Abstract
The vanB2 operon encoding glycopeptide resistance is an integral part of the putative conjugative transposon Tn5382. Characterization of clinical glycopeptide resistant derivatives from an epidemic ampicillin-resistant Enterococcus faecium strain showed precise chromosomal or plasmid insertions of a vanB2-containing Tn5382-like element. Conjugative transposition of the Tn5382-like element was not demonstrated in retransfer studies.
The 34-kb vanB-containing transposon Tn1549, which is similar to the Tn916-Tn1545 family of conjugative transposons, was recently described (11). Tn1549 appears to be identical to the previously described and partially sequenced transposon Tn5382 (4). It is therefore designated a Tn5382-like element. The vanB2 operon seems to be universally linked to the Tn5382-like element and is the most widespread vanB subtype (7, 8, 15, 16, 17, 19). Conjugative transposons encode the functions for excision to a circular intermediate and intercellular transfer (5). The staggered cleavage of Tn916-Tn1545 during transposition results in the transfer of a coupling sequence from the previous target. The circular intermediates are joined at the ends by mismatched strands that are repaired in enterococci (18), representing sequences from flanking regions on either side of the previous insertion. Transposon-flanking regions thus represent the target sequence on one side and one of the mismatched strands on the other (3, 21). Evidence of Tn5382 transposition has been described previously (4, 11). We present additional data for precise transposition of Tn5382-like elements under natural conditions describing the arrival of the Tn5383-like element in a Norwegian ampicillin-resistant Enterococcus faecium outbreak strain (TUH2-21).
Relevant characteristics of the strains used are listed in Table 1. Isolation of DNA (2, 22), pulsed-field gel electrophoresis (PFGE), Southern hybridization, matings, PCRs, and DNA sequencing (6, 7) were performed as described previously. Amplicons and primer sequences are given in Table 2. Various bacteria were used as templates for PCR probe synthesis: 16S rRNA gene (rDNA) probe from Enterococcus faecalis DS16C2 (10), 23S rDNA from E. faecium ATCC 19434, IS1216V from E. faecium BM4147 (14), Tn5382 from E. faecium C68 (4), and vanB from E. faecalis V583 (9). A chromosomal vanB location was assessed by sequential vanB, 16S rDNA, and 23S rDNA hybridizations of PFGE-separated, I-CeuI (New England Biolabs)-digested DNA. Plasmid vanB localization was examined by sequential vanB and 16S rDNA hybridizations of plasmid DNA. Inverse PCR was performed to characterize Tn5382-flanking sequences. The Tn5382 left and right ends were amplified with internal divergent Tn5382 primers after ligation of DraI-, HinfI-, or HphI/BfaI- and AluI-, DdeI-, or Sau3AI-digested total DNA (1 to 3 μg), respectively. The Sequence Navigator Software Package (Perkin-Elmer) and the Blastn and Blastx local alignment search tools (1) were used for sequence analyses.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristicsa | Reference(s) |
|---|---|---|
| E. faecium | ||
| TUH2-21 | Epidemic human clinical strain; Apr Vms | 12 |
| TUH2-18 | Clonally related to TUH2-21; chromosomal vanB2; Apr Vmr | 6, 12 |
| TUH2-19 | Clonally related to TUH2-21; plasmid vanB2; Apr Vmr | 12 |
| TUH2-20 | Clonally related to TUH2-21; chromosomal vanB2; Apr Vmr | 12 |
| TUH2-55 | Clonally related to TUH2-21; plasmid vanB2; Apr Vmr | 12 |
| BM4105-RF | Recipient strain in transfer studies; Vms Rifr Fusr | 20 |
| E. faecalis | ||
| JH2-2 | Recipient strain in transfer studies; Vms Rifr Fusr | 13 |
| UV202 | Recombination-deficient derivative of JH2-2 | 23 |
Apr, ampicillin resistant; Vms, vancomycin susceptible; Vmr, vancomycin resistant; Rifr, rifampicin resistant; Fusr, fucidic acid resistant. The TUH strains were isolated at Haukeland University Hospital, Bergen, Norway (12).
TABLE 2.
PCR primers used in this study
| Amplicon | Primer sequence (5′ - 3′) | Size of amplicon (bp) | Reference |
|---|---|---|---|
| vanB | CAAAGCTCCGCAGCTTGCATG | 484 | 6 |
| TGCATCCAAGCACCCGATATAC | |||
| vanB long | GTTTGATGCAGAGGCAGACGACT | 5,959 | 6 |
| ACAAGTTCCCCTGTATCCAAGTGG | |||
| Tn5382 | GTTCTTATTCCGCAGGTGGTGATT | 311 | 4 |
| ACGCCATGCTATTTACTTCCGGC | |||
| Tn5382 left inverse | GCTATGGCAGTTTTCCGTGTG | Variable | This study |
| TCGCCTCCTTTCTCTATTTGG | |||
| Tn5382 right inverse | GAGGGGGAAATGGTGAGAGGT | Variable | This study |
| AACGCTTCTTCATGGCTCTTG | |||
| TUH2-18 Tn5382 left | TACTGCCAATGATGTCAACCC | 721 | This study |
| GTTCTTATTCCGCAGGTGGTGATT | |||
| TUH2-18 Tn5382 right | GAGGGGGAAATGGTGAGAGGT | 413 | This study |
| ATCCTTTGACGATCATCTTGG | |||
| TUH2-18 Tn5382 target | TACTGCCAATGATGTCAACCC | 439 | This study |
| ATCCTTTGACGATCATCTTGG | |||
| TUH2-19 Tn5382 left | CCGCAAGGGGATTTTAGTA | 677 | This study |
| GTTCTTATTCCGCAGGTGGTGATT | |||
| TUH2-19 Tn5382 right | GAGGGGGAAATGGTGAGAGGT | 248 | This study |
| CCACGGCTACAATAATCACA | |||
| TUH2-19 Tn5382 target | CCGCAAGGGGATTTTAGTA | 224 | This study |
| CCACGGCTACAATAATCACA | |||
| Tn5382-like junction | GAGGGGGAAATGGTGAGAGGT | 701a | This study |
| GTTCTTATTCCGCAGGTGGTGATT | |||
| IS1216V | AAAGCAATTTCAGCAGGATG | 456 | This study |
| GTACGATGTTCTGTCCCTTG | |||
| 16S rDNA | TGCATTAGCTAGTTGGTGAGG | 726 | 7 |
| TCGAATTAAACCACATGCTCC | |||
| 23S rDNA | CGCATGTACAGGATAGGTAGG | 669 | This study |
| AGGTGGGCTTCACACTTAGAT |
Including 6-bp coupling sequence.
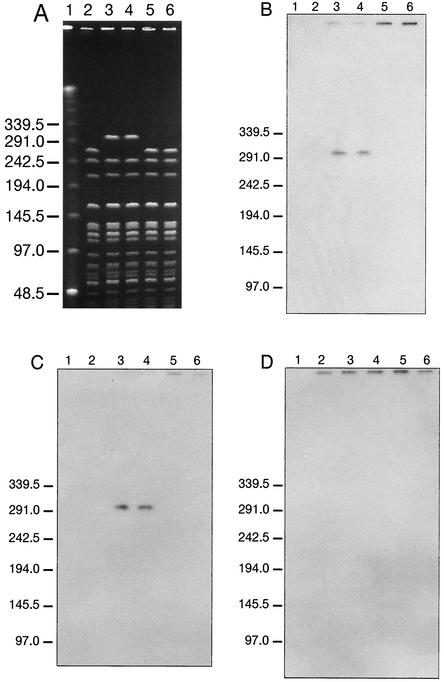
The outbreak due to the ampicillin-resistant strain (TUH2-21) and the four VanB-type strains (TUH2-18, TUH2-19, TUH2-20, and TUH2-55) has been reported previously (12). The vanB2 genotype was confirmed as previously described (6). SmaI PFGE patterns of TUH2-18 and -20 (Fig. 1A, lanes 3 and 4) were closely related to that of TUH2-21 (Fig. 1A, lane 2). The only difference was the replacement of a 270-kb SmaI fragment in TUH2-21 with an approximately 300-kb fragment in TUH2-18 and -20 that hybridized with the vanB (Fig. 1B, lanes 3 and 4) and Tn5382 (Fig. 1C, lanes 3 and 4) probes. Cohybridization of vanB and rDNA probes to I-CeuI PFGE fragments confirmed the chromosomal vanB location (data not shown). Inverse PCR and sequencing showed insertion of the Tn5382-like element in a chromosomal region of TUH2-18 between the putative genes for ribonucleotide reductase and membrane proteins (GenBank accession no. AF289471 and AF289472). TUH2-18 flanking sequences were identical to TUH2-21 and TUH2-19 chromosomal sequences except for six additional base pairs (ATAATT) at the right extremity of the transposon (GenBank accession no. AF289469 to AF289472; Fig. 2A).
FIG. 1.
PFGE of SmaI-digested total DNAs of four vancomycin-resistant strains and the prototype ampicillin-resistant E. faecium outbreak strain (TUH2-21) (A) and corresponding Southern hybridization with a vanB probe (B), a Tn5382 probe (C), and an IS1216V probe (D). Lanes: 1, low-range PFGE marker (New England Biolabs); 2, TUH2-21; 3, TUH2-20; 4, TUH2-18; 5, TUH2-19; 6, TUH2-55.
FIG. 2.
Target sequence at the site of integration of the Tn5382-like transposon into the chromosome of TUH2-18 and in a plasmid in TUH2-19. The nucleotide sequences of the Tn5382-like elements chromosomally located in TUH2-18 (A) and plasmid borne in TUH2-19 (B) were aligned with the target sequence in TUH2-21. Tn5382-like element inverted repeats are in bold letters. The 6-bp coupling sequence is underlined.
Comparative PFGE analysis of TUH2-21, -19, and −55 showed indistinguishable SmaI patterns (Fig. 1A, lanes 2, 5, and 6, respectively). Hybridization of plasmid DNA and I-CeuI PFGE fragments (data not shown) revealed a plasmid-located vanB-containing Tn5382-like element in TUH2-19 and −55, consistent with the observed colocalization of vanB and Tn5382 hybridization signals corresponding to the PFGE wells (lanes 5 and 6, Fig. 1B and C, respectively). Sequencing of inverse PCR products from TUH2-19 revealed insertion of the Tn5382-like element in an IS1216V element (GenBank accession no. AF289475 and AF289476). Hybridization of plasmid DNA (data not shown) provided evidence of the presence of a vanB- and IS1216V-containing plasmid in TUH2-19 and −55. Comigrating IS1216V probe-positive plasmid DNA fragments were observed in TUH2-18, −20, and −21, consistent with an IS1216V-containing plasmid. These data are compatible with the IS1216V signals corresponding to the locations of the wells of the four VanB strains and TUH2-21 (Fig. 1D). The Tn5382-like element-flanking sequences in TUH2-19 were identical to TUH2-21 and −18 plasmid sequences, except for six additional base pairs (AAATTA) at the left extremity of the Tn5382-like element (GenBank accession no. AF289473 to AF289476; Fig. 2B). Taken together, these observations indicate transposition of the Tn5382-like element into a preexisting IS1216V-containing plasmid or a 270-kb SmaI chromosomal fragment in TUH2-21, generating the TUH2-19 and TUH2-18 strains, respectively. The six different additional base pairs of the Tn5382-like element in TUH2-18 and TUH2-19 are linked to the opposite ends of the transposon, indicating introduction of the Tn5382-like element into TUH2-21 by two separate events. Knowing the target sequence for integration in TUH2-21, the additional six base pairs most likely represent flanking sequences from the prior insertion rather than target duplication upon insertion, as earlier hypothesized (4).
Tn5382 junction PCR analysis (Table 2) of TUH2-18 and TUH2-19 revealed the expected 701-bp product indicating the presence of an excised free Tn5382 circular intermediate (results not shown). Unique insertions of the Tn5382-like element (4, 11; this study) and the presence of circular intermediates confirmed by PCR amplification (4, 11; this study) thus suggest that the Tn5382-like element is a functional mobile genetic element in enterococci. However, direct evidence of conjugative transposition of the Tn5382-like element in enterococci has not been demonstrated. Secondary transfer of the Tn5382-like element was therefore attempted with vanB donors (TUH2-18 and TUH2-19) and the recipient E. faecium BM4105-RF or E. faecalis JH2-2 or UV202. The plasmid-associated vanB2 operon (TUH2-19) showed a transfer frequency of about 2 × 10−5 transconjugants per donor with E. faecium BM4105-RF. Low-frequency (6 × 10−9) transfer of the chromosomal vanB2 element (TUH2-18) was obtained only with E. faecium BM4105-RF. Molecular analyses of five transconjugants from each experiment did not reveal conjugative transposition of the Tn5382-like element. Rather, the chromosomal Tn5382-like element in TUH2-18 is transferred as an integral part of variable-size DNA elements and the TUH2-19 Tn5382-like element is transferred as part of a mobilizable plasmid (data not shown). The lack of firm evidence that the Tn5382-like element is a conjugative transposon in enterococci stands in contrast to the molecular data presented in references 4 and 11 and in this study, as well as the finding of several open reading frames with significant homology to proteins involved in conjugative transfer at the left end of the transposon (11). Further studies are needed to address the mechanisms of intracellular transposition and conjugative mobilization of the Tn5382-like element in order to understand and control the spread of such determinants.
Acknowledgments
This work was supported by grants from the Norwegian Research Council.
We thank Stig Harthug, Asbjørn Digranes, and Claire Poyart for providing strains. We also thank Bjørg C. Haldorsen and Mette S. Wesmajervi for excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bickley, J., and R. J. Owen. 1995. Preparation of bacterial genomic DNA. Methods Mol. Biol. 46:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Caparon, M. G., and J. R. Scott. 1989. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell 59:1027-1034. [DOI] [PubMed] [Google Scholar]
- 4.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 6.Dahl, K. H., G. S. Simonsen, Ø. Olsvik, and A. Sundsfjord. 1999. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 43:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl, K. H., E. W. Lundblad, T. P. Røkenes, Ø. Olsvik, and A. Sundsfjord. 2000. Genetic linkage of the vanB2 gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiology 146:1469-1479. [DOI] [PubMed] [Google Scholar]
- 8.Demertzi, E., M. F. Palepou, M. E. Kaufmann, A. Avlamis, and N. Woodford. 2001. Characterisation of VanA and VanB elements from glycopeptide-resistant Enterococcus faecium from Greece. J. Med. Microbiol. 50:682-687. [DOI] [PubMed] [Google Scholar]
- 9.Evers, S., P. E. Reynolds, and P. Courvalin. 1994. Sequence of the vanB and ddl genes encoding d-alanine:d-lactate and d-alanine:d-alanine ligases in vancomycin-resistant Enterococcus faecalis V583. Gene 140:97-102. [DOI] [PubMed] [Google Scholar]
- 10.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 12.Harthug, S., A. Digranes, O. Hope, B. E. Kristiansen, A. G. Allum, and N. Langeland. 2000. Vancomycin resistance emerging in a clonal outbreak caused by ampicillin-resistant Enterococcus faecium. Clin. Microbiol. Infect. 6:19-28. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, L. B., P. Ahrens, L. Dons, R. N. Jones, A. M. Hammerum, and F. M. Aarestrup. 1998. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J. Clin. Microbiol. 36:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawalec, M., M. Gniadkowski, U. Zielinska, W. Klos, and W. Hryniewicz. 2001. Vancomycin-resistant Enterococcus faecium strain carrying the vanB2 gene variant in a Polish hospital. J. Clin. Microbiol. 39:811-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, W. G., J. A. Jernigan, J. K. Rasheed, G. J. Anderson, and F. C. Tenover. 2001. Possible horizontal transfer of the vanB2 gene among genetically diverse strains of vancomycin-resistant Enterococcus faecium in a Korean hospital. J. Clin. Microbiol. 39:1165-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, J. J., C. L. Perng, M. F. Ho, T. S. Chiueh, and W. H. Lee. 2001. High prevalence of VanB2 vancomycin-resistant Enterococcus faecium in Taiwan. J. Clin. Microbiol. 39:2140-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manganelli, R., S. Ricci, and G. Pozzi. 1997. The joint of Tn916 circular intermediates is a homoduplex in Enterococcus faecalis. Plasmid 38:71-78. [DOI] [PubMed] [Google Scholar]
- 19.McGregor, K. F., C. Nolan, H. K. Young, M.-F. I. Palepou, L. Tysall, and N. Woodford. 2001. Prevalence of the vanB2 gene cluster in VanB glycopeptide-resistant enterococci in the United Kingdom and the Republic of Ireland and its association with a Tn5382-like element. Antimicrob. Agents Chemother. 45:367-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poyart, C., and P. Trieu-Cuot. 1994. Heterogeneric conjugal transfer of the pheromone-responsive plasmid pIP964 (IncHlyI) of Enterococcus faecalis in the apparent absence of pheromone induction. FEMS Microbiol. Lett. 122:173-180. [DOI] [PubMed] [Google Scholar]
- 21.Poyart-Salmeron, C., P. Trieu-Cuot, C. Carlier, and P. Courvalin. 1989. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 8:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner, G., I. Klare, and W. Witte. 1997. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol. Lett. 155:55-61. [DOI] [PubMed] [Google Scholar]
- 23.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]