. Even after a 20-min chase, the mature 18S was nearly undetectable, while there was a strong accumulation of 20S pre-rRNA.
FIG. 5.
Depletion of Nob1p inhibits 18S rRNA synthesis. GAL::HA-Nob1 and the isogenic wild-type (WT) strain were grown at 30°C in SDGal-URA medium and then transferred to SDGlu-URA for 8 h. The cells were pulse-labeled with [5,6-3H]uracil for 1 min and then chased with an excess of cold uracil. Total RNA was extracted from cell samples harvested at the indicated time points and resolved on 1.2% agarose-formaldehyde (A) and 6% acrylamide-urea (B) gels. The positions of mature rRNAs, pre-rRNAs and tRNA species are indicated.
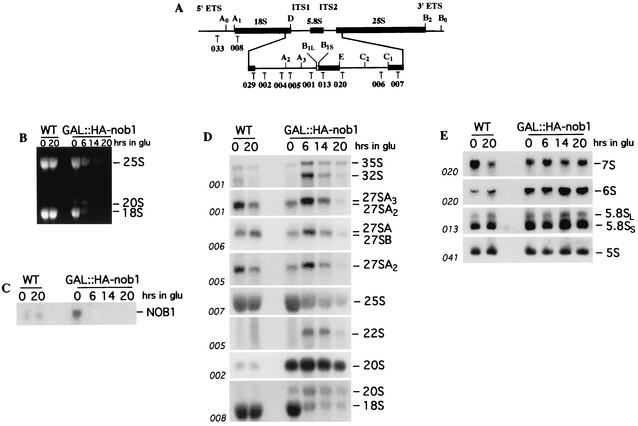
To further characterize pre-rRNA processing in the GAL::HA-nob1 mutant, steady-state levels of mature and precursor rRNA molecules were assessed by Northern hybridization with oligonucleotides specific for the pre-rRNAs and mature rRNAs (Fig. 6A). Strikingly, ethidium bromide staining of the agarose gel detected strong accumulation of an RNA migrating at the position of the 20S pre-rRNA (Fig. 6B). Northern hybridization with probes complementary to mature 18S (probe 008) and the 5′ region of ITS1 (probe 002) confirmed that this molecule was indeed the 20S pre-rRNA.
FIG. 6.
Depletion of Nob1p impairs pre-rRNA processing. (A) Structure and processing sites of the 35S pre-rRNA. This precursor contains the sequences for the mature 18S, 5.8S, and 25S rRNAs, which are separated by the two internal transcribed spacers ITS1 and ITS2 and flanked by the two external transcribed spacers 5′ETS and 3′ETS. The positions of the oligonucleotides probes are indicated. (B to E) Northern analyses of pre-rRNA processing. The GAL::HA-nob1 and wild-type (WT) strains were growth at 30°C in YPGal and then shifted to YPD. The cells were harvested at the times indicated, and total RNA was extracted. Equal amounts of RNA (5 μg) were resolved on a 1.2% agarose-formaldehyde gel (B to D) or 6% acrylamide-urea gel (E) and transferred to a nylon membrane. (B) Ethidium bromide staining of the gel. (C) Probe complementary to NOB1 mRNA. (D and E) Probes specific for the mature rRNAs and pre-rRNAs as indicated.
Substantial accumulation of 20S pre-rRNA was detected even in the permissive galactose medium (0-h time point of Nob1p depletion), when growth (Fig. 4) and the level of mature 18S (Fig. 6B and D) were unaffected. To determine whether this pre-rRNA accumulation is due to the presence of the HA tag, a nontagged version of the GAL-regulated mutant was constructed. Similar 20S pre-rRNA accumulation was seen in this strain on galactose medium (data not shown). In the GAL::HA-nob1 strain grown in galactose medium, the level of the NOB1 mRNA was strongly increased compared to that of the wild-type strain (Fig. 6C). It therefore seems likely that the delay in 20S processing is due to overexpression of Nob1p.
Following transfer of the GAL::HA-nob1 strain to glucose medium, the mature 18S rRNA was strongly depleted. In addition, some accumulation of 35S and 32S pre-rRNAs was detected. An aberrant 22S RNA was detected with probes 005 (Fig. 6D) and 002 (data not shown) but not with probe 033 (data not shown), showing that it extends from site A0 to site A3 and results from cleavage of the 32S pre-rRNA at site A3 in the absence of cleavages at sites A1 and A2 (40). Accumulation of the 22S RNA was also seen in strains depleted for the 18S dimethylase Dim1p (21), consistent with these proteins acting within the same preribosomal particle.
Reduced levels of the 60S subunit precursors, 27SA2, 27SA3, and 27SB pre-rRNA, and the 25S rRNA were observed at late times after transfer to glucose medium (Fig. 6D), although this was less evident for low-molecular-weight RNAs, 7S and 6S pre-rRNA, or the 5.8S rRNA (Fig. 6E). These defects may be secondary consequences of growth inhibition. The levels of most pre-rRNA species showed the effects of the nutritional upshift 6 h after transfer to glucose medium.
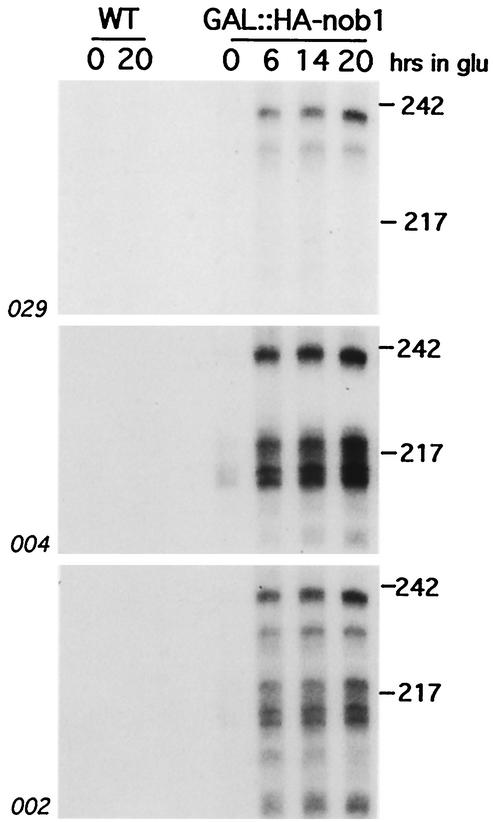
Further analyses identified short RNA species in the Nob1p-depleted strain, which were detected by probes complementary to positions from within the 3′ end of 18S to site A2 (Fig. 7). These RNAs were present at low levels in galactose medium and were strongly accumulated following depletion of Nob1p. Shorter RNA fragments, extending from site D to site A2, have been previously described in strains lacking the 5′ → 3′ exonuclease Xrn1p due to stabilization of the normal, excised ITS1 fragment (32). The species detected in the Nob1p-depleted strain have not previously been reported and presumably represent intermediates in 20S pre-rRNA degradation. Comparison of the different probes used in Fig. 7 shows that they are subject to both 5′ and 3′ truncation. We speculate that they may be stabilized by the binding of a nonproductive processing complex to the 5′ region of ITS1 and the 3′ end of the 18S rRNA.
FIG. 7.
Accumulation of pre-rRNA fragments on Nob1p depletion. Northern hybridization of RNA extracted from the GAL::HA-nob1 and wild-type (WT) strains following glucose shift is shown. The membranes were consecutively hybridized with the probes indicated (see Fig. 6A for probe locations). Molecular weight markers (MspI-digested pBR322) are indicated on the right in thousands.
From these data, we conclude that Nob1p is required for the cytoplasmic conversion of the 20S pre-rRNA to the mature 18S rRNA.
Nob1p localizes to both nucleus and cytoplasm.
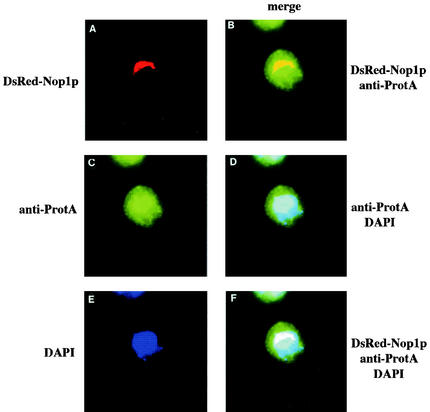
The protein A moiety of the TAP tag was used to localize the fusion Nob1p protein by indirect-immunofluorescence microscopy with anti-protein A antibodies (Fig. 8C). To allow visualization of the nucleolus, the Nob1-TAP strain was additionally transformed with a plasmid expressing DsRed-tagged Nop1p (26). This decorated the crescent-shaped region of the nucleus (Fig. 8A) that is characteristic of the yeast nucleolus. The cells were also treated with DAPI to visualize DNA in the nucleoplasm (Fig. 8E). Superimposition of the different signals (Fig. 8B, D, and F) showed that the Nob1-TAP signal was detected in the nucleoplasm and nucleolus, without clear nucleolar enrichment, and was also present in the cytoplasm. The otherwise isogenic wild-type strain, which was utilized as control, gave no clear signals in the channels used to detect DsRed-Nop1p or Nob1-TAP (data not shown). These data indicate that Nob1p is localized to the nucleolus, nucleoplasm and cytoplasm. This result would be consistent with the model that Nob1p associates with nucleolar 90S or pre-40S particles and accompanies the pre-40S particles during their transport through the nucleoplasm to the cytoplasm.
FIG. 8.
Nob1p is localized to the nucleus and cytoplasm. Indirect immunofluorescence was performed on cells expressing TAP-tagged Nob1p and DsRed-tagged Nop1p. (A) Localization of DsRed-Nop1p. (C) Indirect immunofluorescence of Nob1-TAP with rabbit anti-protein A. (E) DNA stained with DAPI. (B) Superimposition of the signals from DsRed-Nop1p and Nob1-TAP. (D) Superimposition of the signals from Nob1-TAP and DAPI staining. (F) Superimposition of the signals from Nop1p, Nob1-TAP, and DAPI staining.
A large-scale analysis of gene fusions (18) reported the localization of the Nob1-V5 fusion to both the nucleoplasm and nucleolus. This analysis did not report a cytoplasmic signal, but this may be due to the fusion protein used and/or its overproduction in the presence of the wild-type Nob1p.
Nob1p is not required for the export of the pre-40S ribosomal particles.
Depletion of some components of the 90S preribosomes inhibits export of the pre-40S particles to the cytoplasm (12). This raised the possibility that Nob1p was similarly required for pre-40S export to the cytoplasm, with a consequent defect in the (normally cytoplasmic) 20S pre-rRNA cleavage. To test this, we analyzed the cellular localization of the 20S pre-rRNA by fluorescent in situ hybridization with a probe complementary to the 5′ region of ITS1. Localization of the 20S pre-rRNA was determined in the GAL::HA-nob1 strain after growth in galactose medium and at various time points after transfer to glucose medium (Fig. 9). In the wild type, the 20S pre-rRNA signal was predominantly nuclear with a weaker cytoplasmic signal that showed perinuclear enrichment. In the Nob1p-depleted strain, the signal was substantially stronger and predominantly cytoplasmic, although perinuclear enrichment was still visible.
FIG. 9.
Nob1p is not required for nuclear export of the small ribosomal subunit. (A to C) The ITS1-5′ probe preferentially deccrates the nuclei of wild-type cells. (D to F) ITS1-5′ decorates both the nuclei and cytoplasm of GAL::HA-nob1 cells growth in galactose. (G to I) ITS1-5′ strongly decorates the cytoplasm of GAL::HA-nob1 cells after 6 h of depletion in glucose. DNA was labeled with DAPI to visualize the nucleus.
We conclude that depletion of Nob1p does not lead to nuclear accumulation of pre-40S ribosomes and that reduced nuclear export is therefore not responsible for the defect in cleavage of the 20S pre-rRNA.
DISCUSSION
Based on the results of a high-throughput proteomic analysis, we speculated that Nob1p might be present in pre-40S ribosomes, and we show here that this is indeed the case. On sucrose gradients, the major peak of Nob1-TAP cosedimented with pre-40S particles, with a minor peak at the position of 90S preribosomes. Northern and primer extension analyses of immunoprecipitated RNAs confirmed the association of Nob1p with the 20S pre-rRNA, as well as a weaker association with the 35S pre-rRNA and U3 small nucleolar RNA components of the 90S preribosomes. Moreover, the precipitated 20S pre-rRNA was shown to be dimethylated within the 3′ region of the 18S rRNA. This modification is reported to occur in the cytoplasm (4), indicating that Nob1p accompanies the 20S pre-rRNA from the nucleus to the cytoplasm. Consistent with this proposal, Nob1-TAP was localized in both the nucleus and cytoplasm.
Depletion of Nob1p led to the loss of 18S rRNA synthesis, accompanied by the accumulation of remarkably high levels of the 20S pre-rRNA. In the absence of correct ribosome synthesis, the preribosomal particles are generally degraded rapidly. Only in a very few mutant strains do the pre-rRNAs accumulate to a level that represents a significant fraction of the abundance of the mature rRNAs. The ready detection of the 20S pre-rRNA by ethidium staining in the Nob1p-depleted strain is therefore an unusual feature, which might be more consistent with the specific inhibition of pre-rRNA cleavage than with a general defect in pre-40S structure. Together with the presence of the PIN domain in Nob1p, this would be consistent with a direct role in pre-rRNA processing.
A partial processing defect was seen in GAL::nob1 strains grown on permissive, galactose medium. We speculate that excess Nob1p competes for binding to other components of the 20S processing complex, delaying its productive assembly onto the pre-rRNA.
In the Nob1p-depleted strain, 20S pre-rRNA degradation products were also readily observed. The accumulation of such readily detectable degradation intermediates is also unusual in strains that are not compromised in RNA turnover systems. The major species extended from within the 3′ region of the 18S rRNA to the 3′ end of the 20S pre-rRNA (site A2 in ITS1). It is unclear whether the 5′ ends of these species are generated by inaccurate 18S cleavage in the absence of Nob1p or by pausing of 5′ → 3′ exonucleases upstream of site D. In either case, we speculate that the accumulation of these fragments may be a consequence of the assembly of a nonproductive processing complex at the 3′ end of the 18S rRNA and on the 5′ region of ITS1. The size of the largest of these species indicates that the 5′ end is positioned close to the site of m26Am26A dimethylation in the 3′-terminal loop of the 18S rRNA. This modification imposes a complete block on the progression of reverse transcriptase. The effects of RNA modification on the activity of Xrn1p have not been assessed, but pausing at this site during 5′ degradation of the 20S pre-rRNA is an alternative possibility.
Nob1p was identified as a protein that interacted with Nin1p, a regulatory component of the 26S proteosome (34), and it is reported to be required for proteosome biogenesis (35). Consistent with this, Nob1p was reported to sediment at 26S on a sucrose gradient, based on Western analyses with anti-Nob1p (34). We did not, however, detect TAP-tagged Nob1p at this gradient position, and inspection of the published data suggests that Nob1p might have been misidentified in the previous analysis. An additional protein band was decorated by anti-Nob1p that showed gel mobility appropriate for Nob1p and sedimented at approximately 40S on the gradient (34). The data therefore do not support a stable interaction between Nob1p and the 26S proteosome, consistent with the report that Nob1p is degraded following proteosome synthesis (35).
A possible functional interaction between Nob1p and the proteosome can be envisaged in ribosome synthesis. The pre-rRNAs present in defective preribosomal particles are normally very rapidly degraded, but the fate of the associated ribosomal proteins is not clear. The Nob1p-Nin1p interaction might promote the recruitment of the proteosome to ribosomal subunits that are stalled in maturation, in order to bring about r-protein degradation.
Acknowledgments
We thank Ed Hurt for plasmid pUN100-DsRed-Nop1.
M.D. is a Special Fellow of the Leukemia & Lymphoma Society. This work was supported by the Wellcome Trust.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg, D. C., A. L. Goldstein, and C. N. Cole. 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6:1173-1189. [DOI] [PubMed] [Google Scholar]
- 3.Baßler, J., P. Grandi, O. Gadal, T. Leßmann, E. Petfalski, D. Tollervey, J. Lechner, and E. C. Hurt. 2001. Identification of a pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 4.Brand, R. C., J. Klootwijk, T. J. M. van Steanbergen, A. J. de Kok, and R. J. Planta. 1977. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 75:311-318. [DOI] [PubMed] [Google Scholar]
- 5.Clissold, P. M., and C. P. Ponting. 2000. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 10:R888-R890. [DOI] [PubMed] [Google Scholar]
- 6.Fatica, A., A. D. Cronshaw, M. Dlakić, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 7.Fatica, A., and D. Tollervey. 2002. Making ribosome. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 8.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bonwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 9.Gelperin, D., L. Horton, J. Beckman, J. Hensold, and S. K. Lemmon. 2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7:1268-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodstadt, L., and C. P. Ponting. 2001. CHRCMA: consensus-based colouring of multiple alignments for publication. Bioinformatics 17:345-846. [DOI] [PubMed] [Google Scholar]
- 11.Grandi, P., V. Doyl, and E. C. Hurt. 1993. Purification of NSP1 reveals complex formation with "GLFG' nucleporins and a novel nuclear pore protein NIC96. EMBO J. 12:3061-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 13.Harnpicharnchai, P., J. Jakovijevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford. 2001. Composition and functional characterization of yeast 66s ribosome assembly intermediates. Mol. Cell 8:663-670. [DOI] [PubMed] [Google Scholar]
- 14.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Meore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 15.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 229:499-520. [DOI] [PubMed] [Google Scholar]
- 16.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta 517:378-389. [DOI] [PubMed] [Google Scholar]
- 17.Kufel, J., C. Allmang, G. Chanfreau, E. Petfalski, D. L. J. Lafontaine, and D. Tollervey. 2000. Precursors to the U3 snoRNA lack snoRNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20:5415-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafontaine, D., J. Delcour, A.-L. Glasser, J. Desgrès, and J. Vandenhaute. 1994. The DIM1 gene responsible for the conserved m26 Am26 A dimethylation in the 3′-terminal loop of 18S rRNA is essential in yeast. J. Mol. Biol. 24:492-497. [DOI] [PubMed] [Google Scholar]
- 20.Lafontaine, D., and D. Tollervey. 1996. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24:3469-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafontaine, D., J. Vandenhaute, and D. Tollervey. 1995. The yeast 18S rRNA dimethylase Dim1p is required for pre-rRNA processing. Genes Dev. 9:2470-2481. [DOI] [PubMed] [Google Scholar]
- 22.Lafontaine, D., T. Preiss, and D. Tollervey. 1998. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol. Cell. Biol. 18:2360-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Aditional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 24.Makarova, K. S., L. Aravind, M. Y. Galperin, N. V. Grishin, R. L. Tatusov, Y. I. Wolf, and E. V. Koonin. 1999. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. PG. Genome Res. 9:608-628. [PubMed] [Google Scholar]
- 25.Makarova, K. S., V. A. Ponomarev, and E. V. Koonin. 2001. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tsehochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 18:499-509. [DOI] [PubMed] [Google Scholar]
- 27.Moy, T. I., and P. A. Silver. 1999. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 13:2118-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigaut, G., A. Shevchenko, B. Ruts, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 30.Roos, J., J. M. Luz, S. Centoducati, R. Sternglanz, and W. J. Lennarz. 1997. ENP1, an essential gene encoding a nuclear protein that is highly conserved from yeast to humans. Gene 185:137-146. [DOI] [PubMed] [Google Scholar]
- 31.Saveanu, C., D. Bienvenu, A. Namane, P. Gleizes, N. Nicole Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens, A., C. L. Hsu, K. R. Isham, and F. W. Larimer. 1991. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′ → 3′ exoribonuclease 1. J. Bacteriol. 173:7024-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tollervey, D., H. Lehtonen, R. Jansen, H. Kern, and E. C. Hurt. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72:443-457. [DOI] [PubMed] [Google Scholar]
- 34.Tone, Y., N. Tanahashi, K. Tanaka, M. Fujimuro, H. Yokosawa, and A. Toh-e. 2000. Nob1p, a new essential protein, associates with the 26S proteasome of growing Saccharomyces cerevisiae cells. Gene 245:37-45. [DOI] [PubMed]
- 35.Tone, Y., and A. Toh-e. 2002. Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Saccharomyces cerevisiae. Genes Dev. 16:3142-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapman, J., J. Retèl, and R. J. Planta. 1975. Ribosomal precursor particles from yeast. Exp. Cell Res. 90:95-104. [DOI] [PubMed] [Google Scholar]
- 37.Udem, S. A., and J. R. Warner. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248:1412-1416. [PubMed] [Google Scholar]
- 38.Udem, S. A., and J. R. Warner. 1972. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J. Mol. Biol. 65:227-242. [DOI] [PubMed] [Google Scholar]
- 39.Vanrobays, E., P. E. Gleizes, C. Bousquet-Antonelli, J. Noaillac-Depeyre, M. Caizergues-Ferrer, and J. P. Gelugne. 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 20:4204-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 41.Warner, J. R. 2001. Nascent ribosomes. Cell 107:133-136. [DOI] [PubMed] [Google Scholar]