Abstract
Protease inhibitors represent some of the most potent agents available for therapeutic strategies designed to inhibit human immunodeficiency virus type 1 (HIV-1) replication. Under certain circumstances the virus develops resistance to the inhibitor, thereby negating the benefits of this therapy. We have carried out selections for high-level resistance to each of three protease inhibitors (indinavir, ritonavir, and saquinavir) in cell culture. Mutations accumulated over most of the course of the increasing selective pressure. There was significant overlap in the identity of the mutations selected with the different inhibitors, and this gave rise to high levels of cross-resistance. Virus particles from the resistant variants all showed defects in processing at the NC/p1 protease cleavage site in Gag. Selections with pairs of inhibitors yielded similar patterns of resistance mutations. A virus that could replicate at near-toxic levels of the three protease inhibitors combined was selected. The pro sequence of this virus was similar to that of the viruses that had been selected for high-level resistance to each of the drugs singly. Finally, a molecular clone carrying the eight most common resistance mutations seen in these selections was characterized. The sequence of this virus was relatively stable during selection for revertants in spite of displaying poor processing at the NC/p1 site and having significantly reduced fitness. These results reveal patterns of drug resistance that extend to near the limits of attainable selective pressure with these inhibitors and confirm the patterns of cross-resistance for these three inhibitors and the attenuation of virion protein processing and fitness that accompanies high-level resistance.
The evolution of resistance to human immunodeficiency virus type 1 (HIV-1) protease inhibitors (PI) represents a significant limitation to antiviral therapy. Resistance to protease inhibitors was initially shown by selection experiments carried out in vitro to be attributable to well-defined mutations in the pro gene encoding the viral protease. To a large extent, resistance mutations that were identified in the selections in cell culture overlap the mutations seen in subjects failing therapy (reviewed in reference 71).
Therapeutic doses of PI are given at near-toxic levels to provide the maximal inhibitory effect. Even under these circumstances the number of resistance mutations present at the first time of apparent virus rebound is relatively small, although more mutations can accumulate over time under this constant level of selective pressure (11, 13, 22, 45, 50, 62, 74). Thus, the potential limit of selective pressure for these drugs has likely not been explored.
One strategy for achieving higher selective pressure has been to use two PI together. This approach has three potential advantages. First, nonoverlapping toxicities allow for a higher combined inhibitory effect without the associated higher toxicity. Second, one PI can enhance the pharmacokinetic properties of a second inhibitor to provide a higher and more stable drug level between doses (12, 36, 44). Third, PI that are able to select for unique resistance mutations could be paired. These potential advantages have been explored in a number of clinical trials (for examples, see references 8, 10, 16, 17, 23-28, 32, 33, 38, 41, 49, 54, 57, 58, 72, and 78). Some information is available about resistance profiles selected by pairs of PI from PI-naïve subjects failing such therapy (41), although in most cases these subjects had previously failed therapy that included a single PI (16, 25, 32, 38).
Subjects treated with potent PI, either singly or multiply, for an extended period of time can accumulate many mutations. There is an association between the accumulation of multiple mutations and the acquisition of cross-resistance to other PI (11, 13, 15, 19, 20, 29, 35, 45, 46, 55, 64, 73, 76). The functional significance of this cross-resistance is seen in the association between therapy failure with the presence of resistance mutations in the protease or with direct measurements of phenotypic cross-resistance (2, 4, 5, 7, 10, 15-17, 19, 21, 25, 28, 32, 34, 38-40, 47-49, 51, 54, 60, 65, 67, 75, 78).
We have explored the question of high-level selection by using a cell culture-based system to select for high-level resistance to three clinically approved PI (indinavir [IDV], saquinavir [SQV], and ritonavir[RTV]) either separately or in pairs. In addition, we have taken resistant virus pools and selected for resistance to all three inhibitors together at near-toxic drug levels. Most of these selections showed a similar pattern of accumulation of mutations with two active-site mutations at codon positions 82 and 84 in the pro gene and with a partially overlapping set of non-active-site mutations. Finally, we have created an infectious molecular clone carrying many of these mutations and generated virus from this clone to examine the stability of these mutations and their effects on viral fitness. These studies explore the limits of resistance that can be selected by these widely used protease inhibitors.
MATERIALS AND METHODS
PI.
SQV was provided by Ian Duncan, Roche Research Center, RTV was provided by Dale Kempf, Abbott Laboratories, and IDV was provided by Emilio Emini, Merck Research Laboratories.
Cell lines.
CEMx174 cells were maintained in RPMI 1640 medium with 10% fetal calf serum and penicillin-streptomycin. HeLa-CD4-LTR-β-gal (MAGI) cells (37) were grown in Dulbecco's modified Eagle-H medium supplemented with 10% fetal calf serum, 0.1 mg of active G418 per ml, and 0.1 mg of hygromycin B per ml. HeLa and 293T cells were maintained in Dulbecco's modified Eagle-H medium supplemented with 10% fetal calf serum and penicillin-streptomycin. CEMx174 and MAGI cell lines were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program.
Selections.
An aliquot of 3 × 106 CEMx174 cells was incubated at 37°C for 2 to 3 h with 250 μl of a virus stock generated from the HIV-1 infectious molecular clone NL4-3 (1). The culture volume was then brought to 10 ml with medium. Each flask received one of the following inhibitors at the indicated concentration: 8 nM SQV, 45 nM RTV, or 24 nM IDV. The multiple-PI flasks received one of the following concentration combinations: 6 nM SQV-22.5 nM RTV, 30 nM RTV-16 nM IDV, or 16 nM IDV-8 nM SQV. The selection data with SQV have been described in part previously (74) and are included here for comparison. After 48 h and daily thereafter, the cells were pelleted by centrifugation and 10 ml of fresh medium and inhibitors were added. When the culture had undergone extensive cytopathic effect (CPE), the supernatant medium and the cells were harvested separately and stored at −70°C. The virus-containing supernatant was used to start the next round of infection, and after four rounds at the initial concentration, the inhibitor concentration was increased 1.5-fold at each subsequent round of virus growth as described in Results. The level of resistance (50% inhibitory concentration [IC50]) of the single inhibitor-selected virus pools was determined by a modified MAGI cell assay (37), as described previously (74), in which the protease inhibitor is added to productively infected cells and the titers of supernatant virus made in the presence of the inhibitor are determined for infectious virus in the MAGI cell assay. For selection with pairs of inhibitors, the same initial inhibitor concentration cited above was used in the first four rounds of selection, with each inhibitor increased 1.5-fold at each subsequent passage. After one of the inhibitors reached a concentration of >5 μM, both inhibitors in the culture were fixed at 5 μM for a final 10 passages. To select in the presence of all three inhibitors, the third inhibitor was added at 5 μM to each of the three cultures that had been selected to 5 μM each with the pairs of inhibitors and passaged 10 times with each of the three inhibitors present at 5 μM. The three cultures that had been passaged with the three inhibitors were pooled, and the concentrations of the inhibitors were raised 1.5-fold until the concentration of each inhibitor was 17 μM. Attempts to use higher inhibitor concentrations resulted in cell toxicity.
DNA preparation and amplification of the protease-encoding region.
Total cellular DNA was isolated from infected cell pellets by using the QIAamp blood kit (Qiagen). The protease-coding domain of viral DNA was amplified by nested PCR. The PCR conditions are available on request. PCR products were purified by using QIAquick PCR purification kit (Qiagen) and directly sequenced or cloned into the pT7Blue vector (Novagen) and sequenced.
Construction of infectious 8mut clone.
Cellular DNA from SQV round 27 selection (after selection at 5 μM inhibitor for 10 rounds) was used as the source of a protease gene fragment containing eight mutations (L10I, G48V, I54V, L63P, A71V, V82T, I84V, L90M). Nested PCR was performed to create an amplicon spanning the ApaI site in the nucleocapsid (NC)-coding domain to the 5′ end of the pol gene, where an RsrII site was introduced. The second-round PCR product was cloned into the pT7Blue vector, and cloned inserts were sequenced. The Bsp120I (isoschizomer of ApaI)/RsrII fragment from one clone that contained the eight protease mutations was ligated into the Bsp120I/RsrII sites of the pCadNΔBclBsp clone (S. Pettit, unpublished data). This molecular clone contains the XbaI (5′ of gag) to EcoRV (5′ end of reverse transcriptase [RT]) fragment from HXB within the NL4-3 virus backbone. The resulting recombinant molecular clone, 8mut, was prepared by use of a Qiagen plasmid maxikit and transfected into HeLa cells (9). A virus stock was generated by passage in CEMx174 cells of the supernatant medium from the transfected HeLa cells.
Virus coculture assay.
The 8mut molecular clone virus and a wild-type protease virus were used in a coculture-growth competition assay experiment to measure relative fitness. For each of the virus stocks, the titer was determined by the MAGI cell assay to yield the number of infectious units per milliliter. Ratios of 10 to 1 and 20 to 1 of infectious 8mut virus to a wild-type protease virus were used in the competition experiment. For the 10:1 ratio, 2,000 infectious units of the 8mut virus and 200 infectious units of NL4-3 were used to infect 3 × 106 CEMx174 cells. Infections were done in a volume of 0.5 ml for 2 h at 37°C. The cell-virus mixture was centrifuged, and the pellet was resuspended in 1 ml of phosphate-buffered saline. The cells were centrifuged again, and the pellet was resuspended in 0.5 ml of 1× trypsin (Sigma) and incubated for 5 min at 37°C. The cells were centrifuged again and resuspended in medium to make a 10-ml culture. Every day, the cells were pelleted and the supernatant medium was aliquoted and frozen at −70°C. The cell pellet was resuspended in 10 ml of fresh medium and maintained at 37°C. This procedure was repeated until the cultures showed widespread CPE. An RT assay (6) was performed with the supernatants to determine the peak of virus production. Viral RNA was extracted from the collected supernatant samples starting with the day 3 sample for all daily samples until RT values represented at most 30% of the maximum value. RNA extraction and PCR amplification were as described by Resch et al. (56). By day 3 under these conditions, the input viral RNA is diminished, such that the PCR signal represents newly replicating viral genomes, and stopping the cultures prior to the maximal RT activity ensures target cell excess during the course of virus spread. The heteroduplex tracking assay was used to assess the ratio of the variants in the culture supernatant. Relative fitness was calculated as the change (n-fold) of the ratio of numbers of mutant virus to numbers of wild-type virus per day (w). To obtain the change per day, the model in the equation ln (Mt/Wt) = ln (w)*t + ln (M0/W0), where Mt/Wt is the ratio of mutant to wild-type virus at time t and M0/W0 is the ratio of mutant to wild-type virus at time zero, was employed (30). This model states that the logarithm of the ratio of mutant to wild type changes linearly over time and that the slope of the line is the logarithm of the change per time unit. Linear regression was used to determine the slope.
HTA.
Heteroduplex tracking assay (HTA) analysis was performed on some of the rounds of the selections and on the coculture-growth competition assay samples. The assay was performed as described by Resch et al. (56). The probe used to resolve multiple genotypes present in the selections was the multiple-site-specific probe 6.1. This probe was designed to be sensitive to nucleotide changes at codon positions 46, 48, 54, 82, 84, and 90, although there is some sensitivity to changes at other nearby positions. In some cases a subtype A pro-HTA probe was used, derived from the protease-coding region from a subtype A primary HIV-1 isolate (9327.5), obtained from John Mascola.
Cleavage site analysis.
PCR was used to amplify from total cellular DNA a 585-bp product that included the region of the gag gene encoding the protease cleavage sites NC/p1 and p1/p6, as well as the protease-coding region. The PCR products were purified by use of QIAquick PCR purification columns, and the PCR products were sequenced.
Western blot analysis.
Virions from 200 to 500 μl of selection culture supernatants were pelleted in a microcentrifuge at 21,000 × g for 1.5 h. The pellets were resuspended in radioimmunoprecipitation assay buffer in 1/10 volume. For the 8mut and N432S virions, 10 ml of transfection supernatants were pelleted in an ultracentrifuge at 70,000 × g for 30 min. The virus pellets were resuspended in 50 μl of radioimmunoprecipitation assay (Boehringer Mannheim) buffer. The samples were mixed with an equal volume of sample buffer (Novex), heated to 85°C for 2 min, and then run in a 16% acrylamide-Tricine gel (Novex). Proteins in the gel were transferred to a Hybond-P polyvinylidene difluoride membrane (Amersham Pharmacia Biotech). Viral proteins were detected with a polyclonal anti-p15 (a gift from Susan Erickson-Viitanen). This was followed by incubation in the presence of the secondary antibody, anti-rabbit-horseradish peroxidase (Promega). Finally, the blot was incubated for 5 min with ECL Plus (Amersham Pharmacia Biotech) detection reagents and then placed on film for visualization. This p15 antibody appears to be specific for NC, since its reactivity is identical to an anti-NC antibody (data not shown). An N432S mutant of the NC-p1 processing site, which fails to process at the NC-p1 site (53), was used as a marker for the NC-p1 intermediate. In a control experiment we showed that the use of the anti-p15 antibody gave a dose response in signal over a range of a 30-fold dilution of the target proteins (NC and NC-p1) and that the apparent ratio of NC-p1 to NC was maintained over this range of signal.
Nucleotide sequence accession numbers.
The DNA sequences determined in this study have been deposited in the GenBank nucleotide sequence database under accession no. AY203611 to AY203726.
RESULTS
Selection for high-level resistance in vitro.
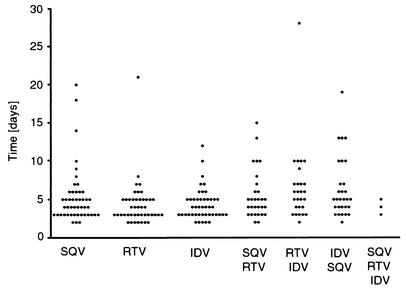
We selected for high-level resistance to each of three HIV-1 protease inhibitors (indinavir, ritonavir, and saquinavir) to evaluate the potential of each drug to select for mutations that would confer high-level resistance and to determine the extent to which these mutations would overlap. Selections were done with each of the inhibitors starting with concentrations in the low nanomolar range and escalating inhibitor concentration by a factor of 1.5 with each subsequent virus passage. When the inhibitor concentration reached 5 μM, the culture was passaged 10 times to enhance replication rates, and then it was increased to 10 μM for 10 passages and then to 15 μM for 10 passages. No cell toxicity was observed at the highest inhibitor concentration used. The amount of time for each passage (i.e., from the time of infection to massive CPE) for each of the inhibitors is shown in Fig. 1. The average time required for the virus to spread through the culture was 4 to 6 days per passage for each of the selections, although several of the passages took longer than 10 days (Fig. 1).
FIG. 1.
Time to extensive CPE during PI selections with increasing inhibitor concentration. Virus generated from the NL4-3 molecular clone was passaged in the presence of increasing inhibitor concentration either with a single inhibitor or with multiple inhibitors as described in the text. The period of time (in days) until the virus-infected culture displayed maximal CPE (massive syncytia) was recorded and is shown as a filled circle for each passage.
HTA analysis after selection in vitro for high-level resistance.
Substitution mutations in the protease-coding domain were examined at various points in the selection protocol by using two approaches. First, an estimate of total heterogeneity was made by using an HTA to display potential multiple genotypes (Fig. 2). In this analysis, the labeled HTA probe was derived from a mutated version of the pro gene designed to have enhanced sensitivity to substitutions at positions 46, 48, 54, 82, 84, and 90, although other nearby substitutions can also affect the migration of the probe (56). The HTA conveniently shows the appearance of new genotypes when assessing a large number of samples and gives accurate quantitation of the relative abundance of multiple species, including minor variants representing as little as 3 to 5% of the population (56). In this way the HTA analysis can complement and guide the sequence analysis.
FIG. 2.
HTA analysis of the pro gene during PI selection. PCR products representing most of protease coding domain were generated as described by Resch et al. (56). The PCR product was annealed to the 6.1 HTA probe (56), and the heteroduplexes were assessed by native polyacrylamide gel electrophoresis. The inhibitor used in the selection is shown above the gel, and the round of selection analyzed is shown at the bottom of each lane. The bracket labeled ht indicates the position in the gel of the heteroduplexes formed between the labeled probe and the unlabeled PCR product. The position labeled hm represents the probe homoduplex. The inhibitor concentration at each round of passage is shown in Table 1.
HTA analysis of the IDV selection revealed no variants with round 1 and 4 samples. Multiple variants were seen at rounds 10, 13, and 16, followed by a homogeneous single population through the rest of the time points, with a small shift upwards in the last sample. With the RTV selection, no new variants were detected in the round 1 or 4 sample, but a doublet that persisted from round 10 to round 15 was seen. By round 24, there was a single band, and in rounds 34 and 44 a single band that shifted slightly higher was apparent. The SQV selection showed a new variant at round 4 and two different pairs of doublets appearing during rounds 15 through 18, followed by a final shift at round 27. This HTA analysis is able to detect only a subset of the resistance-relevant mutations, although the ones that are detected are important for the selecting drugs. These results show that the accumulation of mutations detected by this HTA probe occurred at specific times during the period of selection. In several cases a variant was stable for many rounds of selection, and in two cases (RTV and SQV) the extensive passage at a 5 μM drug concentration reduced the apparent complexity of the culture from two species to one.
Sequence analysis after selection in vitro for high-level resistance.
We next carried out sequence analysis of the viral genome either by direct sequencing of a PCR product spanning the protease-coding domain or by sequencing individual molecular clones derived from the PCR product. In some cases, the sequence of the clone was also linked to the migration of the band in the HTA analysis. Each of the selections showed a similar pattern of increasing numbers of mutations after selection with increasing inhibitor concentration (Table 1). Selection with SQV resulted in the early appearance of mutations at positions 48, 71, and 84, followed by the addition of a mutation at position 90 by round 15. The doublet that was apparent in the HTA analysis at round 17 (Fig. 2) represented the addition of a mutation at position 54. Selection at the highest drug concentrations resulted in the addition of a V82T substitution and mutations at positions 10, 63, 73, and 77. The mutation at position 77 coincided with a reversion at position 71 back to the parental sequence.
TABLE 1.
Accumulation of mutations in inhibitor-treated virus pools
| PI and round no. (PI concn in μM)a | Mutation observed at indicated drug resistance-associated protease position (amino acid)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 (L) | 24 (L) | 46 (M) | 48 (G) | 54 (I) | 63 (L) | 71 (A) | 73 (G) | 77 (V) | 82 (V) | 84 (I) | 90 (L) | |
| SQV | ||||||||||||
| 4 (0.016) | V | V | V | |||||||||
| 10 (0.188) | V | V | V | |||||||||
| 15 (1.86) | V | V | V | L/M | ||||||||
| 17 (4.2) | V | V | V | V | M | |||||||
| 17-ub | V | V | V | V | M | |||||||
| 17-lb | V | V | V | M | ||||||||
| 18 (5)b | V | V | V | S | V | M | ||||||
| 18 (5)b | V | V | V | S | V | M | ||||||
| 27 (5)c | I | V | V | P | V | T | V | M | ||||
| 37 (10) | I | V | V | P | S | I | T | V | M | |||
| 47 (15) | I | V | V | P | S | I | T | V | M | |||
| RTV | ||||||||||||
| 4 (0.06) | ||||||||||||
| 10 (0.687) | V | A | ||||||||||
| 12 (1.55) | L | V | P | A | ||||||||
| 12-ub | V | P | A | |||||||||
| 12-lb | L | V | A | |||||||||
| 14 (3.48) | I | L | V | P | A | |||||||
| 15 (5)b | I | V | P | A | ||||||||
| 15 (5)b | I | L | V | P | A | |||||||
| 24 (5)c,d | I | L | V | P | A | |||||||
| 34 (10) | I | L | V | P | A | V | ||||||
| 44 (15)d,e | I | I | L | V | P | A | V | |||||
| IDV | ||||||||||||
| 4 (0.032) | ||||||||||||
| 10 (0.365) | V | P | A | |||||||||
| 13 (1.233) | V | V | P | A | ||||||||
| 13-ub | V | V | P | A | ||||||||
| 13-mb | V | P | A | |||||||||
| 13-lb | I | V | P | V | A | |||||||
| 16 (4.2) | I | V | P | V | A | |||||||
| 17 (5)b | I | V | P | V | A | |||||||
| 17 (5)b | I | V | P | V | A | |||||||
| 25 (5)c,d | I | I | V | P | V | A | ||||||
| 35 (10) | I | I | V | P | V | A | ||||||
| 45 (15)d | I | I | V | P | V | A | ||||||
ub, upper HTA band; mb, middle HTA band, lb, lower HTA band.
The products from two PCRs were sequenced for this round of selection.
At this round, the products from three PCRs per selection were cloned, at least three clones were sequenced for each PCR product, and the consensus sequence was determined.
Presence of p1/p6 P5′ position mutation P to L.
Presence of NC/p1 P2 position mutation A to V. Additional mutations occurred sporatically at positions 34, 41, 43, and 66. With the exception of E34Q in the RTV selection, none of these other mutations were fixed.
Selection with RTV showed a similar pattern of mutations over time, although in this case a mutation at position 82 appeared early and a mutation at position 84 appeared late. The early appearance of mutations at positions 82, 54, 63, and 46 is consistent with the ordered appearance of mutations that occurs in subjects failing RTV therapy (22, 45). The mixture of species present at round 12 (Fig. 2) represented the presence or absence of a mutation at position 46, although other sites not detected by the HTA probe were also incompletely fixed at that point (i.e., 34 and 63). Overall, mutations became fixed during the selection and were not lost.
Selection with IDV initially resulted in a series of mutations that were similar to those present in the RTV culture (V82A, I54V, L63P). However, the HTA analysis revealed a period of instability followed by apparent stability. Sequence analysis revealed that the mixture present at round 13 (Fig. 2) included the transient appearance of the 54-63-82 constellation of mutations with either M46I or G48V and the appearance of a new species consisting of M46I, I54V, L63P, A71V, and I84A (the latter substitution requiring two nucleotide changes). The 84A-containing genome persisted through the remaining selection period, with L10I being added as the only obvious resistance-associated mutation.
Viral sequences recovered after growth in the presence of the highest inhibitor concentrations had between six and nine mutations within the protease-coding domain that are well recognized as being associated with resistance. There was significant overlap in the mutations that were seen, especially at the higher levels of selection. In each case, approximately 50% of the mutations seen after selection to 15 μM with any one inhibitor were shared among the viral genomes present in another selection, with mutations at positions 10, 46, 54, 63, 82, and 84 appearing in at least two of the selected cultures. One of the two active-site mutations, at positions 82 and 84, appeared early in each of the selections. Mutations at both of these positions played a role in all three of the selections, although the mutations were added in distinct ways for each of the three selections. The active-site mutations appear to influence at least some of the non-active-site mutations, with L63P appearing with or closely after a mutation at position 82, and A71V in two of three cases appearing with a mutation at position 84.
Evolution of drug susceptibility.
Virus pools selected to various levels of resistance were tested for their susceptibility to the inhibitor used in the selection and to the other two PI. Loss in susceptibility (i.e., an increase in the IC50) to the selecting inhibitor after selection for growth in the presence of 15 μM inhibitor was approximately 3,000-fold for SQV, 1,000-fold for RTV, and 250-fold for IDV (Fig. 3). As expected, there was an increase in the level of resistance with increasing selective pressure.
FIG. 3.
Drug susceptibility of selected virus pools. Inhibitor-selected virus pools were tested for resistance to the selecting inhibitor and for cross-resistance to the other two inhibitors. The increase (n-fold) in IC50 is shown on the ordinate. The level of selection for the virus pool tested is shown on the abscissa. Drug sensitivity to SQV is shown in black, that to RTV is shown in stripes, and that to IDV is shown in white.
There was a similar pattern for cross-resistance in all of the virus pools. At the highest level of selection, the loss in susceptibility to the other inhibitors was equivalent to the loss in susceptibility to the selecting inhibitor, with the exception of the susceptibility to SQV of the RTV-selected virus, which was only 200-fold less susceptible to SQV (Fig. 3). As with resistance, cross-resistance increased with increasing selective pressure. Taken together, these results show that the mutations that confer loss in susceptibility to one inhibitor also confer loss in susceptibility to the other two inhibitors and that the high-level selection leads to increased resistance and cross-resistance.
Effects on protein processing.
Amino acid substitutions in the NC/p1 and p1/p6 processing sites of the Gag precursor have been detected in vitro and in vivo as compensatory mutations for the protease (3, 14, 18, 42, 77). We examined these sites in the pools of selected viruses at the levels of 5 and 15 μM selection. No changes were seen in these processing sites in the selection with SQV. However, the NC/p1 site had a change of alanine to valine at the P2 position of the cleavage site at the 15 μM selection level with RTV, and this pool along with those selected at both the 5 and the 15 μM levels with IDV had a P5′ proline-to-leucine change in the p1/p6 cleavage site.
Western analysis of pelleted virus particles was performed from an early time point and from time points adjacent to the time points that were used for sequence analysis. The virion proteins were probed with a polyclonal antibody against p15 (the NC-p1-p6 Gag intermediate) in an effort to determine the extent of processing of the NC-p1 intermediate. As can be seen in Fig. 4, all of the viruses from the early passages showed a reduction in the extent of NC/p1 processing. In the case of the SQV selection, the level of under-processing increased with the accumulation of mutations in pro and in the absence of cleavage site mutations. In the RTV selection, there was a small improvement in the extent of cleavage with the later-passage virus during the time when the NC-p1 cleavage site mutation had evolved. The biggest improvement in processing was seen in the IDV selection, in which the NC-p1 and NC species appeared equal in abundance in virus from rounds 24 and 44. Surprisingly, this corresponded to the appearance of a mutation at the P5′ position of p1/p6 and also with a change of active-site mutations from 82A in round 9 to 84A later.
FIG. 4.
Western analysis of NC/p1 processing in selected pools of virus. Virus selected with the indicated inhibitor at the indicated round of selection was pelleted, and the viral proteins were separated in a polyacrylamide gel under denaturing and reducing conditions. The proteins were transferred out of the gel onto a membrane, and the membrane was probed with a polyclonal rabbit antibody to the HIV-1 NC protein. The position of NC/p1 is shown from virus with NC/p1 processing-site mutation N432S (53). The protease active-site mutation D25A is included as a negative control.
Selection for resistance to multiple PI.
We repeated the selections for resistance with pairs of the three inhibitors (SQV-RTV, SQV-IDV, RTV-IDV). The inhibitors were used initially at a ratio that was roughly equivalent to their IC50s, and the sequential increase in inhibitor concentration was the same as with the individual inhibitors. At the final round of selection, each inhibitor was used at 5 μM. The time of selection for each of the passages is summarized in Fig. 1, and overall these times were similar to those seen with the individual inhibitors.
Sequence analysis of PCR products spanning the pro gene revealed a similar pattern for the accumulation of mutations as seen with the selections with the individual inhibitors (Table 2). Overall, the selection with SQV-RTV was similar to the selection with SQV, with the initial appearance of 48V and 84V mutations and the late appearance of 82T. Most of the other resistance-associated mutations were also shared. Notably, sequence analysis of individual clones from round 27 revealed a mixture of clones having either 82T and 84V or just 84A, with mutations at positions 46 and 48 appearing in both of these backbones, again suggesting an incompatibility between a position 82 mutation and 84A.
TABLE 2.
Accumulation of mutations in multiple PI-treated virus pools
| PI and round no. (PI concn in μM) | Mutation observed at indicated drug resistance-associated protease position (amino acid)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 (L) | 24 (L) | 32 (V) | 36 (M) | 46 (M) | 48 (G) | 54 (I) | 63 (L) | 71 (A) | 77 (V) | 82 (V) | 84 (I) | |
| SQV-RTV | ||||||||||||
| 4 (0.006-0.0225) | V | |||||||||||
| 10 (0.072-0.260) | V | V | ||||||||||
| 15 (0.563-1.98) | V | V | I | A | V | |||||||
| 16 (1.126-3.96) | V | V | I | A | V | |||||||
| 17 (1.689-5.94)a | I | I | V | V | P | V | T | V | ||||
| 17 (1.689-5.94)a | I | I | V | V | P | V | T | V | ||||
| 27 (5.0-5.0)b | I | I | I/L | V | V | P | V | T | V/A | |||
| RTV-IDV | ||||||||||||
| 4 (0.03-0.016) | ||||||||||||
| 10 (0.345-0.183) | I | V | A | |||||||||
| 14 (1.755-0.93) | I | I | V | V | A | |||||||
| 16 (4.0-2.1) | V | V | V | T | V | |||||||
| 17 (6.0-3.2)a | I | I | V | V | P | V | T | V | ||||
| 17 (6.0-3.2)a | I | I | V | V | P | V | T | V | ||||
| 27 (5.0-5.0)b | I | I | I | V | V | P | V | T | V | |||
| IDV-SQV | ||||||||||||
| 4 (0.008-0.016) | ||||||||||||
| 11 (0.093-0.183) | V | V | V | |||||||||
| 15 (0.473-0.93) | V | V | V | |||||||||
| 18 (2.13-4.2) | V | V | P | V | T | V | ||||||
| 19 (3.2-6.3)a | I | I | V | V | P | V | T | V | ||||
| 19 (3.2-6.3)a | I | I | V | V | P | V | T | V | ||||
| 29 (5.0-5.0)b | I | I | V | V | P | V | T | V | ||||
The products of two PCRs were sequenced for this round of selection.
At this round, the products from three PCRs per selection were cloned, at least three clones were sequenced for each PCR product, and the consensus sequence was determined. Additional mutations not shown: R41I at round 10 of the RTV-IDV selection. Also, mutations that appeared in at least two of the clones were M36I of round 27 of the RTV-IDV selection, and M36I, G51R, G48E, and F53Y of round 27 of the SQV-RTV selection.
The overall pattern in the selection with SQV-IDV was very similar to that seen with the SQV-RTV selection, sharing eight resistance-associated mutations and showing the early appearance of a position 84 mutation and the later appearance of a position 82 mutation. In contrast, the selection with RTV-IDV initially paralleled the selection with RTV, with the early appearance of a position 82 mutation followed by the appearance of an 84 mutation. In this culture the G48V mutation also became fixed. At the highest levels of selection, the two cultures that included SQV shared all eight resistance-associated mutations and shared seven of eight mutations with the RTV-IDV culture. Similar trends of association between mutations at positions 63 and 82 and between positions 71 and 84 were seen in the IDV-SQV selection but not in the RTV-IDV selection. In the SQV-RTV selection V77I appeared initially, and only when it reverted did mutations at 63 and 71 appear together after mutations at both 82 and 84.
In order to detect a limit level of resistance to these inhibitors, the three cultures that had been grown in the presence of pairs of inhibitors were further passaged 10 times in the presence of all three inhibitors with 5 μM of each inhibitor. Sequence analysis revealed only small differences between these cultures and the cultures selected with the pairs of inhibitors (compare Tables 2 and 3). Finally, these three cultures were pooled and grown in the presence of increasing inhibitor concentration until the total inhibitor concentration was 51 μM. These passages occurred over fairly brief time periods (Fig. 1), indicating that the virus had attained the ability to replicate in the presence of these high levels of inhibitor. Sequence analysis of the pro gene in this culture revealed a total of 10 resistance-associated mutations that remained similar to those seen with the higher levels of selection with the single- and dual-inhibitor schemes (Table 3). Higher levels of the pooled inhibitors resulted in cell toxicity. This constellation of mutations appears to be sufficient to allow virus replication at the highest levels of inhibitor concentration that can be achieved in culture with this pooling of inhibitors.
TABLE 3.
Accumulation of mutations in triple-protease inhibitor-treated virus pools
| Inhibitors (concn in μM) | Mutation observed at indicated drug resistance-associated protease position (amino acid)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 (L) | 24 (L) | 46 (M) | 48 (G) | 54 (I) | 63 (L) | 71 (A) | 73 (G) | 77 (V) | 82 (V) | 84 (I) | |
| SQV-RTV-IDV (5-5-5)a | I | I | I | V | P | V | A | ||||
| RTV-IDV-SQV (5-5-5)a | I | I | L | V | V | P | V | S | T | V | |
| IDV-SQV-RTV (5-5-5)a | I | I | V | V | P | V | S | I | T | V | |
| SQV-RTV-IDV (17-17-17) | I | I | L | V | V | P | V | S | T | V | |
The supernatants from these three selections were combined to start the next round of selection. Additional mutations not shown are F53Y for SQV-RTV-IDV and I66F for RTV-IDV-SQV.
Characterization of a highly resistant clone.
The results described above were obtained with pools of viruses that likely contained some level of sequence heterogeneity and that had also been adapted to cell culture growth. We created a molecular clone of HIV-1 containing the eight mutations present at round 27 of selection with SQV (Table 1). The virus generated from this clone, referred to as 8mut, had IC50 increases of 1,400-fold for SQV, 350-fold for RTV, and at least 500-fold for IDV. These assays were repeated by using the ViroLogic semiautomated phenotypic sensitivity assay (52). Similar values of IC50 increase were obtained for SQV (>500-fold) and RTV (150-fold), although a lower value was obtained for IDV with this alternative assay (53-fold increase). In addition, this assay was used to detect cross-resistance to nelfinavir (29-fold increase in the IC50) and to a lesser extent to amprenavir (6-fold increase). Thus, the broad and high level of cross-resistance can be associated with a specific set of mutations in the protease.
We next examined the stability of the 8mut virus. The virus was passaged 20 times in the absence of selection and evaluated for reversion of the resistance mutations. HTA analysis revealed the presence of a mixture of two distinct pro genotypes by round 10, followed by the slow outgrowth of the second genotype until it represented approximately one-half of the virus population by round 20 (Fig. 5A). Bulk sequence analysis of the PCR product of the protease region through round 11 showed no changes. From round 12 through round 20, the sequence analysis showed the emergence of a new species representing a reversion at position 48 to the parental glycine codon. Thus, overall, this constellation of mutations appeared to be rather stable during this level of passage in the absence of drug selection. Analysis of the bulk sequence revealed no mutations within the NC/p1 or p1/p6 cleavage sites. This lack of evolution of a processing-site mutation was in spite of the fact that virus from the 8mut showed significantly underprocessed NC/p1 (Fig. 5B).
FIG. 5.
Analysis of the highly resistant cloned virus 8mut. (A) Stability of the 8mut genotype during passage in culture. Successive rounds of passage of 8mut in the absence of inhibitor selection were analyzed by HTA of the protease region. The passage number is shown above each lane. The positions of the 8mut genotype and the V48G reversion to the parental glycine codon are shown, as is the position of the probe homoduplex (hm). (B) Western analysis of NC/p1 processing of 8mut. Western analysis was carried out as described in the legend to Fig. 4. The positions of unprocessed Pr55 Gag, NC-p1, and NC are indicated. The virus analyzed in each lane is indicated at the top of the lane. (C) Virus coculture experiment to measure the fitness of the 8mut virus relative to that of the wild-type virus. Protease HTA was used to distinguish the relative proportions of the two genotypes on a daily basis. The positions of the 8mut and wild-type (WT) genotypes are shown, as is the position of hm. The days of culture are indicated above each lane. (D) Fitness calculation for 8mut. The ln (M/W) ratio (open squares) is plotted versus days in culture, with the slope of the line representing ln (w) (w is the change [n-fold] of the ratio of mutant to wild type per day). Also shown are the abundance of 8mut (open circle) and that of the parental strain (filled circle). dpi, days postinfection.
The growth properties of the 8mut virus were compared directly with a virus devoid of resistance mutations in a virus coculture-growth competition assay. The relative ratio of the coinfecting viruses was monitored daily by using the pro-HTA (Fig. 5C). The parental wild-type virus rapidly outgrew the 8mut virus, indicating that 8mut is significantly attenuated for growth. A calculation of the relative fitness value (30) of 8mut showed that, in the cocultures, the 8mut-to-wild-type ratio changed 0.5- and 0.6-fold per day in two independent determinations (Fig. 5D). Thus, in spite of its apparent relative genetic stability with passage 8mut is significantly attenuated for growth.
DISCUSSION
We initiated these lengthy selections to explore the nature of high-level resistance to several widely used HIV-1 PI used either alone or in combination. Initial virologic failure and resistance to a single PI typically does not lead to the large number of mutations observed in our selections (11, 13, 22, 31, 45, 50, 62, 74). However, larger numbers of mutations are seen on occasion, especially after prolonged exposure to drug or after the use of multiple PI (61, 64, 70, 76). A major difference between the results of our selections and what is seen with patients is that we used increasing drug concentrations over a prolonged period of time while the dosing scheme for patients remains constant. We do not know how this difference affects the combinations of mutations seen or whether it may cause potential differences in the mix of selective pressures for resistance versus fitness.
One significant limitation in cell culture experiments of this type is that the virus pool in which the selection takes place is relatively small, introducing the possibility that stochastic effects may mask a more generalizable observation. There were several places in the selection course in which multiple genotypes coexisted over several passages (Fig. 2), demonstrating the ability of the system to generate multiple viruses with similar selection coefficients. Also, there was considerable sequence overlap in the six independent selections that were done, suggesting that the mutations observed represent common pathways to high-level resistance. This is especially true as many of the mutations likely represent compensatory mutations that affect enzyme activity rather than a direct effect on inhibitor binding (43, 59, 66).
Several previous studies employing either selection in cell culture or the analysis of rebound virus in patients have noted the similarity of mutations and at least partial cross-resistance to one protease inhibitor with viruses selected for resistance to other protease inhibitors (13, 45, 73, 76). Our results confirm and extend those observations. Two of the inhibitors that we used, RTV and IDV, typically initially select for a position 82 mutation as the first active-site mutation (11, 13, 22, 45, 62, 73), as was seen in our selections. In contrast, SQV more often initially selects for position 48 and 90 mutations in vivo (31, 63), although the selection of the active-site I84V mutation can occur in culture (73, 74) and both position 82 and 84 mutations can be seen with a subset of subjects with higher drug exposure (48, 64, 74). Thus, in spite of initial differences in the selected mutations, the subsequent mutations can have significant overlap, and in our selections this included the selection for mutations at two active-site residues, positions 82 and 84. In this way the evolution of resistance was closely paralleled by the evolution of coresistance.
In examining the pattern of appearance of mutations in these selections, we noted several distinctive trends. There were two instances in which the appearance of an I84A mutation was concurrent with the reversion of a position 82 mutation to the wild type. This suggests both that I84A conferred a higher level of resistance and that its presence was incompatible with a position 82 mutation. The absence of I84A from clinical isolates does not allow us to evaluate this potential linkage in sequence databases, and this absence suggests an important, but undefined, difference in the in vitro and in vivo selections. Other potential linkages were examined in the database (69). In two of our selections there was a switch back to the wild-type sequence between a position 71 mutation and a position 77 mutation, suggesting an antagonism between these positions. However, there is no evidence for such antagonism in the larger database (N. Hoffman and R. Swanstrom, unpublished observation). Thus, this appears to be a case where a virus with an altered constellation of mutations replaced a preexisting dominant strain, rather than a choice between mutually exclusive sequence pathways.
Processing-site mutations associated with resistance are seen at the processing sites flanking p1 (NC/p1 and p1/p6) (3, 18, 42, 77) but largely not at other sites in Gag, Pro, or Pol (14). When we examined the extent of processing at NC/p1 we found that this site was significantly underprocessed in budded virions isolated from a wide range of points in the selection scheme, suggesting that attenuation of cleavage at this site is a common feature of viruses with 82-84 mutations (Fig. 4). The NC/p1 position P2 alanine-to-valine mutation appeared late and only in the RTV selection and was associated with a slight apparent improvement in the extent of processing. This was the only selection of the single-drug selections that maintained a V82A mutation in the protease, consistent with the idea that NC/p1 processing site mutations are more strongly selected by position 82 mutations (3). An improvement in the extent of NC/p1 processing occurred in the IDV selection over a period of time when 84A was replacing 82A, and there was no processing-site mutation in the SQV selection in cases where an 84 mutation appeared first. We also observed p1/p6 mutations at P5′ (P to L) but not the more common P1′ L to F (18, 77). We did not observe a build-up of a p15 (NC-p1-p6) intermediate and have been unable to assess the impact of this mutation. Bally et al. (3) have reported an association between this P5′ mutation and a protease position 84 mutation in treated subjects, an association that was somewhat consistent with our studies (Table 1). Given the persistent attenuation of NC/p1 processing, it seems likely either that the virus has limited capability to revert this phenotype or that a high level of underprocessing can be tolerated.
Overall, the patterns of resistance mutations seen in the selections with pairs of PI paralleled those seen with the single inhibitors. In the selections that contained SQV (SQV-RTV and SQV-IDV), the pattern was more similar to the selection with SQV alone, suggesting that the selective pressure from SQV was stronger in these cultures in spite of an initial adjustment in inhibitor concentrations to reflect their relative IC50. In one small study, several PI-naïve subjects who failed SQV-RTV dual therapy had a wide range of mutations that included substitutions at positions 48, 82, 84, and 90 (41). In general, subjects with preexisting PI mutations who fail subsequent dual PI therapy retain the preexisting mutations and add several new mutations (16, 32).
We extended the selection with multiple PI by including all three inhibitors and increasing the inhibitor concentration to levels that approached cell toxicity. No new or distinctive mutations appeared, but rather the similar pattern of accumulation of the many familiar mutations was apparent. Since these selections were initiated with a cloned virus, we could identify all mutations as new sequence variants that were presumably selected. This included substitutions at positions 10, 63, 71, 73, and 77, which are known to be polymorphic in the absence of PI selection (68). Position 10 (59) and position 63 (43, 66) mutations are known to serve as compensatory mutations for deleterious resistance mutations. The fact that these sites are polymorphic in PI-naïve subjects suggests that protease activity may be regulated over a specific range rather than the maximal possible activity.
Finally, we explored the characteristics of virus generated from a molecular clone that had most of the observed mutations. This virus recapitulated the resistance, cross-resistance, and protein processing patterns seen with the pools of selected viruses. In addition, we were able to document a significant fitness decrease in this virus compared to the wild-type parent. Several features of this fitness loss were unexpected. Many of the mutations in this virus are considered compensatory; however, even their presence was not able to increase replication rates to anywhere near that of wild type. Also, when this virus was passaged extensively in culture in the absence of protease inhibitors it was largely stable. Over 20 passages, we detected only the loss of the 48V mutation. There are three explanations for the apparent stability. First, the improved fitness by reversion of other single positions may be sufficiently small not to allow their outgrowth over the period of time during which the virus was cultured. Second, it may be that the compensatory mutations are deleterious alone, as are the resistance mutations, thus, to a certain extent locking the constellation of mutations into a pattern where all single reversions are less fit, although this possibility was not tested. Third, this level of passage in vitro may not provide a sufficient evaluation of genetic stability (although 20 passages in the presence of inhibitor permitted the selection of numerous resistance mutations [Tables 1 and 2]). Distinguishing between these three possibilities is an important question in understanding the nature of the interactions between the mutations that occur during the evolution of high-level resistance to PI.
Acknowledgments
This work was supported through NIH grant R01-AI25321 and the UNC Center For AIDS Research (NIH P30-AI50410).
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HeLa-CD4-LTR-β-gal (MAGI) cells from Michael Emerman, CEMx174 cells from Peter Cresswell, and H9 cells from Robert Gallo. The 293T cell line was graciously provided by John Olsen. We thank Susan Erickson-Viitanen for providing the anti-NC antibody, Steve Pettit for the N432S mutant, and Noah Hoffman for assistance with the database analysis. In addition, we thank Neil Parkin and Rainer Ziermann for carrying out the Phenosense assay with 8mut.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, B., J. Isaacson, M. Knowles, E. Mazabel, and A. K. Patick. 2000. Correlation between human immunodeficiency virus genotypic resistance and virologic response in patients receiving nelfinavir monotherapy or nelfinavir with lamivudine and zidovudine. J. Infect. Dis. 182:420-427. [DOI] [PubMed] [Google Scholar]
- 3.Bally, F., R. Martinez, S. Peters, P. Sudre, and A. Telenti. 2000. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res. Hum. Retrovir. 16:1209-1213. [DOI] [PubMed] [Google Scholar]
- 4.Balotta, C., A. Berlusconi, A. Pan, M. Violin, C. Riva, M. C. Colombo, A. Gori, L. Papagno, S. Corvasce, R. Mazzucchelli, G. Facchi, R. Velleca, G. Saporetti, M. Galli, S. Rusconi, and M. Moroni. 2000. Prevalence of transmitted nucleoside analogue-resistant HIV-1 strains and pre-existing mutations in pol reverse transcriptase and protease region: outcome after treatment in recently infected individuals. Antivir. Ther. 5:7-14. [PubMed] [Google Scholar]
- 5.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, T. C. Merigan, and the CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. AIDS 14:F83-F93. [DOI] [PubMed]
- 6.Buckheit, R. W., Jr., and R. Swanstrom. 1991. Characterization of an HIV-1 isolate displaying an apparent absence of virion-associated reverse transcriptase activity. AIDS Res. Hum. Retrovir. 7:295-302. [DOI] [PubMed] [Google Scholar]
- 7.Call, S. A., M. S. Saag, A. O. Westfall, J. L. Raper, S. V. Pham, J. M. Tolson, N. S. Hellmann, G. A. Cloud, and V. A. Johnson. 2001. Phenotypic drug susceptibility testing predicts long-term virologic suppression better than treatment history in patients with human immunodeficiency virus infection. J. Infect. Dis. 183:401-408. [DOI] [PubMed] [Google Scholar]
- 8.Casado, J. L., F. Dronda, K. Hertogs, A. Antela, and S. Moreno. 2000. Failure of a ritonavir plus saquinavir-based rescue regimen precludes the use of protease inhibitors. AIDS 14:466-468. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Churchill, D. R., A. S. Pym, S. Galpin, R. Foxall, C. Stainsby, J. R. Clarke, S. Kaye, S. Bloor, B. A. Larder, B. Wills, E. Sun, A. G. Babiker, D. J. Back, and J. N. Weber. 1999. The rabbit study: ritonavir and saquinavir in combination in saquinavir-experienced and previously untreated patients. AIDS Res. Hum. Retrovir. 15:1181-1189. [DOI] [PubMed] [Google Scholar]
- 11.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condra, J. H., C. J. Petropoulos, R. Ziermann, W. A. Schleif, M. Shivaprakash, and E. A. Emini. 2000. Drug resistance and predicted virologic responses to human immunodeficiency virus type 1 protease inhibitor therapy. J. Infect. Dis. 182:758-765. [DOI] [PubMed] [Google Scholar]
- 13.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Teppler, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 14.Cote, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cozzi Lepri, A., C. A. Sabin, S. Staszewski, K. Hertogs, A. Muller, H. Rabenau, A. N. Phillips, and V. Miller. 2000. Resistance profiles in patients with viral rebound on potent antiretroviral therapy. J. Infect. Dis. 181:1143-1147. [DOI] [PubMed] [Google Scholar]
- 16.Deeks, S. G., R. M. Grant, G. W. Beatty, C. Horton, J. Detmer, and S. Eastman. 1998. Activity of a ritonavir plus saquinavir-containing regimen in patients with virologic evidence of indinavir or ritonavir failure. AIDS 12:F97-F102. [DOI] [PubMed] [Google Scholar]
- 17.Deeks, S. G., N. S. Hellmann, R. M. Grant, N. T. Parkin, C. J. Petropoulos, M. Becker, W. Symonds, M. Chesney, and P. A. Volberding. 1999. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J. Infect. Dis. 179:1375-1381. [DOI] [PubMed] [Google Scholar]
- 18.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dronda, F., J. L. Casado, S. Moreno, K. Hertogs, I. Garcia-Arata, A. Antela, M. J. Perez-Elias, L. Ruiz, and B. Larder. 2001. Phenotypic cross-resistance to nelfinavir: the role of prior antiretroviral therapy and the number of mutations in the protease gene. AIDS Res. Hum. Retrovir. 17:211-215. [DOI] [PubMed] [Google Scholar]
- 20.Dulioust, A., S. Paulous, L. Guillemot, A. M. Delavalle, F. Boue, and F. Clavel. 1999. Constrained evolution of human immunodeficiency virus type 1 protease during sequential therapy with two distinct protease inhibitors. J. Virol. 73:850-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durant, J., P. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 22.Eastman, P. S., J. Mittler, R. Kelso, C. Gee, E. Boyer, J. Kolberg, M. Urdea, J. M. Leonard, D. W. Norbeck, H. Mo, and M. Markowitz. 1998. Genotypic changes in human immunodeficiency virus type 1 associated with loss of suppression of plasma viral RNA levels in subjects treated with ritonavir (Norvir) monotherapy. J. Virol. 72:5154-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eron, J. J., R. Haubrich, W. Lang, G. Pagano, J. Millard, J. Wolfram, W. Snowden, L. Pedneault, and M. Tisdale. 2001. A phase II trial of dual protease inhibitor therapy: amprenavir in combination with indinavir, nelfinavir, or saquinavir. J. Acquir. Immune Defic. Syndr. 26:458-461. [DOI] [PubMed] [Google Scholar]
- 24.Fatkenheuer, G., R. M. Hoetelmans, N. Hunn, A. Schwenk, C. Franzen, M. Reiser, A. Jutte, J. Rockstroh, V. Diehl, and B. Salzberger. 1999. Salvage therapy with regimens containing ritonavir and saquinavir in extensively pretreated HIV-infected patients. AIDS 13:1485-1489. [DOI] [PubMed] [Google Scholar]
- 25.Gatanaga, H., S. Aizawa, Y. Kikuchi, N. Tachikawa, I. Genka, S. Yoshizawa, Y. Yamamoto, A. Yasuoka, and S. Oka. 1999. Anti-HIV effect of saquinavir combined with ritonavir is limited by previous long-term therapy with protease inhibitors. AIDS Res. Hum. Retrovir. 15:1493-1498. [DOI] [PubMed] [Google Scholar]
- 26.Gulick, R. M., X. J. Hu, S. A. Fiscus, C. V. Fletcher, R. Haubrich, H. Cheng, E. Acosta, S. W. Lagakos, R. Swanstrom, W. Freimuth, S. Snyder, C. Mills, M. Fischl, C. Pettinelli, and D. Katzenstein. 2000. Randomized study of saquinavir with ritonavir or nelfinavir together with delavirdine, adefovir, or both in human immunodeficiency virus-infected adults with virologic failure on indinavir: AIDS clinical trials group study 359. J. Infect. Dis. 182:1375-1384. [DOI] [PubMed] [Google Scholar]
- 27.Hall, C. S., C. P. Raines, S. H. Barnett, R. D. Moore, and J. E. Gallant. 1999. Efficacy of salvage therapy containing ritonavir and saquinavir after failure of single protease inhibitor-containing regimens. AIDS 13:1207-1212. [DOI] [PubMed] [Google Scholar]
- 28.Harrigan, P. R., K. Hertogs, W. Verbiest, R. Pauwels, B. Larder, S. Kemp, S. Bloor, B. Yip, R. Hogg, C. Alexander, and J. S. Montaner. 1999. Baseline HIV drug resistance profile predicts response to ritonavir-saquinavir protease inhibitor therapy in a community setting. AIDS 13:1863-1871. [DOI] [PubMed] [Google Scholar]
- 29.Hertogs, K., S. Bloor, S. D. Kemp, C. Van den Eynde, T. M. Alcorn, R. Pauwels, M. Van Houtte, S. Staszewski, V. Miller, and B. A. Larder. 2000. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS 14:1203-1210. [DOI] [PubMed] [Google Scholar]
- 30.Holland, J. J., J. C. de la Torre, D. K. Clarke, and E. Duarte. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobsen, H., M. Hanggi, M. Ott, I. B. Duncan, S. Owen, M. Andreoni, S. Vella, and J. Mous. 1996. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J. Infect. Dis. 173:1379-1387. [DOI] [PubMed] [Google Scholar]
- 32.Karmochkine, M., A. Si Mohamed, C. Piketty, C. Ginsburg, G. Raguin, V. Schneider-Fauveau, L. Gutmann, M. D. Kazatchkine, and L. Belec. 2000. The cumulative occurrence of resistance mutations in the HIV-1 protease gene is associated with failure of salvage therapy with ritonavir and saquinavir in protease inhibitor-experienced patients. Antivir. Res. 47:179-188. [DOI] [PubMed] [Google Scholar]
- 33.Kaufmann, G. R., C. Duncombe, P. Cunningham, A. Beveridge, A. Carr, D. Sayer, M. French, and D. A. Cooper. 1998. Treatment response and durability of a double protease inhibitor therapy with saquinavir and ritonavir in an observational cohort of HIV-1-infected individuals. AIDS 12:1625-1630. [DOI] [PubMed] [Google Scholar]
- 34.Kaufmann, G. R., K. Suzuki, P. Cunningham, M. Mukaide, M. Kondo, M. Imai, J. Zaunders, and D. A. Cooper. 2001. Impact of HIV type 1 protease, reverse transcriptase, cleavage site, and p6 mutations on the virological response to quadruple therapy with saquinavir, ritonavir, and two nucleoside analogs. AIDS Res. Hum. Retrovir. 17:487-497. [DOI] [PubMed] [Google Scholar]
- 35.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kempf, D. J., K. C. Marsh, G. Kumar, A. D. Rodrigues, J. F. Denissen, E. McDonald, M. J. Kukulka, A. Hsu, G. R. Granneman, P. A. Baroldi, E. Sun, D. Pizzuti, J. J. Plattner, D. W. Norbeck, and J. M. Leonard. 1997. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministration with ritonavir. Antimicrob. Agents Chemother. 41:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lallemand, F., N. Adda, V. Schneider, C. Jacomet, Y. Salhi, and W. Rozenbaum. 1999. Prospective follow-up of 67 indinavir-experienced human immunodeficiency virus-infected and AIDS patients treated with the ritonavir/saquinavir combination. Clin. Infect. Dis. 28:1184-1185. [DOI] [PubMed] [Google Scholar]
- 39.Lawrence, J., J. Schapiro, M. Winters, J. Montoya, A. Zolopa, R. Pesano, B. Efron, D. Winslow, and T. C. Merigan. 1999. Clinical resistance patterns and responses to two sequential protease inhibitor regimens in saquinavir and reverse transcriptase inhibitor-experienced persons. J. Infect. Dis. 179:1356-1364. [DOI] [PubMed] [Google Scholar]
- 40.Lorenzi, P., M. Opravil, B. Hirschel, J. P. Chave, H. J. Furrer, H. Sax, T. V. Perneger, L. Perrin, L. Kaiser, S. Yerly, and the Swiss HIV Cohort Study. 1999. Impact of drug resistance mutations on virologic response to salvage therapy. AIDS 13:F17-F21. [DOI] [PubMed] [Google Scholar]
- 41.Lorenzi, P., S. Yerly, K. Abderrakim, M. Fathi, O. T. Rutschmann, J. von Overbeck, D. Leduc, L. Perrin, B. Hirschel, and the Swiss HIV Cohort Study. 1997. Toxicity, efficacy, plasma drug concentrations and protease mutations in patients with advanced HIV infection treated with ritonavir plus saquinavir. AIDS 11:F95-F99. [DOI] [PubMed]
- 42.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merry, C., M. G. Barry, F. Mulcahy, M. Ryan, J. Heavey, J. F. Tjia, S. E. Gibbons, A. M. Breckenridge, and D. J. Back. 1997. Saquinavir pharmacokinetics alone and in combination with ritonavir in HIV-infected patients. AIDS 11:F29-F33. [DOI] [PubMed] [Google Scholar]
- 45.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 46.Palmer, S., R. W. Shafer, and T. C. Merigan. 1999. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS 13:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paolucci, S., F. Baldanti, R. Maserati, F. Castelli, F. Suter, F. Maggiolo, A. Pan, and G. Gerna. 2000. Quantification of the impact of HIV-1 reverse transcriptase and protease mutations on the efficacy of rescue HAART. Antivir. Res. 45:101-114. [DOI] [PubMed] [Google Scholar]
- 48.Para, M. F., D. V. Glidden, R. W. Coombs, A. C. Collier, J. H. Condra, C. Craig, R. Bassett, R. Leavitt, S. Snyder, V. McAuliffe, and C. Boucher. 2000. Baseline human immunodeficiency virus type 1 phenotype, genotype, and RNA response after switching from long-term hard-capsule saquinavir to indinavir or soft-gel-capsule saquinavir in AIDS clinical trials group protocol 333. J. Infect. Dis. 182:733-743. [DOI] [PubMed] [Google Scholar]
- 49.Paredes, R., T. Puig, A. Arno, E. Negredo, M. Balague, A. Bonjoch, A. Jou, A. Tuldra, C. Tural, G. Sirera, A. Veny, J. Romeu, L. Ruiz, and B. Clotet. 1999. High-dose saquinavir plus ritonavir: long-term efficacy in HIV-positive protease inhibitor-experienced patients and predictors of virologic response. J. Acquir. Immune Defic. Syndr. 22:132-138. [DOI] [PubMed] [Google Scholar]
- 50.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez-Elias, M. J., R. Lanier, V. Munoz, I. Garcia-Arata, J. L. Casado, P. Marti-Belda, A. Moreno, F. Dronda, A. Antela, S. Marco, and S. Moreno. 2000. Phenotypic testing predicts virological response in successive protease inhibitor-based regimens. AIDS 14:F95-F101. [DOI] [PubMed] [Google Scholar]
- 52.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettit, S. C., G. J. Henderson, C. A. Schiffer, and R. Swanstrom. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J. Virol. 76:10226-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piketty, C., E. Race, P. Castiel, L. Belec, G. Peytavin, A. Si-Mohamed, G. Gonzalez-Canali, L. Weiss, F. Clavel, and M. D. Kazatchkine. 1999. Efficacy of a five-drug combination including ritonavir, saquinavir and efavirenz in patients who failed on a conventional triple-drug regimen: phenotypic resistance to protease inhibitors predicts outcome of therapy. AIDS 13:F71-F77. [DOI] [PubMed]
- 55.Race, E., E. Dam, V. Obry, S. Paulous, and F. Clavel. 1999. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS 13:2061-2068. [DOI] [PubMed] [Google Scholar]
- 56.Resch, W., N. Parkin, E. L. Stuelke, T. Watkins, and R. Swanstrom. 2001. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc. Natl. Acad. Sci. USA 98:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhone, S. A., R. S. Hogg, B. Yip, C. Sherlock, B. Conway, M. T. Schechter, M. V. O'Shaughnessy, and J. S. Montaner. 1998. The antiviral effect of ritonavir and saquinavir in combination amongst HIV-infected adults: results from a community-based study. AIDS 12:619-624. [DOI] [PubMed] [Google Scholar]
- 58.Rockstroh, J. K., M. Altfeld, B. Kupfer, R. Kaiser, G. Fatkenheuer, B. Salzberger, K. E. Schneweis, and U. Spengler. 1999. Failure of double protease inhibitor therapy as a salvage therapy for HIV-infected patients resistant to conventional triple therapy. Eur. J. Med. Res. 4:271-274. [PubMed] [Google Scholar]
- 59.Rose, R. E., Y. F. Gong, J. A. Greytok, C. M. Bechtold, B. J. Terry, B. S. Robinson, M. Alam, R. J. Colonno, and P. F. Lin. 1996. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc. Natl. Acad. Sci. USA 93:1648-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ross, L., Q. Liao, H. Gao, S. Pham, J. Tolson, K. Hertogs, B. Larder, and M. S. Saag. 2001. Impact of HIV type 1 drug resistance mutations and phenotypic resistance profile on virologic response to salvage therapy. AIDS Res. Hum. Retrovir. 17:1379-1385. [DOI] [PubMed] [Google Scholar]
- 61.Rousseau, M. N., L. Vergne, B. Montes, M. Peeters, J. Reynes, E. Delaporte, and M. Segondy. 2001. Patterns of resistance mutations to antiretroviral drugs in extensively treated HIV-1-infected patients with failure of highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 26:36-43. [DOI] [PubMed] [Google Scholar]
- 62.Ruiz, L., M. Nijhuis, C. Boucher, T. Puig, A. Bonjoch, J. Martinez-Picado, S. Marfil, D. de Jong, D. Burger, A. Arno, M. Balague, and B. Clotet. 1998. Efficacy of adding indinavir to previous reverse transcriptase nucleoside analogues in relation to genotypic and phenotypic resistance development in advanced HIV-1-infected patients. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:19-28. [DOI] [PubMed] [Google Scholar]
- 63.Schapiro, J. M., J. Lawrence, R. Speck, M. A. Winters, B. Efron, R. W. Coombs, A. C. Collier, and T. C. Merigan. 1999. Resistance mutations to zidovudine and saquinavir in patients receiving zidovudine plus saquinavir or zidovudine and zalcitabine plus saquinavir in AIDS clinical trials group 229. J. Infect. Dis. 179:249-253. [DOI] [PubMed] [Google Scholar]
- 64.Schapiro, J. M., M. A. Winters, J. Lawrence, and T. C. Merigan. 1999. Clinical cross-resistance between the HIV-1 protease inhibitors saquinavir and indinavir and correlations with genotypic mutations. AIDS 13:359-365. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt, B., H. Walter, B. Moschik, C. Paatz, K. van Vaerenbergh, A. M. Vandamme, M. Schmitt, T. Harrer, K. Uberla, and K. Korn. 2000. Simple algorithm derived from a geno-/phenotypic database to predict HIV-1 protease inhibitor resistance. AIDS 14:1731-1738. [DOI] [PubMed] [Google Scholar]
- 66.Schock, H. B., V. M. Garsky, and L. C. Kuo. 1996. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J. Biol. Chem. 271:31957-31963. [DOI] [PubMed] [Google Scholar]
- 67.Sevin, A. D., V. DeGruttola, M. Nijhuis, J. M. Schapiro, A. S. Foulkes, M. F. Para, and C. A. Boucher. 2000. Methods for investigation of the relationship between drug-susceptibility phenotype and human immunodeficiency virus type 1 genotype with applications to AIDS clinical trials group 333. J. Infect. Dis. 182:59-67. [DOI] [PubMed] [Google Scholar]
- 68.Shafer, R. W., P. Hsu, A. K. Patick, C. Craig, and V. Brendel. 1999. Identification of biased amino acid substitution patterns in human immunodeficiency virus type 1 isolates from patients treated with protease inhibitors. J. Virol. 73:6197-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shafer, R. W., D. R. Jung, B. J. Betts, Y. Xi, and M. J. Gonzales. 2000. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 28:346-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shafer, R. W., M. A. Winters, S. Palmer, and T. C. Merigan. 1998. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV-1 isolates from heavily treated patients. Ann. Intern. Med. 128:906-911. [DOI] [PubMed] [Google Scholar]
- 71.Swanstrom, R., and J. Eron. 2000. Human immunodeficiency virus type-1 protease inhibitors: therapeutic successes and failures, suppression and resistance. Pharmacol. Ther. 86:145-170. [DOI] [PubMed] [Google Scholar]
- 72.Tebas, P., A. K. Patick, E. M. Kane, M. K. Klebert, J. H. Simpson, A. Erice, W. G. Powderly, and K. Henry. 1999. Virologic responses to a ritonavir-saquinavir-containing regimen in patients who had previously failed nelfinavir. AIDS 13:F23-F28. [DOI] [PubMed] [Google Scholar]
- 73.Tisdale, M., R. E. Myers, B. Maschera, N. R. Parry, N. M. Oliver, and E. D. Blair. 1995. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob. Agents Chemother. 39:1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaillancourt, M., D. Irlbeck, T. Smith, R. W. Coombs, and R. Swanstrom. 1999. The HIV type 1 protease inhibitor saquinavir can select for multiple mutations that confer increasing resistance. AIDS Res. Hum. Retrovir. 15:355-363. [DOI] [PubMed] [Google Scholar]
- 75.Walter, H., B. Schmidt, A. Rascu, M. Helm, B. Moschik, C. Paatz, M. Kurowski, K. Korn, K. Uberla, and T. Harrer. 2000. Phenotypic HIV-1 resistance correlates with treatment outcome of nelfinavir salvage therapy. Antivir. Ther. 5:249-256. [PubMed] [Google Scholar]
- 76.Winters, M. A., J. M. Schapiro, J. Lawrence, and T. C. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J. Virol. 72:5303-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]