Abstract
Mycobacterium tuberculosis KatG catalyzes the activation of the antitubercular agent isoniazid to yield an inhibitor targeting enoyl reductase (InhA). However, no firm biochemical link between many KatG variants and isoniazid resistance has been established. In the present study, six distinct KatG variants identified in clinical Mycobacterium tuberculosis isolates resistant to isoniazid were generated by site-directed mutagenesis, and the recombinant mutant proteins (KatGA110V, KatGA139P, KatGS315N, KatGL619P, KatGL634F, and KatGD735A) were purified and characterized with respect to their catalase-peroxidase activities (in terms of kcat/Km), rates of free-radical formation from isoniazid oxidation, and, moreover, abilities to activate isoniazid. The A110V amino acid replacement did not result in significant alteration of KatG activities except that the peroxidase activity was enhanced. The other mutations, however, resulted in modestly reduced catalase and peroxidase catalytic efficiencies and, for the four mutants tested, significantly lower activities to oxidize isoniazid. Compared to the wild-type enzyme, the ability of the KatGL634F, KatGA139P, and KatGD735A variants to activate isoniazid decreased by 36%, 76%, and 73%, respectively, whereas the KatGS315N and KatGL619P variants completely lost their abilities to convert isoniazid into the InhA inhibitor. In addition, the inclusion of exogenous Mn2+ to the isoniazid activation reaction mix significantly improved the ability of wild-type and KatG mutants to produce the InhA inhibitor.
Tuberculosis has afflicted mankind for thousands of years, and it is still the single most deadly infectious disease in developing countries. It is estimated that one-third of the world's population is or has been infected with Mycobacterium tuberculosis (24). In 2000, nearly 9 million new tuberculosis cases were reported, and 3 million people died from this disease (41). Current treatment for tuberculosis involves chemotherapy over the course of 6 months with several first-line antitubercular agents, isoniazid (isonicotinic acid hydrazide), rifampin, pyrazinamide, streptomycin, and ethambutol. Although proper prescription with full patient compliance is highly effective in curing pulmonary tuberculosis, the emergence of M. tuberculosis strains that are resistant to the first-line drugs represents a serious challenge to tuberculosis control. In the United States, about 13% of isolates from new tuberculosis cases are resistant to one or more of the first-line antitubercular drugs, and 1.6% of cases are resistant to both isoniazid and rifampin, defined as multidrug-resistant tuberculosis (22, 40).
Since its discovery five decades ago (2, 5, 27, 29), isoniazid has been commonly used to treat and prevent tuberculosis. Despite its importance, only recently has insight been gained into the molecular mechanism of isoniazid's action. It is now known that isoniazid is a prodrug (15) which is converted into a biologically active form by an M. tuberculosis catalase-peroxidase, KatG (44). Mycolic acid synthesis is the primary pathway inhibited by the action of isoniazid (4, 33, 34, 39). Mycolic acids are long-chain α-alkyl β-hydroxy fatty acids containing up to 90 carbons and are the major component of mycobacterial cell walls. Two enzymes involved in the biosynthesis of mycolic acids have been suggested to be the targets of KatG-activated isoniazid: the NADH-dependent enoyl-acyl carrier protein reductase (InhA) (1) and the β-ketoacyl acyl carrier protein synthase (designated KasA) (21).
The crystal structure of isoniazid-inhibited InhA revealed that an isonicotinic acyl-NADH was bound to the active site of InhA (31). It was later shown that the same isonicotinic acyl-NADH InhA inhibitor can be generated by either KatG- or Mn2+-mediated activation of isoniazid in vitro (16). InhA binds this inhibitor with an affinity much higher than that for NADH, resulting in complete inactivation of InhA in vitro (16). The interaction between KasA and isoniazid was implied by the recent discovery of a covalent complex of isoniazid, KasA, and its acyl carrier protein (AcpM) from isoniazid-treated M. tuberculosis (21).
Although overexpression of the inhA gene and mutations of the InhA protein have been associated with isoniazid resistance (9, 25, 26), the majority of clinical isolates of isoniazid-resistant M. tuberculosis have mutations in the katG gene that do not occur in isoniazid-susceptible strains (7-9, 20, 23, 25, 26). Many KatG mutations have been shown to be associated with a decrease in or abolition of catalase and peroxidase activities (8, 29, 30, 38, 43). The most frequently encountered KatG mutation in clinical M. tuberculosis isolates is the substitution of serine for threonine at amino acid 315. This mutation of KatG resulted in about a 70% decrease in the catalase and peroxidase activities and a compromised ability to oxidize isoniazid to isonicotinic acid (32, 38).
Catalase and peroxidase activities were commonly measured to assess the functional consequences of KatG mutations. However, a direct determination of the enzymatic activities of various KatG variants in generating the InhA inhibitor from isoniazid has rarely been carried out. Hence, many aspects of the role of KatG mutations in isoniazid resistance are not clear. In the work reported here, six katG mutations uniquely represented in isoniazid-resistant clinical isolates were introduced into the wild-type katG gene by site-directed mutagenesis. The corresponding mutant enzyme variants were expressed in Escherichia coli, purified, and characterized with respect to their catalase and peroxidase activities, rates of isoniazid oxidation, and, moreover, their abilities to activate isoniazid. The effect of Mn2+ on isoniazid activation was also investigated.
MATERIALS AND METHODS
Materials.
Isoniazid, NADH, MnCl2, nitro blue tetrazolium (NBT), glucose oxidase (type II), δ-aminolevulinic acid, and Sephadex G-25 were obtained from Sigma. H2O2 and t-butyl hydroperoxide were products of Aldrich. DEAE-Sepharose Fast Flow, Q-Sepharose Fast Flow, phenyl-Sepharose Fast Flow, MonoQ HR 10/10, and Sephacryl S-300 HR were purchased from Amersham Pharmacia Biotech. All phosphate buffers were used at pH 7.0 and consisted of phosphate at mole fractions of 0.39 sodium monobase and 0.61 potassium dibase.
Site-directed mutagenesis.
The katG coding region of pKAG3 (16) was modified by the QuickChange site-directed mutagenesis kit (Stratagene) with the primers 5′-CCGAGCAACACCCACCCATTACAGAAACCACCACC-3′ and 5′-GGTGGTGGTTTCTGTAATGGGTGGGTGTTGCTCGG-3′ to generate pKAG4. Single mutations of the wild-type katG gene in pKAG4 were obtained with the same kit. The mutations, mutation positions, and mutagenesis primers are summarized in Table 1. All mutations were confirmed by DNA sequencing.
TABLE 1.
M. tuberculosis KatG mutations and primers
| Position | Mutation | Primera |
|---|---|---|
| 110 | A → V | 5′-CGGTAGGTGCCGACAGCGTGCCACG-3′ |
| 139 | A → P | 5′-CTTGTCCAAGCTGGGGTTGTCGGGCCAGC-3′ |
| 315 | S → N | 5′-GACCTCGATGCCGTTGGTGATCGCGTC-3′ |
| 619 | L → P | 5′-CTCAGGGGCACTGGGCGTAAGCAGGTTC-3′ |
| 634 | L → F | 5′-GTAGTTTGCGCCGAAGACGCGCAGGCCAC-3′ |
| 735 | D → A | 5′-GCACGTCGAACCTGGCGAGGTTCATCACC-3′ |
Mutated nucleotides are underlined.
Protein expression and purification.
Recombinant M. tuberculosis InhA was expressed and purified from pINA-transformed E. coli JM109 (16). Wild-type and mutant KatG proteins were purified by fast protein liquid chromatography from KatG-deficient E. coli strain UM262 (recA katG::Tn10 pro leu rpsL hsdR endI lacY) (17) harboring the desired plasmid by the method of Lei et al. (16) with slight modifications. One modification was the supplementation of Luria-Bertani broth with 50 μM δ-aminolevulinic acid, a heme biosynthesis intermediate, which was added to avoid the accumulation of KatG apoprotein (3). Additionally, the protein samples eluted from the DEAE-Sepharose Fast Flow column were applied to a Sephacryl S-300 HR column (2.6 by 60 cm) preequilibrated with 150 mM NaCl in 50 mM phosphate, pH 7.0, and eluted with the same buffer.
Enzyme assays.
Catalase activity was assayed spectrophotometrically by monitoring the decrease in H2O2 concentration at A240 (ɛ240 = 0.0436 mM−1 cm−1) (11). Reactions were initiated by adding KatG to 1 ml of 50 mM phosphate containing various amounts of H2O2. Peroxidase activity was determined by measuring the oxidation rate of 0.1 mM o-dianisidine at A460 (ɛ460 = 11.3 mM−1 cm−1) in the presence of 23 mM t-butyl hydroperoxide in 50 mM phosphate (19). One unit of peroxidase activity catalyzes the oxidation of 1 μmol of o-dianisidine per min at 23°C. In addition, rates of free-radical formation from isoniazid oxidation in the presence of a constant H2O2 flux (generated by glucose and glucose oxidase) were monitored by following the reduction of NBT by the method of Hillar and Loewen (12).
Isoniazid activation.
Isoniazid activation by KatG was determined by the formation of InhA-inhibitor complex as described previously (16). A 1-ml 50 mM phosphate solution containing 1 mM NADH, 1 mM isoniazid, and 0.5 μM KatG was incubated for 3 h at room temperature under aerobic condition with or without 5 μM MnCl2. KatG was separated from the reaction mixture by passing through a Microcon YM-10 filter unit with a molecular weight cutoff of 10,000 (Millipore). The filtrate (0.8 ml) was incubated with InhA (78.5 or 200 μM in 0.2 ml) at room temperature for 30 min and then loaded on a Sephadex G-25 column (1 by 40 cm) preequilibrated and eluted with 50 mM phosphate. The gel filtration was repeated once, and the fractions containing InhA protein were collected.
The UV-visible absorption spectra of InhA-containing fractions were recorded with an SLM 3000 Array spectrophotometer. On the basis of the characteristic absorption spectral properties of the InhA-inhibitor complex, the amounts of InhA-bound inhibitor can be calculated with 7.1 and 7.2 mM−1 cm−1 for ɛ291 and ɛ326, respectively, and concentrations of InhA can be determined by using 26.4 mM−1 cm−1 for ɛ291 (16). There was no detectable absorption at 326 nm for InhA (16).
Miscellaneous measurements.
KatG molar concentrations were calculated according to the heme contents determined by the pyridine hemochrome assay with ɛ418 = 191.5 mM−1 cm−1 (6). InhA protein concentrations were determined by the method of Lowry et al. (18) with bovine serum albumin as a standard. All data presented are the means ± standard deviation of three measurements.
RESULTS
Recombinant KatG mutants.
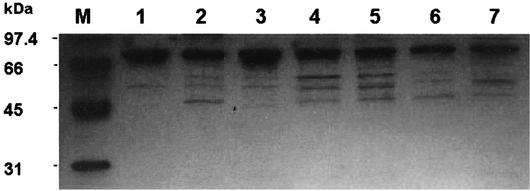
Six katG alleles encoding KatG A110V, A139P, S315N, L619P, L634P, and D735A variants identified uniquely in isoniazid-resistant M. tuberculosis clinical isolates (28) were obtained by site-directed mutagenesis. All KatG variants were expressed in the KatG-deficient E. coli UM262 and purified to 50 to 90% homogeneity, based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 1). The addition of δ-aminolevulinic acid to the cell growth medium greatly enhanced the production of KatG holoenzyme (3). After corrections for the impurity, all KatG enzymes had A408/A280 ratios of 0.59 to 0.74, corresponding to 1.2 to 1.5 hemes per KatG dimer.
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of wild-type and mutant KatGs. Lanes: M, molecular size markers; 1, wild type; 2, D735A; 3, A110V; 4, S315N; 5, A139P; 6, L619P, 7; L634F.
Catalase and peroxidase activities.
Catalase activities of KatG enzymes were determined at various concentrations of H2O2, and values of Km, kcat, and kcat/Km are shown in Table 2. Wild-type KatG displayed a kcat, Km, and kcat/Km similar to values reported previously (32, 38). Compared to the wild-type KatG, all mutants had elevated Km values. KatGA110V had a 60% higher value for kcat, giving rise to a kcat/Km that was about the same as that of the wild-type KatG. All other mutants had kcat values similar to that of the wild-type KatG, all giving lower kcat/Km values (at 50 to 70% of that of the wild-type enzyme) due to their elevated Km values. Moreover, KatGS315N had catalase properties similar to those of KatGS315T (38).
TABLE 2.
Catalase activity of wild-type and mutant KatG
| KatG | kcat (s−1)a | Km (mM) | kcat/Km (M−1 s−1) | kcat/Km ratio, mutant/wild type |
|---|---|---|---|---|
| Wild type | 3,270 ± 260 | 5.9 ± 0.7 | 0.56 × 106 | 1.00 |
| A110V | 5,230 ± 210 | 10.3 ± 0.6 | 0.51 × 106 | 0.91 |
| A139P | 3,330 ± 10 | 10.7 ± 0.8 | 0.31 × 106 | 0.55 |
| S315N | 2,990 ± 70 | 16.8 ± 0.8 | 0.18 × 106 | 0.32 |
| L619P | 3,180 ± 30 | 17.1 ± 0.3 | 0.19 × 106 | 0.34 |
| L634F | 3,450 ± 380 | 14.3 ± 2.9 | 0.24 × 106 | 0.43 |
| D735A | 2,750 ± 70 | 14.8 ± 0.8 | 0.18 × 106 | 0.32 |
kcat is the enzyme turnover rate, a form of specific activity, and is expressed on the basis of dimeric KatG native enzyme and variants. A kcat value of 1 s−1 is equivalent to a specific activity of 0.375 μmol/mg of KatG/min.
KatG also has peroxidase activity with several substrates (14). In this study, the oxidation rates of various levels of o-dianisidine were measured, and the kinetic parameters are shown in Table 3. Compared to wild-type KatG, KatGA110V was 40% higher in kcat and 24% lower in Km, resulting in an 80% increase in kcat/Km. The other mutants had two- to fourfold-lower values of kcat/Km. Changes in Km were rather limited in value. Therefore, the observed decreases in kcat/Km were primarily a consequence of lower kcat values.
TABLE 3.
Peroxidase activity of wild-type and mutant KatGsa
| KatG | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat/Km ratio, mutant/wild type |
|---|---|---|---|---|
| Wild type | 3.48 ± 0.13 | 84.2 ± 7.3 | 4.13 × 104 | 1.00 |
| A110V | 4.85 ± 0.24 | 64.3 ± 1.7 | 7.54 × 104 | 1.83 |
| A139P | 1.69 ± 0.15 | 95.7 ± 18.6 | 1.77 × 104 | 0.43 |
| S315N | 0.86 ± 0.09 | 80.6 ± 14.1 | 1.07 × 104 | 0.26 |
| L619P | 1.06 ± 0.06 | 78.2 ± 9.5 | 1.36 × 104 | 0.33 |
| L634F | 0.99 ± 0.03 | 65.4 ± 5.0 | 1.51 × 104 | 0.37 |
| D735A | 0.99 ± 0.08 | 62.9 ± 10.2 | 1.57 × 104 | 0.38 |
See Table 2, footnote a.
Isoniazid oxidation activity.
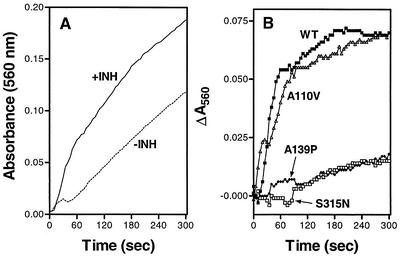
KatG has also been shown to catalyze the oxidation of isoniazid (12, 13). One assay for isoniazid oxidation by KatG in the presence of H2O2 was developed by coupling the free radicals generated in the reaction with NBT reduction (12). When wild-type KatG was tested in this assay, we noted a significant background activity of NBT reduction in the absence of isoniazid. However, the NBT reduction was substantially enhanced in the presence of isoniazid (Fig. 2A), allowing us to obtain the net isoniazid oxidation/NBT reduction time course after subtraction of the background activity (Fig. 2B).
FIG. 2.
KatG-mediated oxidation of isoniazid, followed by NBT reduction. All experiments were carried out at room temperature. To 1 ml of 50 mM phosphate buffer, KatG proteins, NBT, glucose oxidase, and glucose were added at 1 μM, 0.2 mM, 5 mg, and 4 mM, respectively. (A) Time course of NBT reduction with and without the addition of 9 mM isoniazid (INH) with wild-type KatG. (B) Time courses of net isoniazid-dependent NBT reduction by wild-type KatG (WT) and three variants. For each KatG sample tested, NBT reduction in the absence of isoniazid was subtracted from that in the presence of isoniazid to obtain the net isoniazid-dependent NBT reduction over time.
Five KatG variants were also assayed in the absence and presence of isoniazid, and their net isoniazid oxidation/NBT reduction time courses were obtained as described above. The time courses are shown in Fig. 2B for wild-type KatG, KatGA110V, KatGA139P, and KatGS315N. The time courses for the last two KatG mutants were essentially the same as those of KatGL619P and KatGD735A (not shown). The rates of net isoniazid oxidation/NBT reduction were determined with data obtained in the initial 60 s for the wild-type enzyme and KatGA110V and the entire 300-s period for the other KatG mutants. In comparison with the net isoniazid oxidation/NBT reduction rate of the wild-type enzyme, KatGA110V was about 90% as active, whereas the other four KatG variants were about 6 to 9% as active.
Isoniazid activation by KatG.
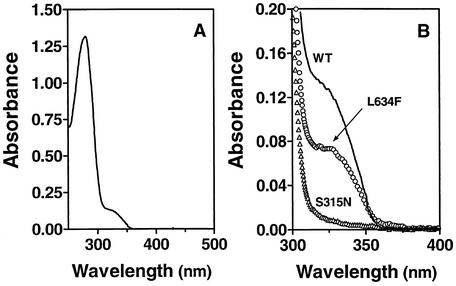
M. tuberculosis KatG catalyzes the activation of isoniazid to yield an InhA inhibitor. Only recently has a method for the direct detection and quantification of this InhA inhibitor been developed by using InhA to trap an inactive enzyme-inhibitor complex (16). This InhA-inhibitor complex has an absorption peak at 278 nm and a pronounced shoulder around 326 nm which can be used as an indicator for detection and quantification of InhA inhibitor formation (16). With the wild-type and all mutant KatG proteins used for the activation of isoniazid, various degrees of formation of the InhA inhibitor (trapped in the form of the inhibitor-InhA complex) were detected, as shown by the various magnitudes of the absorption shoulder at 326 nm (Fig. 3, Table 4). Compared to wild-type KatG, KatGA110V produced nearly the same level of the InhA inhibitor, whereas reduced yields of production were detected for KatGL634F, KatGD735A, and KatGA139P at 64, 27, and 24% of the wild-type enzyme level, respectively. No InhA inhibitor formation was detected for KatGS315N and KatGL619P, as indicated by the absence of absorption at around 326 nm.
FIG. 3.
Isoniazid activation by wild-type and mutant KatG. The reactions were carried out for 3 h at room temperature with a sample of 1 ml of 50 mM phosphate, pH 7.0, containing 1 mM isoniazid, 1 mM NADH, and 0.5 μM wild-type KatG or a A110V, A139P, S315N, L619P, L634F, or D735A KatG mutant. KatG was then removed with Micron YM-10 filter units. One milliliter of sample containing 0.8 ml of filtrate and 0.2 ml of 78.5 μM InhA was incubated at room temperature for 20 min. The mixture was subjected to Sephadex G-25 column filtration twice, and the InhA-inhibitor complex was isolated. The spectra were normalized to a content of 10 μM InhA. (A) Absorption spectrum of the InhA-inhibitor complex with wild-type KatG for isoniazid activation. (B) Absorption spectrum of the same InhA-inhibitor complex derived from wild-type KatG (WT) activation of isoniazid is reproduced for the range of 300 to 400 nm. This spectrum is compared with those obtained under identical conditions with either the L634F or S315N KatG mutant in place of wild-type KatG. The final spectrum obtained for a negative control, in which wild-type KatG was used but no isoniazid was added, under otherwise identical conditions was identical to that shown for the S315N KatG mutant. For simplicity, spectra obtained with the other KatG mutants are not included. The corresponding yields of the inhibitor derived from isoniazid activation by all KatG species tested were calculated from the spectral data so obtained and are presented in Table 4.
TABLE 4.
InhA inhibitor production catalyzed by wild-type and mutant KatGs in the absence and presence of Mn2+
| KatG | Isoniazid activation without Mn2+
|
Isoniazid activation with 5 μM Mn2+
|
||
|---|---|---|---|---|
| Inhibitor yielda (μmol/mg of KatG) | Relative yield by KatGb | Inhibitor yield (μmol/mg of KatG) | Relative yield by KatGc | |
| Wild type | 0.11 | 1.00 | 0.43 | 1.00 |
| A110V | 0.10 | 0.90 | 0.38 | 0.88 |
| A139P | 0.03 | 0.24 | 0.10 | 0.12 |
| S315N | NDd | 0 | 0.12 | 0.18 |
| L619P | ND | 0 | 0.12 | 0.19 |
| L634F | 0.07 | 0.64 | 0.37 | 0.86 |
| D735A | 0.03 | 0.27 | 0.12 | 0.18 |
| None | ND | 0 | 0.05 | 0 (0.13)e |
Inhibitor generated after 3 h.
Ratio of yield by KatG variant to yield by native KatG.
First, the basal yield with 5 μM Mn2+ in the absence of KatG was subtracted from all the observed yields to obtain the net yields due to enzymatic catalysis. The relative yield was then determined as the ratio of net yield by KatG variant to net yield by native KatG.
ND, not detectable.
Ratio of basal yield with 5 μM Mn2+ to net yield with native KatG.
Effect of Mn2+ on isoniazid activation.
Mn2+ is not essential to but can enhance isoniazid activation by M. tuberculosis KatG (16). Here we examined the levels of the InhA inhibitor produced by the mutant KatG enzymes with Mn2+ exogenously added at 5 μM (Table 4). Consistent with earlier reports (16, 31, 43), Mn2+ alone can produce InhA inhibitor in a slow, low-yield process. The concentrations of InhA inhibitor from isoniazid activation reactions catalyzed by wild-type KatG, KatGA110V, KatGA139P, KatGL634F, and KatGD735A were increased by 3.4- to 5.2-fold in the presence of 5 μM Mn2+. The addition of Mn2+ also partially restored the ability of KatGS315N and KatGL619P to produce the InhA inhibitor. However, the KatGA139P, KatGS315N, KatGL619P, and KatGD735A mutants produced the InhA inhibitor at levels of only 12 to 19% of that of wild-type KatG under identical conditions.
DISCUSSION
One of the most commonly occurring KatG variants, KatGL463R, was originally thought to be responsible for isoniazid resistance, but that strain was found to be fully active in isoniazid activation (14), demonstrating the importance of biochemical studies on KatG mutants in relation to isoniazid resistance. However, only a few reports (8, 32, 37, 38, 42) have studied the relationship between isoniazid resistance and the catalase, peroxidase, and isoniazid oxidation activities and isoniazid binding to KatG mutants.
To investigate the biochemical link between specific mutations in the katG gene and isoniazid resistance, we generated, purified, and characterized six KatG variants present uniquely in isoniazid-resistant clinical isolates. Three mutants (KatGA139P, KatGL634F, and KatGD735A) had significantly diminished isoniazid activation activities. Two other mutants (KatGS315N and KatGL619P) failed to convert isoniazid into the InhA inhibitor. Hence, we demonstrated biochemically that certain single-residue amino acid replacements can completely abolish the ability of KatG to produce the InhA inhibitor. On the other hand, KatGA110V was 88% as active as wild-type KatG in isoniazid activation. Therefore, these six KatG mutants had a wide range of abilities to produce the InhA inhibitor.
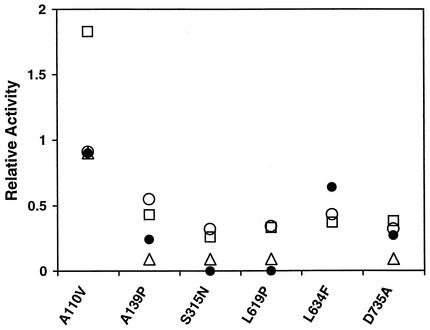
In addition to isoniazid activation, we also compared these KatG variants with wild-type KatG with respect to their catalase, peroxidase, and isoniazid oxidation activities. On the basis of relative activity, defined as the ratio of mutant KatG activity to wild-type KatG activity, a summary of such comparisons is shown in Fig. 4. With the exception of KatGA110V (which showed substantially higher peroxidase activity), the catalase activities of all other KatG mutants correlated quite well with their peroxidase activities. However, no consistent pattern emerged from the correlations of catalase-peroxidase activities with isoniazid activation activities. The relative isoniazid activation activities of KatGA110V and KatGD735A correlated well with their catalase activities and, in the latter case, peroxidase activity. However, in comparison with the catalase-peroxidase activities, KatGL634F showed higher isoniazid activation activity, whereas KatGA139P was less active and KatGS315N and KatGL619P were completely inactive in activating isoniazid for the formation of InhA inhibitor.
FIG. 4.
Comparison of enzyme activities of KatG mutants with wild-type KatG. Results of catalase (○), peroxidase (□), isoniazid oxidation/NBT reduction (▵), and isoniazid activation in the absence of Mn2+ (•) activity determinations described in the text were used to calculate the relative activities, defined as the ratios of KatG mutant activity to wild-type KatG activity.
The relative activities of isoniazid oxidation/NBT reduction were also measured for five KatG variants, which showed a generally better but still inconsistent correlation with their isoniazid activation activities. A good correlation was found for KatGA110V. While KatGA139P and KatGD735A were about 25% as active as the wild type and KatGS315N and KatGL619P were inactive in isoniazid activation, they all showed about 6 to 9% of wild-type activity in isoniazid oxidation/NBT reduction. These findings indicate that, in biochemical characterization of KatG mutants, isoniazid activation activities should be determined directly rather than by inference from catalase, peroxidase, or isoniazid oxidation/NBT reduction activities.
Like other bacterial catalase-peroxidase enzymes (36), mycobacterial KatG is believed to consist of three domains, a 54-residue N-terminal domain, a heme-binding catalytic domain (residues 55 to 423), and a C-terminal domain (residues 424 to 740) (10). The A110, S315, L634, and D735 residues are conserved among KatG catalases from E. coli, Salmonella enterica serovar Typhimurium, Streptomyces reticuli, Bacillus stearothermophilus, Mycobacterium bovis, Mycobacterium intracellulare, Mycobacterium smegmatis, and M. tuberculosis. The L619 residue is also conserved among these species except for M. intracellulare, in which it is replaced by a valine residue. The A139 residue is conserved in the cytochrome c peroxidase of Saccharomyces cerevisiae and KatG of B. stearothermophilus, M. bovis, M. intracellulare, M. semgmatis, and M. tuberculosis, whereas it is replaced by valine in E. coli and S. enterica serovar Typhimurium KatG. Therefore, the six KatG variants from isoniazid-resistant M. tuberculosis isolates all carry a single mutation of a conserved residue. However, three residues (A110V, A139P, and S315N) are located in the catalytic/heme-binding domain, whereas three other residues (L619P, L634F, and D735A) are in the functionally undefined C-terminal domain.
Within the C-terminal domain, mutation R463L or L587M has no detectable effect on KatG enzymatic properties (14, 32) or, in the former case, the ability to activate isoniazid (14). However, the mutation L587P in both M. tuberculosis and M. bovis results in instability and inactivation of the KatG protein (32). We found that the catalase, peroxidase, isoniazid oxidation, and activation activities of KatGL619P, KatGL634F, and KatGD735A were all variously affected by their mutations. These and earlier findings together suggest that the C-terminal domain plays a role in stabilizing subunit-subunit interactions (32) but may also be important for KatG enzyme function.
For the three residues in the catalytic domain, the A110V substitution did not dramatically alter the enzymatic activities, suggesting that A110 is not directly involved in heme or substrate binding. The replacement of serine at position 315 of KatG with threonine, asparagine, arginine, glycine, or isoleucine has been identified in clinical isoniazid-resistant M. tuberculosis isolates. Both the catalase-peroxidase activities and the ability to oxidize isoniazid to isonicotinic acid are compromised in the most frequent variant, S315T (32, 38). KatGS315N has now been found to be inactive in isoniazid activation, suggesting that the isoniazid resistance of the other KatG S315 mutants may also be a consequence of their inability to generate the InhA inhibitor. The M. tuberculosis KatG A139 residue is equivalent to the A83 residue of yeast cytochrome c peroxidase, which is in a loop near the heme-binding site (35). However, the specific role of KatG residue A139 is unclear.
Evidence is available to suggest a functional role of Mn2+ in isoniazid susceptibility and the ability of KatG to generate the InhA inhibitor. M. smegmatis KatG cannot activate isoniazid for InhA inactivation in the absence of Mn2+, but Mn2+ is not essential to InhA inhibitor formation by M. tuberculosis KatG. Correspondingly, most strains of M. tuberculosis are highly susceptible to isoniazid (MIC < 0.02 μg/ml), but M. smegmatis is naturally resistant to high levels of isoniazid (MIC > 30 μg/ml), perhaps related to the low Mn2+ contents of these cells in vivo. Although Mn2+ is not essential to M. tuberculosis KatG-mediated isoniazid activation, it does enhance the production of InhA inhibitor by wild-type KatG (16) and all the mutants tested in this work, including KatGS315N and KatGL619P, which did not activate isoniazid in the absence of Mn2+. Understanding the mechanisms of enhancement of InhA inhibitor production by Mn2+ could be a key to improving drug efficiency and, consequently, combating the drug resistance caused by frequently encountered KatG mutations.
Acknowledgments
We thank Yuqiu Tang and Chi-Hui Li for technical assistance, and we are grateful to Peter Loewen at the University of Manitoba for the generous gift of the KatG-deficient Escherichia coli strain UM262.
S.-C.T. acknowledges the support of this work by Texas Advanced Research Program grant 003652-0070-1999.
REFERENCES
- 1.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, J., W. A. Lott, B. A. Steinberg, and H. L. Yale. 1952. Chemotherapy of experimental tuberculosis V. Isonicotinic acid hydrazide (Nydrazid) and related compounds. Am. Rev. Tuberc. 65:357-364. [DOI] [PubMed] [Google Scholar]
- 3.Chouchane, S., I. Lippai, and R. S. Magliozzo. 2000. Catalase-peroxidase (Mycobacterium tuberculosis KatG) catalysis and isoniazid activation. Biochemistry 39:9975-9983. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, L. A., and K. Takayama. 1979. Isoniazid inhibition of the synthesis of monounsaturated long-chain fatty acids in Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 16:104-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox, H. H. 1952. The chemical approach to the control of tuberculosis. Science 116:129-134. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrhop, J. H., and K. M. Smith. 1975. Laboratory methods, p. 804-807. In K. M. Smith (ed.), Prophyrins and metalloporphyrins. Elsevier Scientific Publishing Co., New York, N.Y.
- 7.Haas, W. H., K. Schilke, J. Brand, B. Amthor, K. Weyer, P. B. Fourie, G. Bretzel, V. Sticht-Groh, and H. J. Bremer. 1997. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob. Agents Chemother. 41:1601-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heym, B., P. M. Alzari, N. Honoré, and S. T. Cole. 1995. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 15:235-245. [DOI] [PubMed] [Google Scholar]
- 9.Heym, B., N. Honore, C. Truffot-Pernot, A. Banerjee, C. Schurra, W. R. Jacobs, J. D. van Embden, J. H. Grosset, and S. T. Cole. 1994. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet 344:293-298. [DOI] [PubMed] [Google Scholar]
- 10.Heym, B., Y. Zhang, S. Poulet, D. Young, and S. T. Cole. 1993. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J. Bacteriol. 175:4255-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hildebrandt, A. G., and I. Roots. 1975. Reduced nicotinamide adenine dinucleotide phosphate (NADPH)-dependent formation and breakdown of hydrogen peroxide during mixed function oxidation reactions in liver microsomes. Arch. Biochem. Biophys. 171:385-397. [DOI] [PubMed] [Google Scholar]
- 12.Hillar, A., and P. C. Loewen. 1995. Comparison of isoniazid oxidation catalyzed by bacterial catalase-peroxidase and horseradish peroxidase. Arch. Biochem. Biophys. 323:438-446. [DOI] [PubMed] [Google Scholar]
- 13.Johnsson, K., and P. G. Schultz. 1994. Mechanistic studies of the oxidation of isoniazid by the catalase peroxidase from Mycobacterium tuberculosis. J. Am. Chem. Soc. 116:7425-7426. [Google Scholar]
- 14.Johnsson, K., W. A. Froland, and P. G. Schultz. 1997. Overexpression, purification, and characterization of the catalase-peroxidase KatG from Mycobacterium tuberculosis. J. Biol. Chem. 272:2834-2840. [DOI] [PubMed] [Google Scholar]
- 15.Johnsson, K., D. S. King, and P. G. Schultz. 1995. The studies on the mechanism of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J. Am. Chem. Soc. 117:5009-5010. [Google Scholar]
- 16.Lei, B., C.-J. Wei, and S.-C. Tu. 2000. Action mechanism of antitubercular isoniazid. Activation by Mycobacterium tuberculosis KatG, isolation, and characterization of inhA inhibitor. J. Biol. Chem. 275:2520-2526. [DOI] [PubMed] [Google Scholar]
- 17.Loewen, P. C., J. Switala, M. Smolenski, and B. L. Triggs-Raine. 1990. Molecular characterization of three mutations in katG affecting the activity of hydroperoxidase I of Escherichia coli. Biochem. Cell. Biol. 68:1037-1044. [DOI] [PubMed] [Google Scholar]
- 18.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 19.Marcinkeviciene, J. A., R. S. Magliozzo, and J. S. Blanchard. 1995. Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid activation. J. Biol. Chem. 270:22290-22295. [DOI] [PubMed] [Google Scholar]
- 20.Marttila, H. J., H. Soini, P. Huovinen, and M. K. Viljanen. 1996. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob. Agents Chemother. 40:2187-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mdluli, K., R. A. Slayden, Y. Zhu, S. Ramaswamy, X. Pan, D. Mead, D. D. Crane, J. M. Musser, and C. E. Barry 3rd. 1998. Inhibition of a Mycobacterium tuberculosis beta-ketoacyl acyl carrier protein synthase by isoniazid. Science 280:1607-1610. [DOI] [PubMed] [Google Scholar]
- 22.Moore, M., I. M. Onorato, E. McCray, and K. G. Castro. 1997. Trends in drug-resistant tuberculosis in the United States, 1993-1996. JAMA 278:833-837. [PubMed] [Google Scholar]
- 23.Morris, S., G. H. Bai, P. Suffys, L. Portillo-Gomez, M. Fairchok, and D. Rouse. 1995. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J. Infect. Dis. 171:954-960. [DOI] [PubMed] [Google Scholar]
- 24.Murray, C. J. L., K. Styblo, and A. Rouillon. 1990. Tuberculosis in developing countries: burden, intervention and cost. Bull. Int. Union Tuberc. Lung Dis. 65:6-24. [PubMed] [Google Scholar]
- 25.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser, J. M., V. Kapur, D. L. Williams, B. N. Kreiswirth, D. van Soolingen, and J. D. van Embden. 1996. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J. Infect. Dis. 173:196-202. [DOI] [PubMed] [Google Scholar]
- 27.Pansy, F., H. Stander, and R. Donovick. 1952. In vitro studies on isonicotinic acid hydrazide. Am. Rev. Tuber. 65:761-764. [PubMed] [Google Scholar]
- 28.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 29.Robitzek, E. H., and I. J. Selikoff. 1952. Hydrazine derivatives of isonicotinic acid (Rimfon, Marsilid) in the treatment of active progressive caseous-pneumonic tuberculosis. Am. Rev. Tuberc. 65:402-428. [DOI] [PubMed] [Google Scholar]
- 30.Rouse, D. A., J. A. DeVito, Z. Li, H. Byer, and S. L. Morris. 1996. Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase-peroxidase activities and isoniazid resistance. Mol. Microbiol. 22:583-592. [DOI] [PubMed] [Google Scholar]
- 31.Rozwarski, D. A., G. A. Grant, D. H. R. Barton, W. R. Jacobs, Jr., and J. C. Sacchettini. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279:98-102. [DOI] [PubMed] [Google Scholar]
- 32.Saint-Joanis, B., H. Souchon, M. Wilming, K. Johnsson, P. M. Alzari, and S. T. Cole. 1999. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase-peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338:753-760. [PMC free article] [PubMed] [Google Scholar]
- 33.Takayama, K., H. K. Schnoes, E. L. Armstrong, and R. W. Boyle. 1975. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J. Lipid Res. 16:308-317. [PubMed] [Google Scholar]
- 34.Takayama, K., L. Wang, and H. L. David. 1972. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2:29-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, J., J. M. Mauro, S. L. Oatley, L. A. Fishel, V. A. Ashford, N. H. Xuong, and J. Kraut. 1990. X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis. Biochemistry 29:7160-7173. [DOI] [PubMed] [Google Scholar]
- 36.Welinder, K. G. 1991. Bacterial catalase-peroxidases are gene duplicated members of the plant peroxidase superfamily. Biochim. Biophys. Acta 1080:215-220. [DOI] [PubMed] [Google Scholar]
- 37.Wengenack, N. L., S. Todorovic, L. Yu, and F. Rusnak. 1998. Evidence for differential binding of isoniazid by Mycobacterium tuberculosis KatG and the isoniazid-resistant mutant KatG(S315T). Biochemistry 37:15825-15834. [DOI] [PubMed] [Google Scholar]
- 38.Wengenack, N. L., J. R. Uhl, A. L. St. Amand, A. J. Tomlinson, L. M. Benson, S. Naylor, B. C. Kline, F. R. Cockerill 3rd, and F. Rusnak. 1997. Recombinant Mycobacterium tuberculosis KatG(S315T) is a competent catalase-peroxidase with reduced activity toward isoniazid. J. Infect. Dis. 176:722-727. [DOI] [PubMed] [Google Scholar]
- 39.Winder, F. G., and P. B. Collins. 1970. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J. Gen. Microbiol. 63:41-48. [DOI] [PubMed] [Google Scholar]
- 40.World Health Organization. 2000. Antituberculosis drug resistance in the world. Report no. 2: prevalence and trends 2. World Health Organization, Geneva, Switzerland.
- 41.World Health Organization. 2001. Global tuberculosis control 2001. World Health Organization, Geneva, Switzerland.
- 42.Yu, S., S. Chouchane, and M. R. S. 2002. Characterization of the W321F mutant of Mycobacterium tuberculosis catalase-peroxidase KatG. Protein Sci. 11:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zabinski, R. F., and J. S. Blanchard. 1997. The requirement for manganese and oxygen in the isoniazid-dependent inactivation of Mycobacterium tuberculosis enoyl reductase. J. Am. Chem. Soc. 119:2331-2332. [Google Scholar]
- 44.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]