Abstract
The zinc finger transcription factor PacC undergoes two-step proteolytic activation in response to alkaline ambient pH. PalA is a component of the fungal ambient pH signal transduction pathway. Its mammalian homologue AIP1/Alix interacts with the apoptosis-linked protein ALG-2. We show that both PalA and AIP1/Alix recognize a protein-protein binding motif that we denote YPXL/I, where Tyr, Pro, and Leu/Ile are crucial for its interactive properties. Two such motifs flanking the signaling protease cleavage site mediate direct binding of PalA to PacC, required for the first and only pH-regulated cleavage of this transcription factor. PalA can bind the “closed” (i.e., wild-type full-length) conformer of PacC, suggesting that PalA binding constitutes the first stage in the two-step proteolytic cascade, recruiting or facilitating access of the signaling protease, presumably PalB. In addition to recognizing YPXL/I motifs, both PalA and AIP1/Alix interact with the Aspergillus class E Vps protein Vps32 homologue, a member of a protein complex involved in the early steps of the multivesicular body pathway, suggesting that this interaction is an additional feature of proteins of the PalA/AIP1/Alix family.
The regulation of gene expression by ambient pH in Aspergillus nidulans involves the zinc finger transcription factor, PacC, and six proteins, PalA, PalB, PalC, PalF, PalH, and PalI, mediating ambient pH signal transduction and denoted the pal signaling pathway components (6, 7, 22, 27, 28). With the sole exception of PalB, which appears to be a calpain-like cysteine protease (7, 12) and a human calpain 7 homologue (11), the amino acid sequences of the pal gene products give few clues to their function (for a review, see reference 33). PalH and PalI appear to be seven-pass and four-pass membrane proteins, respectively, and are likely candidates for membrane pH sensors. PalF and PalC have no apparent homologues outside fungi. Finally, PalA has a mammalian homologue, AIP1/Alix, which interacts physically with the penta-EF hand small calpain family member ALG-2, a protein involved in apoptosis (21, 26, 43, 44). The fact that only PalB has a predictable function underscores the interest in this novel eukaryotic signal transduction pathway, which appears to be mechanistically dissimilar to all other known eukaryotic signal transduction pathways.
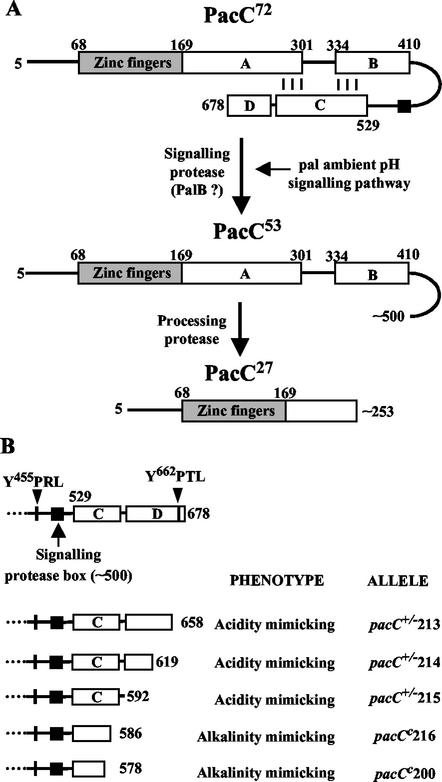
Ambient pH signaling occurs under alkaline conditions. In such circumstances, pH signal transduction enables the activation of the otherwise transcriptionally inactive 674-residue PacC translation product by a two-step proteolysis mechanism which can be compared to regulated intramembrane proteolysis. In the first step, which is crucially regulated by ambient pH, the 72-kDa PacC translation product (PacC72) is converted to a 53-kDa intermediate (PacC53) lacking the ∼180 C-terminal residues (see Fig. 1A). This step is catalyzed by the signaling protease, likely to be PalB. In a second, pH-independent step, this (committed) intermediate is converted to the 27-kDa processed product (PacC27) containing the ∼250 N-terminal residues by an as yet unidentified processing protease (see Fig. 1A) (8, 10, 25, 31). The crucial feature of this regulatory mechanism, which involves changes in PacC nucleocytoplasmic distribution (24), is that PacC72 is protected from activating proteolysis under inappropriate circumstances (i.e., acidic ambient pH and absence of pal signaling) by adopting a closed conformation, which prevents accessibility to the processing protease. This model is supported by mutational analysis of pacC (8, 10, 25, 40). Thus, alkalinity-mimicking pacCc C-terminally truncating mutations (mimicking the signaling protease cleavage) or certain amino acid substitutions disrupt the closed conformation, leading to pH-independent activation of PacC, whereas a subclass of acidity-mimicking pacC+/− mutations prevent the signaling cleavage, resulting in closed PacC72 at any ambient pH.
FIG. 1.
(A) Schematic representation of the two-step mechanism of PacC proteolytic activation. The different regions in PacC are indicated by boxes, with numbers denoting amino acid residues. Region C of Espeso et al. (10) is divided into regions C and D (also see the text). To avoid confusion with previous publications, residues are numbered as if translation proceeded from methionine codon 1, although translation proceeds from methionine codon 5 (25). (B) Subdivision of the 150 C-terminal residues into regions C and D by nested deletion analysis.
In view of the presence of PalA homologues in mammals, we focused our attention on PalA. Studies of the yeast interactome both ex vivo and in vivo have revealed a possible connection between the pH signaling pathway and the class E endocytotic machinery (reviewed in reference 33), and a recent study has shown that the yeast homologues of PalA and PacC (Rim20p and Rim101p, respectively) interact with each other (46), indicating that the role of PalA/Rim20p involves its ability to mediate protein-protein interactions. We show here that PalA recognizes a short YPXL/I amino acid sequence motif and that PacC contains two such motifs flanking the signaling protease cleavage site, which are completely conserved in all members of the PacC/Rim101p family and which are required for the signaling cleavage of PacC72. We also show that PalA interacts in two-hybrid assays with Vps32, a class E endocytotic protein. The ability to recognize YPXL/I motifs and to interact with Vps32p is conserved in the human PalA homologue AIP1/Alix. It is very significant that the YPXL/I PalA/AIP1/Alix binding motif consensus sequence includes the YPDL motif in the late domain of the GAG p9 protein of the equine infectious anemia virus (EIAV) (35), which is required for efficient release of virions from the plasma membrane and possibly connects budding particles to the class E endocytotic machinery.
MATERIALS AND METHODS
A. nidulans methods.
A. nidulans strains used in this work carried markers in standard use (4). Phenotype testing of pH regulatory mutations was done by the method of Tilburn et al. (40). With the exception of pacC+/−207, which was selected as a UV-induced mutation in a diploid with the areAr5 pyroA4 pantoB100/areAr5 inoB2 glrA1 fwA1 genotype to enable the utilization of 10 mM γ-aminobutyrate as a nitrogen source in glucose minimal medium, all other novel pacC alleles (described in Table 1) were constructed in vitro by PCR using mutagenic primers and introduced into A. nidulans by homologous gene replacement of a ΔpacC::pyr4 allele as described previously (8). Candidate strains were confirmed by Southern analysis.
TABLE 1.
Previously undescribed mutant pacC alleles characterized in this work
| Allele | DNA mutation(s)a | Change in coding sequence | Mutant protein residues |
|---|---|---|---|
| pacC+/−207 | T2321A | Y455N | Tyr455Asn |
| pacC+/−211 | T2321G | Y455D | Tyr455Asp |
| pacCc216 | G2717T | E587Stop | 5-586 |
| pacC+/−215 | G2735T, A2736G, C2737A | D593Stop | 5-592 |
| pacC+/−214 | G2816T, T2817G, T2818A, T2819G | V620Stop | 5-619 |
| pacC+/−213 | A2933T, T2935A | S659Stop | 5-658 |
| pacC+/−212 | T2942A | Y662N | Tyr662Asn |
In pH shift experiments, mycelia were grown for 15.5 h at 37°C in 3% sucrose-PPB (penicillin production broth) (9) buffered at pH 4.4 with 50 mM sodium citrate and shifted for 45 min to the same medium buffered at pH 8.9 with 100 mM HEPES-NaOH. Final pH values before and after the shift were approximately 4.3 and 8.5, respectively.
Two-hybrid analysis.
The Saccharomyces cerevisiae strain used for two-hybrid studies was CTY10.5d (Mata ade2-101 his3-Δ200 leu2-Δ1 trp1-Δ901gal4 gal80 URA3::lexAop-lacZ). Standard genetic methods were used. Yeast cells were grown in synthetic dextrose minimal medium (SD) lacking appropriate supplements, to maintain selection for plasmids (36). For β-galactosidase assays, transformants were patched onto selective SD medium and grown for 2 days at 30°C. Filter lift assays for blue color were performed as described previously (47), and the cultures were developed for 1 h. For quantitative assays, four different transformants were grown to mid-log phase in selective SD medium. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units (23). Yeast protein extracts for immunoblot analysis were prepared as described previously (42) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) followed by immunoblotting with a monoclonal anti-HA antibody (Roche).
Plasmids.
The plasmids used in this study are listed in Table 2. pLexA-PalA, pGAD-PalA, and pGST-PalA were constructed by inserting a PCR fragment containing the PalA coding sequence in the polylinker site of pLexA(1-202)+PL, pACTII, and pGEX-2T, respectively. Constructs encoding PacC fusion proteins to the Gal4 activating domain (GAD) were derived from pACTII by inserting the corresponding PacC fragments into the BamHI site of the polylinker. Site-directed pacC mutations were obtained by PCR-directed sequence modification and first introduced into pSpacC, a pBS-SK(+) derivative containing the pacC gene (8). pGST-PacC(529-678), pGST-PacC(169-410), pHis-PacC, and pHis-PacCL340S have been described previously (10). pHis-PacCY455D-Y662N was constructed by replacing a fragment from pHis-PacC with a fragment from a pSpacC derivative containing the corresponding mutations. To construct pLexA-SVMYPTLRGL and mutant derivatives, we inserted a double-stranded oligonucleotide with the corresponding coding sequence followed by a stop codon in the polylinker of pLexA(1-202)+PL. pLexA-Vps32 is a pLexA(1-202)+PL derivative containing the VPS32 coding sequence (our unpublished results) amplified by PCR from A. nidulans genomic DNA. The pACTII derivative, pGAD-AIP1/Alix, was constructed by PCR with a human AIP1/Alix clone (IMAGE 5284284) as template.
TABLE 2.
Plasmids used in this study
| Construct | Vectora |
|---|---|
| LexA-PalA | pLexA(1-202)+PL |
| GAD-PalA | pACTII |
| GST-PalA | pGEX-2T |
| GAD-PacC(341-678) | pACTII |
| GAD-PacC(341-529) | pACTII |
| GAD-PacC(341-529)Y455D | pACTII |
| GAD-PacC(529-678) | pACTII |
| GAD-PacC(529-678)Y662N | pACTII |
| GST-PacC(529-678) | pGEX-2T |
| GST-PacC(169-410) | pGEX-2T |
| His-PacC | pD1 |
| His-PacCL340S | pD1 |
| His-PacCY455D-Y662N | pD1 |
| LexA-SVMYPTLRGL | pLexA(1-202)+PL |
| LexA-SVMAPTLRGL | pLexA(1-202)+PL |
| LexA-SVMYATLRGL | pLexA(1-202)+PL |
| LexA-SVMYPALRGL | pLexA(1-202)+PL |
| LexA-SVMYPTARGL | pLexA(1-202)+PL |
| LexA-SVMNPTLRGL | pLexA(1-202)+PL |
| LexA-Vps32 | pLexA(1-202)+PL |
| GAD-AIP1/Alix | pACTII |
Protein expression and in vitro binding assays.
N-terminally His-tagged PacC proteins (wild-type, double-mutant Y455D-Y662N, and mutant L340S) were synthesized in vitro and labeled with [35S]methionine (1,000 Ci/mmol) by using the Promega TNT coupled transcription-translation system and appropriate templates (Table 2).
Glutathione S-transferase (GST)-PalA, GST-PacC(529-678), GST-PacC(169-410), and GST were expressed from Escherichia coli DH1 transformed with the corresponding plasmids (Table 2). Synthesis of these proteins was induced at 37°C by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to 50-ml cultures, which were incubated for an additional 3 h. Bacterial pellets were resuspended in 7 ml of STE buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA). After addition of 1% (vol/vol) Triton X-100, 5 mM dithiothreitol, and Complete protease inhibitor cocktail (Roche), the cells were lysed in a French press. Extracts were cleared by centrifugation and incubated (1-ml portions) with glutathione-Sepharose 4B beads (Pharmacia) at 4°C for 1.5 h. After being washed six times with 1 ml of STE buffer plus 1% (vol/vol) Triton X-100, beads with bound proteins were split into equal portions. In vitro-synthesized wild-type and mutant PacC proteins (5 to 7 μl) were added to the beads and allowed to bind at 4°C for 1 h in 500 μl of STE buffer with 1% Triton X-100. After five washes with STE-1% Triton X-100 and a further wash with STE buffer, the beads were boiled in sample buffer and proteins were separated by SDS-PAGE (10% polyacrylamide). Bound proteins were detected by autoradiography (labeled preys) or Coomassie staining (baits).
EMSA and immunoblot analysis of A. nidulans protein extracts.
A. nidulans protein extracts were prepared from lyophilized mycelium as described previously (8). Electrophoretic mobility shift assays (EMSA) were performed by the method of Orejas et al. (31), using a32P-labeled double-stranded oligonucleotide containing the IpnA2 PacC binding sequence and 5 μg of A. nidulans protein extracts. Electrophoresis was performed in 4% (wt/vol) polyacrylamide gels. For Western analysis, 50 μg of A. nidulans protein extract was resolved by SDS-PAGE (10 or 12% polyacrylamide) and PacC was detected using a rat anti-PacC(5-265) polyclonal antiserum (1/2,000) (25) and a peroxidase-conjugated goat anti-rat secondary antibody (1/4,000) (Southern Biotechnology). Peroxidase activity was detected by enhanced chemiluminescence with ECL reagents (Amersham).
RESULTS
The most C-terminal sequences in PacC promote, rather than prevent, proteolytic processing activation.
A C-terminal domain in PacC (within residues 529 through 678, interacting region C), is involved in intramolecular interactions with two other regions (A and B) located downstream of the DNA binding domain, thus preventing PacC72 proteolytic activation under acidic growth conditions (Fig. 1A) (10). Consequently, PacC mutant proteins truncated upstream of or within interacting region C resemble the signaling cleavage PacC53 product in that they are committed to processing proteolysis at any ambient pH (8, 31). This pH-independent PacC activation results in an alkalinity-mimicking, gain-of-function phenotype. The strong alkalinity-mimicking pacCc200 mutation, which truncates the protein after residue 578 (40), establishes that downstream sequences are essential for interacting region C (Fig. 1B). Residues 579 through 678 do not show significant sequence conservation among fungal PacC proteins, with the exception of residues 660 through 678, which are largely conserved between A. nidulans and its closest relatives (data not shown). To address their possible role, we constructed by gene replacement a series of nonsense mutations truncating PacC after residues 586, 592, 619, and 658 (Fig. 1B). pacC+/−216 (PacC587stop) leads to an alkalinity-mimicking gain-of-function phenotype, virtually indistinguishable from that of pacCc200 (PacC579fs) and is therefore truncated within or upstream of sequences essential for interacting region C. In contrast, residues 592 through 678 are not required for interacting region C, since their absence does not lead to alkalinity mimicry (Fig. 1B) but instead leads to a weak loss-of-function, acidity-mimicking phenotype, indicating that this C-terminal region promotes, rather than prevents, proteolytic processing. A detailed molecular characterization of pacC+/−213, which truncates the protein after residue 658 (Fig. 1B), is given below. Growth tests showing the weak acidity-mimicking phenotype of pacC+/−213 are discussed below. Mutations truncating PacC after residues 592 or 619 (Fig. 1B) are phenotypically identical to pacC+/−213. We conclude that interacting region C is located within residues 529 to 592 and that residues 593 to 678 (denoted region D in Fig. 1B) contain a functional element which facilitates PacC processing.
Region D contains a YPXL/I motif which is repeated upstream of the signaling protease cleavage site.
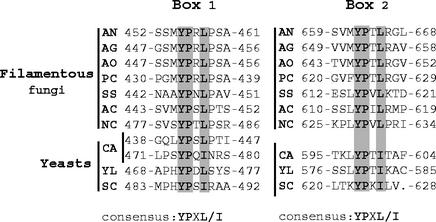
Multiple sequence alignment of fungal PacC proteins and their Rim101p yeast homologues did not reveal large blocks of sequence conservation outside of the DNA binding domain. However, a closer inspection of the alignment of the C-terminal moieties in PacC/Rim101p proteins revealed two short, completely conserved motifs (consensus YPXL/I) at positions 455 and 662 of A. nidulans PacC, which are present in all PacC/Rim101p family members (Fig. 2). The more C-terminal of these motifs (Box 2) is located very close to the C termini of the proteins (Fig. 2), within region D of A. nidulans PacC defined above. The more N-terminal motif (Box 1) overlaps in all cases with an SH3 binding motif, PXXP (either YPXL/IP in filamentous fungi or PXYPXL/I in yeasts [Fig. 2]). In A. nidulans PacC, Box 1 is placed ∼40 residues upstream of the signaling protease cleavage site, located at residue ∼500. Of note, Candida albicans Rim101p contains a duplication of Box 1 (Fig. 2).
FIG. 2.
Conservation of the YPXL/I motifs in the PacC/Rim101p family. Amino acid alignment showing the two completely conserved YPXL/I motifs (Box 1 and Box 2) found in all PacC/Rim101p homologues. Identical and conserved (Leu/Ile) residues are shaded in gray. Sequences of PacC/Rim101p homologues are denoted as follows: AN, A. nidulans; AG, A. niger, AO, A. oryzae; PC, Penicillium chrysogenum; SS, Sclerotinia sclerotium; AC, Acremonium chrysogenum; NC, Neurospora crassa; CA, Candida albicans; YL, Yarrowia lipolytica; SC, Saccharomyces cerevisiae.
The YPXL/I motifs are specifically recognized by PalA.
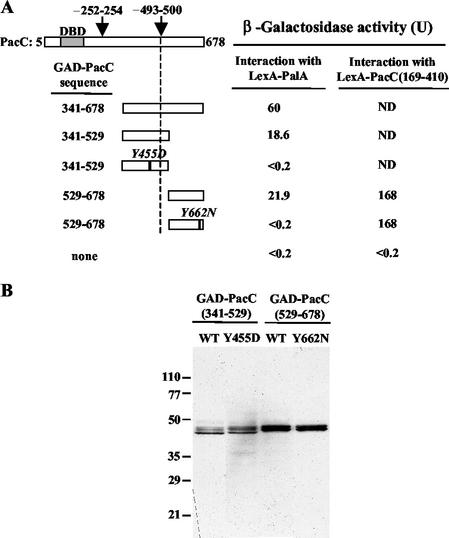
Rim20p, the budding-yeast PalA homologue, interacts with Rim101p, the yeast PacC homologue (46). A two-hybrid assay (data not shown) in which PalA fused to GAD interacts with PacC fused to the Gal4p DNA binding domain shows that this interaction is conserved in Aspergillus. By testing a series of GAD-PacC deletion constructs for two-hybrid interaction with LexA-PalA, we established that the C-terminal half of PacC (residues 341 through 678) is sufficient for the interaction (Fig. 3A). The PacC region interacting with PalA was further delimited to two nonoverlapping PacC polypeptides (residues 341 through 529 and 529 through 678), indicating that PalA interacts with at least two regions in the C-terminal half of PacC.
FIG. 3.
Two-hybrid interaction of PalA with PacC. Yeast strain CTY10-5d was used, and proteins were expressed from plasmids listed in Table 2. (A) GAD-PacC fusions contain the indicated PacC residues. The shaded bar indicates the DNA binding domain (DBD). Arrows mark the approximate position of the signaling-protease (∼493 to 500) (8) and processing-protease (∼252 to 254) (25) cleavage sites. Values are the average β-galactosidase activity of four transformants. Standard errors were <14%. In control experiments, GAD protein fusions did not interact with LexA (<0.4 U). ND, not determined. (B) Western analysis of protein extracts from transformants expressing LexA-PalA and the indicated GAD-PacC protein fusions which were detected with anti-HA antibodies. WT, wild type.
The location of YPXL/I motifs within the regions of PacC that interact with PalA suggested their possible involvement in the PalA-PacC interaction. Therefore, we tested the effect of Tyr substitutions in each of the YPXL/I motifs in the two-hybrid interaction between LexA-PalA and GAD-PacC (Fig. 3A) (see also below). Both the Tyr455Asp and Tyr662Asn substitutions abolished the two-hybrid interaction between PalA and their respective PacC regions, as determined by β-galactosidase assays. In contrast, the Tyr662Asn substitution had no effect on the intramolecular interaction between PacC interacting region C (residues 529 through 678) and residues 169 through 410 (containing regions A and B) described by Espeso et al. (10) (Fig. 3A), showing that PacCTyr662Asn specifically impairs interaction with PalA. Immunoblot analysis (Fig. 3B) demonstrated that the wild-type and mutant fusion proteins are expressed at similar levels and that these single-residue substitutions do not destabilize the GAD fusion proteins. Together, these results indicate that the two YPXL/I motifs in PacC mediate its interaction with PalA.
PalA binding to YPXL/I motifs in PacC is direct and independent of the PacC conformation.
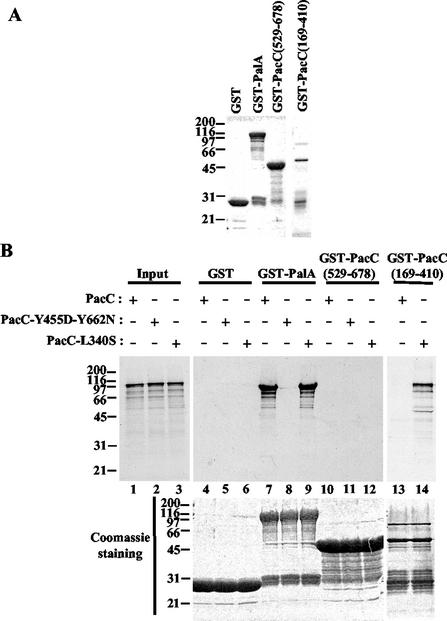
To provide corroborating evidence that PalA and PacC interact and to show that this interaction is direct, we assayed the binding of in vitro-synthesized PacC to a purified, bacterially expressed GST-PalA fusion protein. GST-PalA or GST (Fig. 4A) was immobilized on glutathione-Sepharose beads and incubated with [35S]PacC, synthesized in vitro using a coupled transcription-translation system. Whereas no binding was detected with GST alone, PacC was retained by GST-PalA beads (Fig. 4B, lanes 4 and 7). This binding was prevented when a double-mutant (Y455D-Y662N) PacC protein was used (lane 8), demonstrating its dependence on the integrity of the YPXL/I motifs. Together, these results indicate that PalA and PacC interact directly via the YPXL/I motifs in PacC.
FIG. 4.
In vitro binding of PalA to PacC by using pull-down assays. (A) GST fusion proteins bound to glutathione-Sepharose 4B beads (Pharmacia) used in the binding assays (10% of the total) were separated by SDS-PAGE (10% polyacrylamide) and stained with Coomassie blue. (B) Beads loaded with GST alone (lanes 4 to 6) or with GST fusions to PalA (lanes 7 to 9), PacC(529-678) (lanes 10 to 12), or PacC (169-410) (lanes 13 and 14) were incubated with in vitro-synthesized [35S]PacC (wild type, double-mutant Y455D-Y662N, or mutant L340S, as indicated). After being washed, bound proteins were boiled in sample buffer, separated by SDS-PAGE, and analyzed by autoradiography (top) and by Coomassie staining (bottom). Lanes marked Input contain in vitro-synthesized PacC (wild type [lane 1], double-mutant Y455D-Y662N [lane 2] and mutant L340S [lane 3]) used for binding experiments (20% of the total reaction mixture). Protein markers are in kilodaltons.
In the absence of pH signal transduction, PacC adopts a processing-protease-inaccessible closed conformation (Fig. 1A). This closed conformation is disrupted by a Leu340Ser substitution, which prevents intramolecular interactions (8, 10, 25). Pull-down assays with GST-PalA provided evidence that PalA recognizes YPXL/I motifs irrespective of whether the prey PacC protein is in the closed conformation (wild type) or this conformation has been disrupted by mutation (Leu340Ser substitution) (Fig. 4B, lanes 7 and 9). Control pull-down assays demonstrating that in vitro-synthesized PacC is in the closed conformation and that this conformation is disrupted by the L340S mutation are shown in lanes 13 and 14. These assays demonstrate that GST-PacC(169-410)-loaded beads pull down PacCL340S but not wild-type PacC. This indicates that interacting region C in the prey is not available for interaction with the GST-PacC(169-410) bait when the prey (PacC) is in the closed (wild-type) conformation (10). Finally, an additional control (lanes 10 to 12) demonstrates that a GST-PacC(529-678) bait containing interacting region C does not pull down either a closed PacC prey (where interacting residues 169 through 410 are involved in maintaining the closed conformation) or a PacCL340S prey (where interacting residues 169 through 410 are available but interaction is prevented by the Leu340Ser substitution). We conclude that PalA is able to bind to PacC even if the latter is in the closed conformation.
The two YPXL/I motifs in PacC play a physiological role in vivo.
To address the physiological role of the YPXL/I motif-mediated interactions between PacC and PalA, we introduced by gene replacement the pacC mutations leading to Tyr455Asp and Tyr662Asn substitutions, alleles pacC+/−211 and pacC+/−212, respectively.
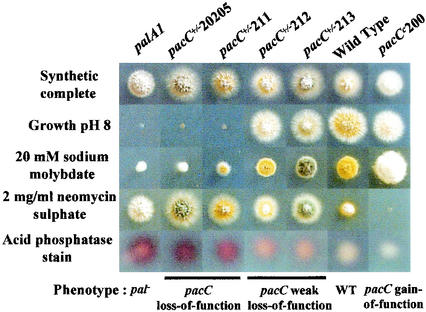
The Tyr455Asp substitution (pacC+/−211) results in a stringent loss-of-function, acidity-mimicking phenotype. A pacC+/−211 mutant does not grow at alkaline pH and is hypersensitive to molybdate, hyperresistant to neomycin, and strongly derepressed for extracellular acid phosphatase at pH 6.8 (Fig. 5). Further proof of the critical involvement of Tyr455 in PacC function is provided by the classically selected, stringently acidity-mimicking pacC+/−207 mutation resulting in Tyr455Asn. pacC+/−207 is phenotypically indistinguishable from pacC+/−211 (data not shown). These data indicated that Y455PXL/I-mediated recognition of PacC by PalA is a major requirement for reception of the pH signal. Of note, the acidity mimicry of pacC+/−211 or pacC+/−207 is only slightly less extreme than that resulting from the pacC+/−20205 loss-of-function mutation (Fig. 5 and data not shown). pacC+/−20205, which prevents the signaling cleavage step (8), phenotypically resembles null mutations in the palA, palB, palC, palF, and palH genes, inactivating the pH signal transduction pathway (25).
FIG. 5.
Phenotypic analysis of pacC mutations affecting the YPXL/I motifs (see Materials and Methods).
In contrast to Tyr455Asp, the Tyr662Asn substitution (pacC+/−212) results in a weak loss-of-function phenotype (Fig. 5). Thus, it also leads to strong resistance to neomycin but differs from the situation for pacC+/−211 (Y455D) in that pacC+/−212 is only partially derepressing for acid phosphatase and only slightly more sensitive to molybdate or alkaline pH than is the wild type. pacC+/−213, truncating PacC after residue 658 within region D (see above), is phenotypically indistinguishable from pacC+/−212 (Fig. 5), strongly suggesting that Tyr662Asn completely inactivates this motif and that the role of the region truncated in pacC+/−213 in promoting PacC proteolytic activation resides in the Y662PXL/I-dependent recognition of PacC by PalA. This weaker phenotype resulting from mutating the Y662PXL/I motif demonstrates that although both PalA binding sites are necessary for normal PacC activation, the Box 1 Y455PXL/I motif plays the major role.
The YPXL/I motifs are required for pH-dependent proteolytic activation of PacC.
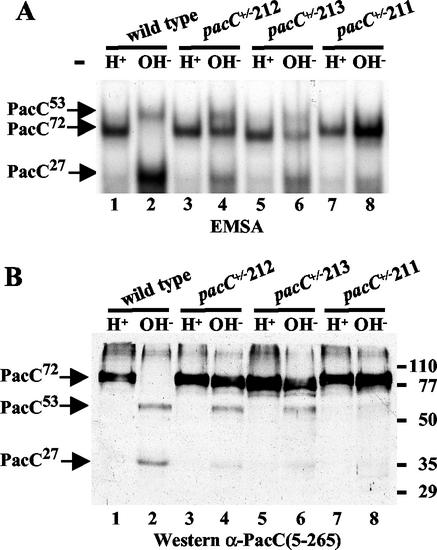
Transduction of the ambient pH signal leads to cleavage of transcriptionally inactive PacC72 by the signaling protease (presumably PalB) to yield PacC53, the substrate of the processing protease. Using EMSA and Western analyses of mycelial extracts, we examined the involvement of the PacC YPXL/I motifs in pH signal reception. Mycelia of the wild-type and pacC mutant strains were grown under acidic conditions and shifted to alkaline conditions for 45 min. Wild-type PacC72 is fully converted to the processing intermediate (PacC53) and processed form (PacC27) within 45 min after the shift (8), as determined both by EMSA (Fig. 6A, lanes 1 and 2; note that the PacC53-DNA complex shows reduced mobility compared to the PacC72 complex due to conformational differences) or by Western blot analysis (Fig. 6B, lanes 1 and 2). The pacC+/−211 (Tyr455Asp) product was severely impaired in the signaling protease step and therefore was poorly processed (Fig. 6, lanes 7 and 8). These and the above results unambiguously demonstrate that PalA binding to the PacC Box 1 Y455PXL/I motif is required for the signaling cleavage step. In contrast, and as expected from its much weaker acidity-mimicking phenotype, pacC+/−212 (Tyr662Asn), inactivating the more C-terminal Box 2 Y662PXL/I motif, was only partially deficient in pH signal reception (Fig. 6, lanes 3 and 4). pacC+/−212 appeared to impair the signaling cleavage to a certain extent, since it resembled pacC+/−211 in leading to a marked accumulation of PacC72 but, in contrast, did not prevent the formation of the intermediate (lanes 3 and 4). In agreement with phenotypic tests, the effect of pacC+/−213, truncating PacC immediately upstream of the Y662PXL/I motif, is indistinguishable from that of pacC+/−212 (lanes 5 and 6), demonstrating that the Tyr662Asn substitution fully inactivates the motif. These and the above results indicate that the Box 1 and Box 2 motifs are physiologically involved in the role of PalA in pH signal transduction.
FIG. 6.
Impairment of pH-dependent processing of PacC in mutants lacking the YPXL/I motifs. The wild type and the indicated mutant strains were grown under acidic conditions (H+) and shifted for 45 min to alkaline conditions (OH−). Protein extracts were analyzed by EMSA (A) and Western blot analysis with anti-PacC(5-205) antiserum (B). The positions of the different PacC polypeptides (B) or of the corresponding complexes with DNA (A) are indicated by arrows. Protein markers are shown in kilodaltons.
PalA and its human homologue AIP1/Alix specifically recognize the YPXL/I motif.
Multiple sequence alignment of A. nidulans PacC with Rim101p family members revealed that amino acid sequence similarity outside the conserved zinc finger region appears to be restricted to the YPXL/I PalA binding motifs. This strongly suggested that these motifs would be fully functional in an isolated sequence context and that a short sequence might be sufficient for interaction with PalA. Figure 7 shows that a decapeptide containing the PacC Y662PXL/I motif fused to the LexA DNA binding domain strongly interacted with GAD-PalA in the two-hybrid system, as determined by the intense β-galactosidase staining detected in a filter assay. Conversely, a fusion of the YPXL/I decapeptide to the Gal4p activation domain (GAD) also interacted with the PalA C-terminal region (residues 375 to 799) (data not shown). A Tyr-to-Asn substitution mimicking that resulting from pacC+/−207 (Y455N) or pacC+/−212 (Y662N) abolished this YPXL/I-PalA interaction (data not shown), demonstrating that it requires a functionally critical residue. Taken together, these data show that this YPXL/I-containing decapeptide is sufficient for interaction with PalA.
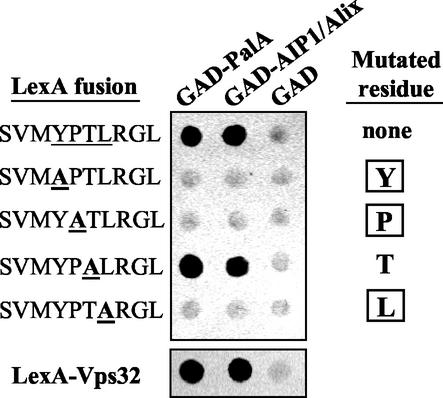
FIG. 7.
Recognition of the YPXL/I motif and Vps32 by PalA family proteins is conserved from fungi to humans. At the top is shown alanine-scanning mutagenesis of the YPXL/I motif. Two-hybrid interactions between GAD-PalA or GAD-AIP1/Alix and wild-type LexA-SVMYPTLRGL or the indicated mutant decapeptides carrying Ala substitutions affecting residues within the consensus motif are shown. Interaction of LexA-SVMYPTLRGL and its derivatives with GAD-PalA or GAD-AIP1/Alix was tested in S. cerevisiae strain CTY10-5d. Filter lift assay products were developed for 1 h. In each case, seven independent transformants were tested with identical results. The bottom panel is a similar two-hybrid experiment showing the interaction between PalA or AIP1/Alix and Vps32. The Vps32-PalA and Vps32-AIP1/Alix interactions have been confirmed in the reverse bait-prey arrangement (data not shown).
Like the Tyr-to-Asn substitution, Ala substitutions involving Tyr, Pro, and Leu within the consensus YPXL/I sequence abolished the interaction (Fig. 7). In contrast, a Thr-to-Ala substitution involving the variable residue in the motif had no effect. These two-hybrid assays in conjunction with the pacC+/−207, pacC+/−211 and pacC+/−212 phenotypes provide a functional validation of the consensus sequence deduced by sequence comparison.
PalA homologues are widespread in the eukaryotic world (27, 46), including vertebrates, where the PalA family members include the apoptosis-related protein AIP1/Alix (26, 44). This raised the possibility that the ability of PalA to interact with the YPXL/I motif might be conserved in AIP1/Alix proteins. To address this point, we tested the human AIP1/Alix protein (GenBank accession no. AF151793 [45]; 26 % identity to PalA over 798 residues) in a two-hybrid assay. It interacted strongly with the Y662PXL/I-containing decapeptide, with a sequence specificity indistinguishable from that of PalA, as determined by Ala scanning mutagenesis (Fig. 7). Thus, the ability of PalA and its homologues to recognize the YPXL/I motif is conserved from fungi to humans.
Conserved two-hybrid interaction between PalA/AIP1/Alix family members and Vps32, a class E Vps protein.
Vacuolar-protein sorting (Vps) proteins are largely conserved from fungi to humans (18). For example, homologues of yeast Vps32p showing ∼38% amino acid sequence identity are found in both the A. nidulans and human genomes (our unpublished results). In view of biochemical and genetic evidence which indicates an interaction between yeast PalA family members and Vps32 (reviewed in reference 33) (see Discussion; Fig. 8), we tested by two-hybrid analysis whether this interaction is conserved in A. nidulans. Figure 7 shows that PalA strongly interacts with the putative A. nidulans Vps32. Remarkably, this ability to interact with A. nidulans Vps32 is conserved in human AIP1/Alix (Fig. 7). Since fungal and human Vps32p homologues do not contain YPXL/I motifs, these two-hybrid assay results, together with yeast biochemical and genetic data (14, 15, 41), suggest that the ability to interact with this class E Vps protein is an additional feature of PalA/AlP1/Alix family members.
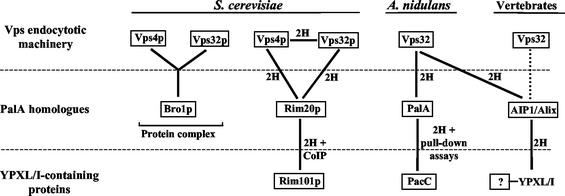
FIG. 8.
Conserved interactions between PalA/AIP1/Alix family members, YPXL/I-containing proteins, and the Vps endocytotic machinery. 2H indicates two-hybrid, and CoIP indicates coimmunoprecipitation (46). The dotted line indicates that interaction is deduced from heterologous interaction between human AIP1/Alix and the A. nidulans Vps32 homologue. The Bro1p-Vps4p-Vps32p complex was isolated by Gavin et al. (14) and agrees with two-hybrid assay data (15, 41).
DISCUSSION
We show here that PalA, a component of the ambient pH signaling pathway in fungi, binds directly to YPXL/I motifs in the transcription factor PacC and that these motifs mediate a protein-protein interaction which is crucial for ambient pH signal transduction. PalA is a homologue of the apoptosis-related AIP1/Alix vertebrate proteins. A YPXL/I-containing decapeptide is sufficient for binding by either fungal PalA or human AIP1/Alix, which strongly suggests that recognition of YPXL/I-containing protein partners is central to the different physiological roles of proteins in this family.
The ambient pH-dependent step in the proteolytic activation of the fungal PacC zinc finger protein is the cleavage of PacC72 (the translation product) to yield the PacC53 intermediate, which is committed to proteolytic activation (Fig. 1A). This conversion of PacC72 to PacC53 has been denoted the signaling protease step and is probably mediated by the PalB calpain-like cysteine protease, a homologue of human calpain 7. By exploiting the ease with which A. nidulans can be manipulated genetically, we show that PalA binding to the two YPXL/I motifs in PacC is required for the action of the signaling protease. Although the strength of the interaction between each site and PalA appears to be rather similar, as determined by a two-hybrid assay (Fig. 3), the contributions of each YPXL/I motif to pH signal reception appear to be markedly different. Substitution of the critical Tyr residue in the more N-terminal motif severely impairs the signaling protease step and therefore leads to a tight pacC loss-of-function phenotype. In contrast, deletion or tyrosine substitution in the more C-terminal motif has a less pronounced effect and leads to a weak loss-of-function phenotype. The molecular basis for the differing physiological contributions of the two YPXL/I motifs to ambient pH signal-dependent proteolytic activation is unknown, although we note the presence of an SH3 binding site overlapping the most N-terminal motif (Box 1 in Fig. 2) contained in all members of the PacC/Rim101p family of zinc finger proteins. This might suggest that PalA binding to this site antagonizes the binding of a negatively acting SH3 domain-containing protein.
Although the pH signal transduction pathway is widespread in both yeast and filamentous ascomycetes (33), proteolytic activation of PacC/Rim101p proteins has been demonstrated only in A. nidulans (31) and S. cerevisiae (20). However, the strict conservation of the two YPXL/I motifs in all members of the family (Fig. 2), together with extant mutational evidence (reviewed in reference 33), strongly suggests that all these proteins undergo at least one common proteolytic activation step in which protein-protein recognition through YPXL/I motifs plays a crucial role. In agreement, two YPXL/I motifs in the S. cerevisiae PacC homologue, Rim101p, are located in two regions of this protein that were previously found to interact with Rim20p, the PalA homologue (46).
PalA is able to bind YPXL/I motifs in PacC72, i.e., the translation product in the closed conformation. This would be consistent with PalA playing a role in recruiting the signaling protease, in disrupting the intramolecular interactions maintaining the closed conformation, or both. The calpain-like cysteine protease PalB is likely to be the signaling protease (8). In yeast, Rim20p and Rim13p, the respective PalA and PalB orthologues, interact with Snf7p/Vps32p, an endosome-associated class E Vps protein (15) (see below), which led to the suggestion that Rim20p would recruit Rim13p to its substrate Rim101p (46). Functional interaction between PalA homologues and cysteine proteases extends beyond the fungal world. For example, mammalian AIP1/Alix interacts with ALG-2, a member of the calpain small-subunit superfamily (reviewed in reference 38), which is involved in apoptosis and is a potential calpain regulatory subunit (21, 26, 43, 44). In view of the strength and complete conservation in amino acid sequence specificity of the interaction between AIP1/Alix or PalA and the YPXL/I motif, we hypothesize that AIP1/Alix might recruit YPXL/I-containing substrates to the putative ALG-2-associated cysteine protease.
Two PalA homologues, Rim20p and Bro1p, are present in the yeast proteome, but only Rim20p plays a role in pH regulation (29, 46). Data from several laboratories strongly suggest that these proteins physically interact with certain class E Vps protein complexes acting at the cytosolic side of endosomes to mediate the sorting of transmembrane proteins into the multivesicular body (MVB) pathway. In genome-wide two-hybrid screens, Rim20p interacts with two class E Vps proteins, Vps32p and Vps4p (15, 17, 41) (Fig. 8). Snf7p/Vps32p is a component of ESCRT-III (endosomal sorting complex required for transport III) acting at a late stage in the sorting of endosomal cargo into the MVB pathway (1, 2). Vps4p is an AAA ATPase which regulates the dissociation of Snf7p/Vps32p-containing ESCRT-III complexes from the endosomal membrane pathway (1). In agreement with these two-hybrid assay results, large-scale in vivo analysis of yeast protein complexes (14) revealed Bro1p in physical association with Vps4p and Vps32p (Fig. 8). Indeed, BRO1 is allelic to Vps31, another member of the class E vps family (39). We show here that the interactions of Bro1p and Rim20p with Vps32p are conserved in fungal PalA and human AIP1/Alix, suggesting that proteins of the PalA family physically connect YPXL/I-containing proteins to MVB pathway sorting complexes.
The YXXφ endocytotic motif described in higher eukaryotes (where φ is a bulky hydrophobic residue) (30) would appear to resemble the YPXL/I PalA/AIP1/Alix binding motif. However, not only are the consensus sequences of these two motifs different (3) (Fig. 7; see above) but also the motifs are functionally unrelated. Thus, while the YPXL/I motif appears to be connected with late endocytosis/MVB pathway class E protein complexes, YXXφ motifs recruit the early endocytotic machinery to plasma membrane proteins by interacting with the μ2 subunit of the AP2 adapter complex of clathrin-coated pits (30).
In contrast, the sequence specificity of the PalA/AIP1/Alix binding motif (Fig. 7) is indistinguishable from that of the YPDL late-domain motif identified in the GAG p9 protein of the equine infectious anemia virus (EIAV), in which replacement of Tyr, Pro, or Leu by Ala abolishes function (35). Late-domain motifs are short sequence motifs present in GAG p9 proteins of retroviruses which are required for efficient release of budding virions from the plasma membrane of infected cells. These GAG p9 late-domain motifs include, in addition to EIAV YPDL, the human immunodeficiency virus type 1 PTAP and the Rous sarcoma virus PPPY motifs (reviewed in reference 34). Despite their different amino acid sequences, the three GAG p9 late-domain motifs are functionally interchangeable, suggesting that they work by recruiting different subunits of a multiprotein complex involved in budding of all three viral particles (19, 32, 35).
Recent work (13) demonstrated that PTAP and PPPY motif function is dependent on (mammalian) Vps4, the key AAA ATPase required for disassembly of ESCRT-III complexes from the endosomal membrane. In addition, human immunodeficiency virus type 1 PTAP motif function is mediated through interaction with mammalian Tsg101/Vps23 (5, 13), an ESCRT-I complex protein also involved in the MVB pathway (1, 2, 16). These data point to a class E protein complex as the multiprotein machinery mediating GAG p9 late-motif function. Therefore, the likelihood that the EIAV GAG p9 motif and the YPXL/I PalA/AIP1/Alix binding motif not only share their consensus sequences but also share the ability to interact with the class E Vps machinery is very suggestive and may help us understand the thus far elusive function of human AIP1/Alix.
Acknowledgments
We thank T. Suárez for helpful discussions and A. Akintade and E. Reoyo for technical assistance.
We thank the CICYT, the BBSRC, and the EU (through grants BIO2000-920 [to M.A.P.], 60/P11494 [to H.N.A.], and QLK3-CT-1999-00729 [to M.A.P. and H.N.A.]) for support. O.V. held an EMBO long-term fellowship and is currently supported by the Ramón y Cajal Program (MCyT; Spain). L.R. held a BBSRC studentship.
REFERENCES
- 1.Babst, M., D. Katzmann, E. Estepa-Sabal, T. Meerloo, and S. Emr. 2002. Escrt-III. An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., D. Katzmann, W. Snyder, B. Wendland, and S. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 3.Boll, W., H. Ohno, Z. Songyang, I. Rapoport, L. C. Cantley, J. S. Bonifacino, and T. Kirchhausen. 1996. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 15:5789-5795. [PMC free article] [PubMed] [Google Scholar]
- 4.Clutterbuck, A. J. 1993. Aspergillus nidulans, p. 3.71-3.84. In S. J. O'Brien (ed.), Genetic maps. Locus maps of complex genomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denison, S. H., S. Negrete-Urtasun, J. M. Mingot, J. Tilburn, W. A. Mayer, A. Goel, E. A. Espeso, M. A. Peñalva, and H. N. Arst, Jr. 1998. Putative membrane components of signal transduction pathways for ambient pH regulation in Aspergillus and meiosis in Saccharomyces are homologous. Mol. Microbiol. 30:259-264. [DOI] [PubMed] [Google Scholar]
- 7.Denison, S. H., M. Orejas, and H. N. Arst, Jr. 1995. Signaling of ambient pH in Aspergillus involves a cysteine protease. J. Biol. Chem. 270:28519-28522. [DOI] [PubMed] [Google Scholar]
- 8.Díez, E., J. Álvaro, E. A. Espeso, L. Rainbow, T. Suárez, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva. 2002. Activation of the Aspergillus PacC zinc-finger transcription factor requires two proteolytic steps. EMBO J. 21:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espeso, E. A., and M. A. Peñalva. 1992. Carbon catabolite repression can account for the temporal pattern of expression of a penicillin biosynthetic gene in Aspergillus nidulans. Mol. Microbiol. 6:1457-1465. [DOI] [PubMed] [Google Scholar]
- 10.Espeso, E. A., T. Roncal, E. Díez, L. Rainbow, E. Bignell, J. Álvaro, T. Suárez, S. H. Denison, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva. 2000. On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J. 19:719-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futai, E., T. Kubo, H. Sorimachi, K. Suzuki, and T. Maeda. 2001. Molecular cloning of PalBH, a mammalian homologue of the Aspergillus atypical calpain PalB. Biochim. Biophys. Acta 1517:316-319. [DOI] [PubMed] [Google Scholar]
- 12.Futai, E., T. Maeda, H. Sorimachi, K. Kitamoto, S. Ishiura, and K. Suzuki. 1999. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol. Gen. Genet. 260:559-568. [DOI] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 17.Kranz, A., A. Kinner, and R. Kolling. 2001. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell 12:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemmon, S. K., and L. M. Traub. 2000. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12:457-466. [DOI] [PubMed] [Google Scholar]
- 19.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, W. S., and A. P. Mitchell. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo, K. W., Q. Zhang, M. Li, and M. Zhang. 1999. Apoptosis-linked gene product ALG-2 is a new member of the calpain small subunit subfamily of Ca2+-binding proteins. Biochemistry 38:7498-7508. [DOI] [PubMed] [Google Scholar]
- 22.Maccheroni, W., G. S. May, N. M. Martinez-Rossi, and A. Rossi. 1997. The sequence of palF, an environmental pH response gene in Aspergillus nidulans. Gene 194:163-167. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Mingot, J. M., E. A. Espeso, E. Díez, and M. A. Peñalva. 2001. Ambient pH signaling regulates nuclear localization of the Aspergillus nidulans PacC transcription factor. Mol. Cell. Biol. 21:1688-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mingot, J. M., J. Tilburn, E. Díez, E. Bignell, M. Orejas, D. A. Widdick, S. Sarkar, C. V. Brown, M. X. Caddick, E. A. Espeso, H. N. Arst, Jr., and M. A. Peñalva. 1999. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signalling is bypassed. Mol. Cell. Biol. 19:1390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Missotten, M., A. Nichols, K. Rieger, and R. Sadoul. 1999. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 6:124-129. [DOI] [PubMed] [Google Scholar]
- 27.Negrete-Urtasun, S., S. H. Denison, and H. N. Arst, Jr. 1997. Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J. Bacteriol. 179:1832-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negrete-Urtasun, S., W. Reiter, E. Díez, S. H. Denison, J. Tilburn, E. A. Espeso, M. A. Peñalva, and H. N. Arst, Jr. 1999. Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol. Microbiol. 33:994-1003. [DOI] [PubMed] [Google Scholar]
- 29.Nickas, M. E., and M. P. Yaffe. 1996. BRO1, a novel gene that interacts with components of the Pkc1p-mitogen-activated-protein kinase pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2585-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohno, H., J. Stewart, M. C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, and J. S. Bonifacino. 1995. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269:1872-1875. [DOI] [PubMed] [Google Scholar]
- 31.Orejas, M., E. A. Espeso, J. Tilburn, S. Sarkar, H. N. Arst, Jr., and M. A. Peñalva. 1995. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 9:1622-1632. [DOI] [PubMed] [Google Scholar]
- 32.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peñalva, M. A., and H. N. Arst, Jr. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez, O. D., and G. P. Nolan. 2001. Resistance is futile: assimilation of cellular machinery by HIV-1. Immunity 15:687-690. [DOI] [PubMed] [Google Scholar]
- 35.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Ruden, D. M., J. Ma, Y. Li, K. Wood, and M. Ptashne. 1991. Generating yeast transcriptional activators containing no yeast protein sequences. Nature 350:250-252. [DOI] [PubMed] [Google Scholar]
- 38.Sorimachi, H., S. Ishiura, and K. Suzuki. 1997. Structure and physiological functions of calpains. Biochem. J. 328:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Springael, J. Y., E. Nikko, B. André, and A. M. Marini. 2002. Yeast Npi3/Bro1 is involved in ubiquitin-dependent control of permease trafficking. FEBS Lett. 517:103-109. [DOI] [PubMed] [Google Scholar]
- 40.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Peñalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 42.Vincent, O., and M. Carlson. 1999. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 18:6672-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vito, P., E. Lacana, and L. D'Adamio. 1996. Interfering with apoptosis: Ca(2+)-binding protein ALG-2 and Alzheimer's disease gene ALG-3. Science 271:521-525. [DOI] [PubMed] [Google Scholar]
- 44.Vito, P., L. Pellegrini, C. Guiet, and L. D'Adamio. 1999. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J. Biol. Chem. 274:1533-1540. [DOI] [PubMed] [Google Scholar]
- 45.Wu, Y., S. Pan, S. Che, G. He, M. Nelman-Gonzalez, M. M. Weil, and J. Kuang. 2001. Overexpression of Hp95 induces G1 phase arrest in confluent HeLa cells. Differentiation 67:139-153. [DOI] [PubMed] [Google Scholar]
- 46.Xu, W., and A. P. Mitchell. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, X., E. J. Hubbard, and M. Carlson. 1992. A protein kinase substrate identified by the two-hybrid system. Science 257:680-682. [DOI] [PubMed] [Google Scholar]