Abstract
Although the role of Hox genes in patterning the mammalian body plan has been studied extensively during embryonic and fetal development, relatively little is known concerning Hox gene function in adult animals. Analysis of mice with mutant Hoxa9, Hoxb9, and Hoxd9 genes shows that these paralogous genes are required for mediating the expansion and/or differentiation of the mammary epithelium ductal system in response to pregnancy. Mothers with these three mutant genes cannot raise their own pups, but the pups can be rescued by fostering by wild-type mothers. Histologically, the mammary glands of the mutant mothers seem normal before pregnancy but do not develop properly in response to pregnancy and parturition. Hoxa9, Hoxb9, and Hoxd9 are expressed normally in adult mammary glands, suggesting a direct role for these genes in the development of mammary tissue after pregnancy. Because loss-of-function mutations in these Hox genes cause hypoplasia of the mammary gland after pregnancy, it may be productive to look for misexpression of these genes in mammary carcinomas.
Hox genes encode transcription factors belonging to the Antennapedia homeodomain class. These genes may be involved in the establishment of body plans for all metazoans of the animal kingdom. In mammals, the Hox gene complex contains 39 genes arranged into four linkage groups on four separate chromosomes. These four clusters are referred to as HoxA, HoxB, HoxC, and HoxD. Individual genes within each of the four mammalian linkage groups have been classified into 13 paralogous families on the basis of sequence similarity and chromosomal position within each linkage group. Members of the same paralogous family often exhibit similar expression patterns and cofunction to pattern the mammalian embryo.
Studies on mouse gain-of-function and loss-of-function mutations have shown that Hox genes collectively control regionalization of the mouse embryo along the major body axes (1–4). In mammals, Hox genes function not as individual entities but as members of a highly integrated network with paralogous genes, adjacent genes in the same linkage group, and even nonparalogous genes in separate linkage groups, interacting positively, negatively, and in parallel with each other to orchestrate the morphological regionalization of the embryo (5–16). A molecular interpretation of the genetic data suggests that multiple Hox genes concurrently modulate the activity of multiple target genes through binding to common cis regulatory elements and that it is the integration of the multiple Hox gene signals that dictate the final output from these target genes (4, 7, 13, 17).
Studies of single and compound mouse mutants of Hoxa9 and Hoxb9 showed synergistic interactions between these two genes in the patterning of both the lumbosacral axial skeleton and the humerus (12). Synergistic interactions between Hoxa9 and Hoxb9 also have been illustrated by observations of abnormal patterning of the thoracic region in single and compound mutants of Hoxa9 and Hoxb9 (18). Much more extensive abnormalities are found in the formation of the axial and appendicular skeletal structures of Hoxa9, Hoxb9, Hoxd9 triple mutants compared with the double mutants. For example, in the thoracic region of the triple mutants, fusions of the first, second, and third ribs are seen with 100% penetrance; however, in the double mutants, these fusions are restricted to only two ribs (18). Also, in the formation of the forelimbs, rather than observing only malformation of the humerus common to Hoxa9, Hoxd9 double mutants (12), the defects in the triple mutants extend into the formation of the radius and ulna. Articulations between the humerus and the radius and ulna are poorly formed, and the radius is shorter and curved. Even the articulations with the autopod are defective, such that the animals walk on the sides or backs of their palms.
The topic of this manuscript, however, is not the roles of Hoxa9, Hoxb9, and Hoxd9 during mouse embryogenesis but the role of these genes in the adult mouse. The mammary glands of Hoxa9, Hoxb9, Hoxd9 triple-mutant virgin females seem normal. However, triple-mutant females show hypoplasia of the mammary glands during pregnancy and post parturition. These mothers cannot raise their pups because of a failure to produce milk. All three genes are expressed in adult mammary tissue, suggesting a direct role for these genes in mediating the expansion and/or differentiation of the mammary epithelium ductal system in preparation for lactation.
MATERIALS AND METHODS
Generation of Mice Deficient for Hoxa9, Hoxb9, and Hoxd9.
The generation of mice carrying disruptions in their respective homeoboxes for Hoxa9 and Hoxb9 has been described (18). An MC1 neoPoly(A) cassette (19) was inserted into the Eco47IV site of the Hoxd9 homeobox. This mutation disrupts the ability of Hoxd9 protein to bind DNA. Mutant mice carrying this disruption were generated by embryonic-stem-cell-mediated gene targeting and mouse chimera production as described (20, 21). Matings between individual mutants were carried out to generate double mutants and, subsequently, triple mutants. Genotypes of Hoxa9 and Hoxb9 were determined by PCR as described (18).
Histology.
Mammary-gland biopsies were fixed in Carnoy’s fixative or in 4% formaldehyde in PBS overnight at room temperature. Fat was removed from the tissues by three 1-h washes of acetone. For whole-mount preparation, mammary glands were rehydrated through a decreasing ethanol series, then stained in hematoxylin for 10 min, and destained in running deionized water for 10 min. For histological sections, mammary glands were embedded in paraffin after the acetone washes. Sections 10 μm thick were collected and stained regressively with hematoxylin and eosin.
RNA Whole-Mount in Situ Hybridization.
RNA whole-mount in situ hybridization on E9.0–E12.5 embryos was performed as described (22). The templates for Hoxa9, Hoxb9, and Hoxd9 RNA in situ probes were a 0.6-kb BglII–EcoRI fragment from Hoxa9, a 0.5-kb fragment in the 3′ untranslated region of Hoxb9, and a 0.8-kb PstI–EcoRI fragment from the 3′ untranslated region of Hoxd9.
Reverse Transcription–PCR.
Total RNA was isolated from E12.5 embryos (without heads), adult liver biopsies, and adult mammary glands of different stages by using the TRIzol reagents (BRL) according to the manufacturer’s guidelines. The RNA samples were treated with RNase-free DNase (Boehringer Mannheim) at 37°C for 40 min, followed by phenolchloroform extraction and ethanol precipitation. Total RNA (1.5 μg) was used in the reverse transcription reaction with poly(dT)12–18 primers. To demonstrate that the amplification in the reverse transcription–PCR was solely from the cDNA and not from any DNA contamination, a second set of mixtures was prepared with the same amount of RNA as the first set with all the reagents except for the reverse transcriptase. The amount of reverse transcription mixture used in the PCR was equalized by the amplification of the β-actin cDNA. PCR was performed as follows: 5 min at 94°C; then 30 cycles of 95°C for 20 s, 65°C for 20 s, and 72°C for 20 s; and then a 5-min extension at 72°C. The primers used were β-actin (forward, 5′-CTCCATCGTGGGCCGCTCTAG-3′; reverse, 5′-GTAACAATGCCATGTTCAAT-3′; 137-bp band product), Hoxa9 (forward, 5′-CGCTGGAACTGGAGAAGGAGTTTCTG-3′; reverse, 5′-ATCCTGCGGTTCTGGAACCAGATC-3′; 123-bp band product), Hoxb9 (forward, 5′-CAGGGAGGCTGTCCTGTCTAATC-3′; reverse, 5′-CTTCTCTAGCTCCAGCGTCTGG-3′; 177-bp band product), Hoxc9 (forward, 5′-GCAACCCCGTGGCCAACTGGATCC-3′; reverse, 5′-AAGACGGTGGGCTTTTCTCTATCTTGT-3′; 376-bp band product), and Hoxd9 (forward, 5′-AGCGAACTGGATCCACGCTCGCTCCA-3′; reverse, 5′-GACTTGTCTCTCTGTAAGGTTCAGAATCC-3′; 196-bp band product).
RESULTS
The genotype of mutant mice is designated by capital and lower-case letters, where A, B, and D denote the wild-type alleles and a, b, and d denote the loss-of-function alleles of Hoxa9, Hoxb9, and Hoxd9, respectively. Thus, the triple-mutant homozygous mouse is designated aabbdd.
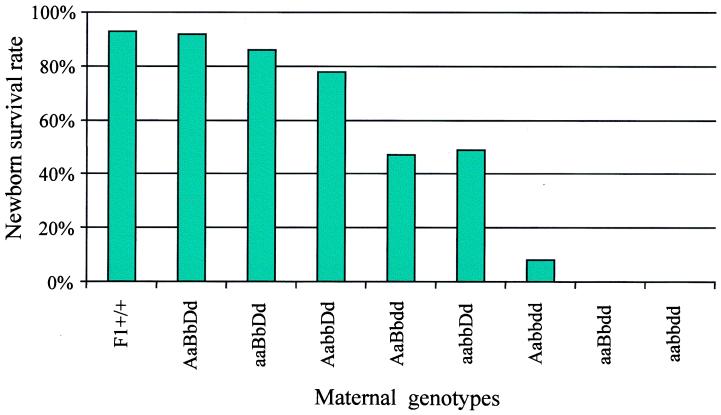
Pups born to Aabbdd, aaBbdd, and aabbdd females usually died within 48 h of birth (Fig. 1). Examination indicated that the recovered dead pups had signs of dehydration and little or no milk in their stomachs. Dead pups recovered from mothers of Aabbdd, aaBbdd, and aabbdd can be of any genotype predicted from the parental genotypes. To show that it is the mother’s genotype that is critical to the viability of offspring, we examined the results of two types of matings. The first involved matings between females with four mutant alleles of Hoxa9, Hoxb9, or Hoxd9 crossed with aabbdd males, and the second involved matings between aabbdd females and males with four mutant alleles in these genes. Pups born to these two types of matings should have the same distribution of mutant alleles. However, the majority of pups from the first type of matings survived to weaning, whereas none of the pups from the second type of matings survived without human intervention.
Figure 1.
The percentage of newborn pups that survived to weaning decreased dramatically when the females carried a greater number of mutant Hox 9 alleles. About 94% of the pups born to control females survived to weaning. The newborn survival rate dropped to 9% for pups of Aabbdd females and to 0% for pups of aaBbdd and aabbdd females.
Pups born to Aabbdd, aaBbdd, and aabbdd mutant females could be rescued if the newborns were fostered with a wild-type, lactating female. Approximately 65% of the pups born to these mutant females thrived under the care of foster mothers. This number may be an underestimate of the actual survival frequency, because it was not always possible to foster pups immediately after birth. A rapid increase in the volume of milk in the pups’ stomachs was observed after fostering. These experiments show that no irreversible damage is inflicted on the pups during pregnancy or parturition. The ability of these newborns to obtain sufficient nutrition from foster females suggests that either the quality or a reduction in the quantity of milk produced by the mutant mothers is responsible for the mortality of the newborns.
In reverse fostering experiments, we removed the pups born to Aabbdd, aaBbdd, and aabbdd females and replaced them with newborn pups (from 1- to 4-days old) from Swiss–Webster mothers. All wild-type pups switched to mutant females were healthy and had stomachs full of milk at the time of fostering. However, after several days, all of them showed signs of dehydration and were runted compared with their littermates who stayed with their own Swiss–Webster mothers. Half of them died within 48 h of being placed with the mutant mothers. This result indicates that the mutant females have difficulty in nursing their pups. Because a portion of the Swiss–Webster pups transferred to the mutant mothers survived to weaning, it does not seem that the milk produced by mutant females is toxic or lacks an essential nutrient for pup survival. Rather, this result suggests that a reduction in the quantity of milk produced is responsible for death of the pups. It is also apparent that the first few days after birth are the most critical to survival. Pups initially nurtured by the Swiss–Webster mother have a greater survival rate when transferred to the mutant mothers than do the mutant mother’s own biological pups reared from birth.
Hypoplasia of Mammary Glands During and After Pregnancy.
The mammary glands of Aabbdd, aaBbdd, and aabbdd virgin females were indistinguishable either by whole-mount or by histological examination of sections from those of wild-type, age-matched control virgin females (data not shown). However, relative underdevelopment of the mammary tissue from the mutant females was evident during pregnancy and post parturition. Fig. 2 A and B compares whole-mount preparations of the fourth mammary gland from a wild-type female and an aabbdd mutant female both at 12.5 days of pregnancy, respectively. In wild-type females at this stage of pregnancy, the mammary gland already occupies almost the entire fat pad. In aabbdd mice, branching of the epithelial ductal system is reduced significantly. At higher magnifications, lobuloalveolar structures of apparent normal cellular morphology are seen in the mutant pregnant female mice but are reduced in number (Fig. 2 C–F). In wild-type mammary tissues, lobuloalveoli already cover the entire length of the epithelial branch, whereas in the mutant mammary glands, lobuloalveoli are restricted to the ends of the bud (evident in Fig. 2 C–F).
Figure 2.
Hypoplasia is evident in the mammary glands of the Aabbdd, aaBbdd, and aabbdd mutant females during pregnancy and around parturition. A and B are whole-mount preparations (×2.5). C, D, G, and H are sections under low magnification (×62). E, F, I, and J are sections under high magnification (×250). A, C, and E are from the fourth mammary glands of wild-type females at 12.5 days of pregnancy. B, D, and F are from the fourth mammary glands of aabbdd females at 12.5 days of pregnancy . G and I are from the fourth mammary gland of a wild-type female shortly post partum. H and J are from the fourth mammary gland of an aabbdd female shortly post partum. The mammary epithelia (e) include mammary ducts (d) and lobuloalveolar structures (a) in pregnant and lactating females. The mammary stroma (s) consist mainly of fat cells (f) and fibroblasts. The arrow in J points to an intracellular oil droplet in an alveolar cell.
After parturition, reduced development of the mammary glands of Aabbdd, aaBbdd, and aabbdd females, compared with age-matched control wild-type females, continues to be apparent (Fig. 2 G–J). In many cases, the morphological characteristics of the mutant mammary glands resemble those of mammary glands of normal mice at midpregnancy. At this stage, not only are lobuloalveolar structures in the mutant females reduced in number, but also large amounts of intracellular fat droplets are apparent (Fig. 2 H and J). The continued hypoplasia of the mutant mammary glands seems to result from a delay in both the growth and differentiation of the mammary epithelium. Such a delay could account for reduced milk production at parturition.
Expression of Hoxa9, Hoxb9, and Hoxd9 in Mammary Glands.
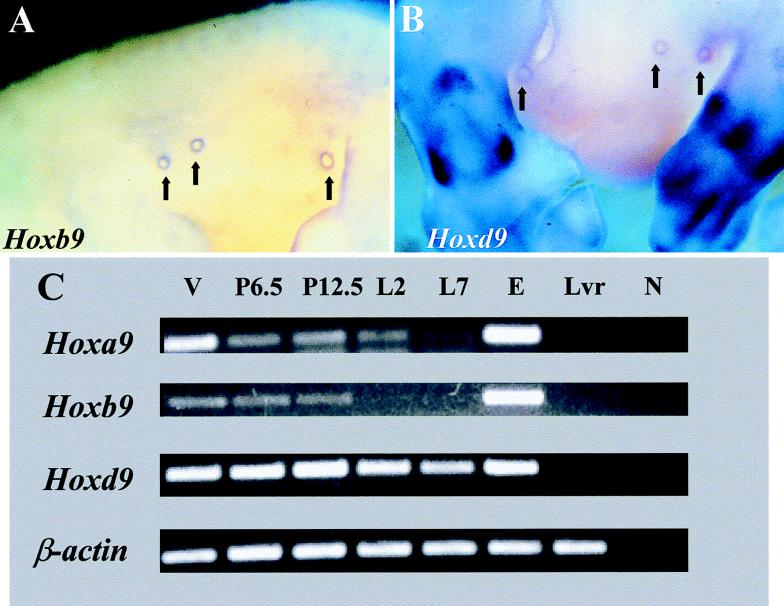
Hoxb9 and Hoxd9 are expressed in the mammary primordia of E12.5 embryos as detected by RNA in situ hybridization (Fig. 3 A and B). By using a Hoxa9 probe, we also observed a very weak signal in the mesenchyme surrounding the primitive nipples (data not shown). Interestingly, the expression pattern of these genes in the mammary primordia is reciprocal to that of Bmp-2, which is expressed in the center of the circle of cells expressing Hoxa9, Hoxb9, and Hoxd9 (data not shown). At this stage, Hoxa9, Hoxb9, and Hoxd9 seem to be restricted to the mesenchyme of the mammary primordium, whereas Bmp-2 expression is observed in the epithelium.
Figure 3.
RNA whole-mount in situ hybridization on E12.5 Hoxb9 (A) and Hoxd9 (B) embryos. Arrows point to the dense mesenchyme surrounding the primitive nipples. (C) Results of reverse transcription–PCR analyses with templates from various sources: the mammary gland of a virgin mouse (V), the mammary gland from a mouse at 6.5 days of pregnancy (P6.5), the mammary gland from a mouse at 12.5 days of pregnancy (P12.5), the mammary gland from a mouse that had been lactating for 2 days (L2), the mammary gland from a mouse that had been lactating for 7 days (L7), E12.5 embryos without heads (E), adult liver (Lvr), and a no-template control (N).
Because of the large amount of fat present in the mammary glands of adult mice, in situ hybridization is not a very effective means of visualizing RNA expression in this tissue. The presence of fat results in nonspecific absorption of the RNA probe and therefore high background. Instead, reverse transcription–PCR was used to investigate expression of these Hox genes in adult mammary glands. The amplification of β-actin was used as a control for quality and quantity of templates in each sample. The expression of Hoxa9, Hoxb9, and Hoxd9 was found in mammary-gland samples from 8-week-old virgin females, females at 6.5 days of pregnancy, females at 12.5 days of pregnancy, females lactating for 2 days, and females lactating for 7 days, as well as E12.5 embryos, but expression of Hoxa9, Hoxb9, and Hoxd9 was not found in the liver, which serves as a negative control (Fig. 3C). The expression of Hoxa9, Hoxb9, and Hoxd9 in adult mammary-gland tissues suggests that they have direct roles, in response to pregnancy, in the development of the mammary gland, which, if disrupted, can lead to hypoplasia of the gland.
It has been well documented that the development of mammary glands is mediated by the production of reproductive hormones, such as estrogen, progesterone, and prolactin. These hormones also are required absolutely for normal pregnancy. Because even aabbdd females can become pregnant and deliver potentially viable pups, the major hormonal circuits in the mutant females must be intact. In support of this hypothesis, measurements of serum-estrogen levels in virgin and pregnant aabbdd females indicate that concentrations of this hormone do not differ significantly from those in age-matched, wild-type female controls (data not shown). Thus, the mammary hypoplasia observed in aabbdd mice is not likely to be caused by a deficit in the production of reproductive hormones.
DISCUSSION
Roles of Hox Genes in Adult Mammary-Gland Development.
Mutations in Hox genes often cause regional hypoplasia resulting from an apparent loss of cell proliferation. This hypoplasia is particularly evident, for example, during limb development, where disruption of 5′ Hox genes does not result in the conversion of one limb bone into another (i.e., a homeotic transformation) but in the reduction in the length of specific bony elements. Addition of multiple 5′ Hox gene mutations to the same mouse results in the exacerbation of the limb hypoplasia. For example, in Hoxa11, Hoxd11 double mutants, only remnants of the radius and ulna are formed, and in Hoxa13 and Hoxd13 double mutants, only remnants of the paws are formed (7, 13). During limb development, Hox genes are not only functional during the initial patterning of the mesenchymal condensations, which lead to bone formation during chondrogenesis, but also at later stages of long-bone growth. During this latter phase, Hox genes are expressed and function in the prehypertrophic cells in the growth plates.
An independent link between Hox genes and cell proliferation is seen in certain neoplasias. Ectopic activation of Hox genes has been observed in a number of human and mouse leukemias. In myeloid leukemias, this correlation includes activation of human HoxA9 and mouse Hoxa9 by chromosomal translocations or proviral insertions (23–25). In addition, a number of Hox genes have been shown to function as oncogenes in tissue-culture transfection assays (26, 27). Given their apparent role in controlling cell proliferation in a number of in vivo and in vitro settings, it is perhaps not unexpected that Hox genes were recruited during mammalian evolution to regulate the expansion and differentiation of mammary tissue during pregnancy and after parturition.
The roles of Hoxa9, Hoxb9, and Hoxd9 in postembryonic development of the mammary gland have not been defined clearly. During pregnancy, the mutant phenotype seems to be a failure of cell expansion. Significantly less branching of the epithelial ductal system is apparent, but the morphology of the lobuloalveolar structures appears normal. After parturition, however, not only is hypoplasia of the mammary tissue evident, but the morphology of the lobuloalveolar structures is also aberrant. These observations suggest that these Hox genes may not be restricted to a single role, but they may be performing multiple roles that are changing with time. Thus, it is possible that Hoxa9, Hoxb9, and Hoxd9 not only influence the proliferation and branching of mammary epithelium but may also affect its differentiation at a later stage. Such multiple roles are apparent in many other structures and organs affected by Hox gene mutations, such as the formation of the long bones of the limb (7, 13, 28).
In embryos, the Hox9 paralogous genes seem to be expressed in the mesenchyme of the newly formed mammary gland. In adults, although the quality of in situ hybridization is poor, the signal is still associated predominantly with the mesenchyme. This association is evident most clearly for Hoxd9. The inductive role of the mammary-gland mesenchyme or stroma in promoting branching of the epithelial ductal system has been documented clearly in organ culture (29). Further, hepatocyte growth factor (HGF), a secreted factor produced by the mammary mesenchyme, has been implicated through interactions with its receptor c-met as a potential inducer of the epithelial branching (30). It might be informative to investigate whether the growth control and/or differentiation function of these paralogous Hox genes operate through the HGF–c-met signaling pathway.
Hoxc9 is also expressed in the adult mammary gland during pregnancy and after parturition (unpublished results). It is anticipated that introduction of Hoxc9 mutant alleles into aabbdd mice would exacerbate mammary-gland hypoplasia further. However, because introduction of Hoxc9 mutant alleles into mice that already have Hoxa9, Hoxb9, and Hoxd9 mutations further reduces their fertility and viability, this approach is not practical. To obviate this problem, mice would need to be generated with conditional mutations in these genes, where the mutation is restricted to adult mesenchymal-derived breast tissue. The generation of such mice would be possible through the use of the CRE-recombinase/LoxP system to mediate site-specific recombination in breast mesenchymal cells (31, 32).
In summary, we have shown that loss-of-function mutations in the Hox9 paralogous genes result in mammary gland hypoplasia. Before pregnancy, the mammary tissue in Hoxa9, Hoxb9, Hoxd9 triple-mutant homozygotes appears normal. However, in response to pregnancy, both expansion and differentiation of the mammary-gland ductal system are defective, leading to deficits in the production of milk. A prediction based on this finding is that gain-of-function mutations that cause ectopic activation of Hox genes could lead to hyperplasia of mammary tissue and thereby contribute to mammary neoplasia. Ectopic expression of a number of Hox genes has been observed in mouse mammary tumors (33). It may be productive to look for misexpression of Hox9 paralogous Hox genes in human mammary carcinomas.
Acknowledgments
We thank O. Chisaka, J. Goddard, L. Reynolds, C. Peterson, and D. Spyropoulos for their contributions in generating the triple-mutant colony for Hoxa9, Hoxb9, and Hoxd9; M. Allen, S. Barnett, C. Lenz, G. Peterson, M. Wagstaff, and J. Hayes for technical assistance; C. Daniel for suggestions; A. Godwin for helpful comments on the manuscript; and L. Oswald for help with the preparation of the manuscript. F.C. was supported, in part, by a research fellowship from the University of Utah.
ABBREVIATION
- En
embryonic day n
Footnotes
A Commentary on this article begins on page 322.
References
- 1.Krumlauf R. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 2.Maconochie M, Nonchev S, Morrison A, Krumlauf R. Annu Rev Genet. 1996;30:529–556. doi: 10.1146/annurev.genet.30.1.529. [DOI] [PubMed] [Google Scholar]
- 3.Capecchi M R. In: Molecular and Cellular Aspects of Neural Development. Cowan W M, Jessell W M, Zipursky S L, editors. New York: Oxford Univ. Press; 1997. pp. 334–355. [Google Scholar]
- 4.Capecchi M R. Cold Spring Harbor Symposia on Quantitative Biology. Vol. 62. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 273–281. [PubMed] [Google Scholar]
- 5.Condie B G, Capecchi M R. Nature (London) 1994;370:304–307. doi: 10.1038/370304a0. [DOI] [PubMed] [Google Scholar]
- 6.Davis A P, Capecchi M R. Development (Cambridge, UK) 1994;120:2187–2198. doi: 10.1242/dev.120.8.2187. [DOI] [PubMed] [Google Scholar]
- 7.Davis A P, Witte D P, Hsieh L H, Potter S S, Capecchi M R. Nature (London) 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 8.Horan G S, Kovacs E N, Behringer R R, Featherstone M S. Dev Biol. 1995;169:359–372. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 9.Rancourt D E, Tsuzuki T, Capecchi M R. Genes Dev. 1995;9:108–122. doi: 10.1101/gad.9.1.108. [DOI] [PubMed] [Google Scholar]
- 10.Davis A P, Capecchi M R. Development (Cambridge, UK) 1996;122:1175–1185. doi: 10.1242/dev.122.4.1175. [DOI] [PubMed] [Google Scholar]
- 11.Favier B, Rijli F M, Fromental-Ramain C, Fraulob V, Chambon P, Dollé P. Development (Cambridge, UK) 1996;122:449–460. doi: 10.1242/dev.122.2.449. [DOI] [PubMed] [Google Scholar]
- 12.Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Dierich A, Dollé P, Chambon P. Development (Cambridge, UK) 1996;122:461–472. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- 13.Fromental-Ramain C, Warot X, Messadecq N, Le Mouellic M, Dollé P, Chambon P. Development (Cambridge, UK) 1996;122:2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- 14.van der Hoeven F, Zákány J, Duboule D. Cell. 1996;85:1025–1035. doi: 10.1016/s0092-8674(00)81303-3. [DOI] [PubMed] [Google Scholar]
- 15.Manley N R, Capecchi M R. Dev Biol. 1997;192:274–288. doi: 10.1006/dbio.1997.8765. [DOI] [PubMed] [Google Scholar]
- 16.Manley N R, Capecchi M R. Dev Biol. 1998;195:1–15. doi: 10.1006/dbio.1997.8827. [DOI] [PubMed] [Google Scholar]
- 17.Zákány J, Fromental-Ramain C, Warot X, Duboule D. Proc Natl Acad Sci USA. 1997;94:13695–13698. doi: 10.1073/pnas.94.25.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F, Capecchi M R. Dev Biol. 1997;181:186–196. doi: 10.1006/dbio.1996.8440. [DOI] [PubMed] [Google Scholar]
- 19.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 20.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 21.Capecchi M R. Sci Am. 1994;270(3):54–61. doi: 10.1038/scientificamerican0394-52. [DOI] [PubMed] [Google Scholar]
- 22.Manley N R, Capecchi M R. Development (Cambridge, UK) 1995;121:1989–2003. doi: 10.1242/dev.121.7.1989. [DOI] [PubMed] [Google Scholar]
- 23.Blatt C, Sachs L. Biochem Biophys Res Commun. 1988;156:1265–1270. doi: 10.1016/s0006-291x(88)80769-1. [DOI] [PubMed] [Google Scholar]
- 24.Borrow J, Shearman A M, Stanton V J, Becher R, Collins T, Williams A J, Dube I, Katz F, Kwong Y L, Morris C, et al. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura T, Largaespada D A, Lee M P, Johnson L A, Ohyashiki K, Toyama K, Chen S J, Willman C L, Chen I M, Feinberg A P, et al. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 26.Perkins A, Kongsuwan K, Visvader J, Adams J M, Cory S. Proc Natl Acad Sci USA. 1991;87:8398–8402. doi: 10.1073/pnas.87.21.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aberdam D, Negreanu V, Sachs L, Blatt C. Mol Cell Biol. 1991;11:554–557. doi: 10.1128/mcb.11.1.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warot X, Fromental-Ramain C, Fraulob V, Chambon P, Dollé P. Development (Cambridge, UK) 1997;124:4781–4791. doi: 10.1242/dev.124.23.4781. [DOI] [PubMed] [Google Scholar]
- 29.Sakakura T, Sakagami Y, Nishizuka Y. Dev Biol. 1979;72:201–210. doi: 10.1016/0012-1606(79)90111-8. [DOI] [PubMed] [Google Scholar]
- 30.Niranjan B, Buluwela L, Yant J, Perusinghe N, Atherton A, Phippard D, Dale T, Gusterson B, Kamalati T. Development (Cambridge, UK) 1995;121:2897–2908. doi: 10.1242/dev.121.9.2897. [DOI] [PubMed] [Google Scholar]
- 31.Gu H, Zou Y-R, Rajewsky K. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 32.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 33.Friedmann Y, Daniel C A, Strickland P, Daniel C W. Cancer Res. 1994;54:5981–5985. [PubMed] [Google Scholar]