Abstract
In vitro susceptibility testing was performed on strains of Chlamydia trachomatis, Chlamydia pneumoniae, and Chlamydia psittaci under various conditions, including the cell line utilized, the time between infection and the addition of an antimicrobial, the concentration of inoculum, and the effect of multiple passage on the minimal chlamydicidal concentrations for the antibiotics doxycycline, azithromycin, erythromycin, ofloxacin, and tetracycline. With macrolides, the MIC varied depending upon the cell line utilized. With all antimicrobials, the MIC was related to the time at which the antimicrobial was added after infection. By an optimized cell culture passage method, all strains of chlamydia tested demonstrated survival after exposure to high levels (>100 times the MIC) of antimicrobials. Furthermore, upon retest, these surviving organisms did not demonstrate increased MICs. Thus, this phenomenon does not reflect selection of antimicrobial-resistant mutants but rather survival of some organisms in high antimicrobial concentrations (heterotypic survival). An additional 44 clinical isolates of C. trachomatis from patients with single-incident infections were tested against those from patients with recurrent or persistent infections, and heterotypic survival was seen in all isolates tested; hence, in vitro resistance did not correlate with the patient's apparent clinical outcome.
Chlamydia trachomatis, an obligate intracellular bacterium, causes a broad spectrum of clinical syndromes ranging from urethritis and epididymitis in men to cervicitis, endometritis, pelvic inflammatory disease, and ectopic pregnancy in women. C. trachomatis infection is the most commonly reported bacterial sexually transmitted disease in the United States (7). Despite a decline in prevalence in geographic areas with screening and treatment programs, an estimated three million individuals still contract chlamydia each year in the United States (6). The estimated annual cost attributable to chlamydia and its complications is approximately two billion dollars (14).
In an era of increasing resistance to antimicrobials in many bacterial species, resistance has not been of great concern in the treatment of chlamydial infections. This may be due in part to the facts that very few patients have chlamydia cultures obtained, that tests of cure evaluations are not routinely done, and that even if cultured, few isolates are tested for antimicrobial susceptibilities due to the laborious and unstandardized methodology. Recently, however, clinical treatment failures have been reported and attributed to multidrug-resistant C. trachomatis strains (15, 27; G. Schmid, B. Van Der Pol, R. B. Jones, and R. Johnson, Abstr. Int. Congr. Sex. Transm. Infect., Int. J. Sex. Transm. Dis. AIDS 12:41, 2001). These strains were reported to demonstrate heterotypic resistance, a pattern in which small numbers of organisms survive antimicrobial concentrations well above the MIC (15, 27; G. Schmid et al., Abstr. Int. Congr. Sex. Transm. Infect.). C. trachomatis strains exhibiting homotypic resistance, a pattern in which the majority of organisms survive antimicrobial concentrations well above the MIC, have not been reported in isolates from humans.
Despite its apparent rarity, there are reasons to suspect that antimicrobial resistance does indeed occur in urogenital chlamydia. For example, C. trachomatis may be repeatedly cultured from some patients despite antimicrobial therapy (15, 18, 27; G. Schmid et al., Abstr. Int. Congr. Sex. Transm. Infect.), suggesting resistance. Same-serovar recurrences of chlamydia are frequent (2, 4, 9, 23), which would also be consistent with resistant strains persisting after therapy (but could be due to reinfection as well). Finally, selection for resistance in the laboratory has been demonstrated for a variety of antimicrobials (10, 17), and recent C. trachomatis isolates from swine (termed Chlamydia suis) chronically exposed to tetracycline demonstrate homotypic resistance (1, 19). Chlamydia pneumoniae responds to treatment with macrolides, tetracyclines and fluoroquinolones, and to date naturally occurring antimicrobial resistance has not been reported in C. pneumoniae or in other Chlamydia species (28).
Although many research groups perform antimicrobial susceptibility testing of Chlamydia organisms worldwide, there is not a standardized methodology or uniformly accepted interpretation of results. Because of the potential impact of antimicrobial resistance on the prevention and treatment of Chlamydia infections, we investigated differences in methodologies, cell lines utilized, and interpretation of antimicrobial susceptibility testing results in order to provide a basis for standardizing the methodologic approach and interpretation of antimicrobial susceptibility testing in the Chlamydia species. As the association of in vitro antimicrobial resistance with treatment failures or recurrences of C. trachomatis infection in humans has not been extensively studied, we also assessed the MIC and minimal chlamydicidal concentration (MCC) of C. trachomatis isolates associated with either treatment failure or same-serovar recurrence in comparison with those of strains associated with apparently successful therapy. As an example of homotypic resistance in our studies, we utilized a C. suis strain for which the tetracycline and doxycycline MICs were elevated (8.0 and 1.0 μg/ml, respectively).
MATERIALS AND METHODS
Organisms.
Laboratory reference strains for C. trachomatis (serovars B, D, E, F, G, H, I, Ia, H, J, K, and L2), C. pneumoniae (TW-183 and CWL-029), and C. psittaci (6BC and GPIC) are maintained and routinely used in our laboratory for research. The tetracycline-resistant C. suis strain R-19 was provided by Arthur A. Andersen, National Animal Disease Center, Ames, Iowa. Clinical strains were selected from a large archive of serotyped clinical isolates obtained from patients who had a culture-documented C. trachomatis genital infection at any of the Seattle-King County Health Department sexually transmitted disease clinics. Single-episode C. trachomatis isolates were selected from patients who had no record in our database of having an additional previous or subsequent chlamydial infection. Persistent isolates were chosen from patients who had three or more same-serovar recurrent C. trachomatis infections over a period of more than 2 years with no intervening infections by a different chlamydial serovar. Treatment failure isolates were selected from patients who had two or more visits with a positive C. trachomatis culture after receiving documented treatment. Four unknown strains without identifiers were provided by the Centers for Disease Control and Prevention (CDC) for susceptibility testing; two isolates were from patients diagnosed as treatment failures and had been determined to be multidrug resistant by CDC laboratories, and two isolates were controls without antimicrobial resistance.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing of C. trachomatis, C. pneumoniae, and C. psittaci strains in six cell lines (HeLa, McCoy, BGMK, HEp-2, HL, and Vero) without passage (MIC testing) on 96-well microtiter plates and by multiple passage (MCC testing) in 48-well microtiter plates and 12-mm glass shell vials was undertaken with doxycycline, azithromycin, erythromycin, ofloxacin, and tetracycline. Antimicrobial susceptibility testing in 48-well microtiter plates and 12-mm glass shell vials with doxycycline, azithromycin, and ofloxacin was undertaken in McCoy cells for the C. suis strain R-19, the reference C. trachomatis and C. psittaci strains, and the clinical isolates of C. trachomatis and in HEp-2 cells for the reference C. pneumoniae strains. Cells were maintained in antimicrobial-free growth medium consisting of minimal essential medium with 10% fetal bovine serum and 220 μg of l-glutamine/liter added. The inoculum size of infectious chlamydial forms for all MIC and MCC comparisons was 10,000 to 50,000 inclusion-forming units (IFU) in each well for 48-well microtiter plates or on 12-mm coverslips in shell vials, resulting in the infection of approximately 10 to 50% of the cells in the monolayers.
MIC determinations were performed on monolayers in 96-well plates. For all centrifugation procedures, a Beckman model J-6 M centrifuge was utilized. Each chlamydial strain was inoculated into three rows of 12 wells and centrifuged at 1,200 × g for 1 h at 37°C, and then supernatants were aspirated. Antimicrobial agents were obtained in powder form, weighed and adjusted for purity, and reconstituted according to the manufacturer's instructions. Dilution schemes were prepared by using twofold dilutions in antimicrobial-free growth medium containing cycloheximide (1.0 μg/ml). For each antimicrobial, 100 μl of each dilution (300 μl for 48-well plates and shell vials) was added to the appropriate wells to give a final concentration range of 0.008 to 128 μg/ml. Passage of tissue culture plates was done by a freeze-thaw method where monolayers were frozen at −70°C, thawed in a 37°C water bath, disrupted with a pipette tip, passed onto fresh monolayers, and centrifuged at 1,200 × g for 1 h at 37°C. Wells were then aspirated and overlaid with appropriate twofold drug dilutions made in antimicrobial-free growth medium with cycloheximide added. Cells were incubated at 37°C in 4% CO2 for 24 to 72 h (48 h for C. trachomatis, 72 h for C. pneumoniae, and 24 h for C. psittaci) and fixed with methanol. Chlamydial inclusions were detected by fluorescence by using a genus-specific monoclonal antibody CF-2 (Washington Research Foundation, Seattle).
The passage of monolayers in shell vials differed from that on culture plates in that after incubation, the medium containing the antimicrobial was aspirated and the cells were rinsed twice with Hanks balanced salt solution. The cells were then disrupted by sonication by using an ultrasonic processor, and the resulting suspension was centrifuged at 300 × g for 10 min at 4°C to remove debris. The supernatant was inoculated onto fresh monolayers in shell vials, centrifuged at 1,200 × g for 1 h at 37°C, aspirated, and overlaid and incubated as described previously. For each passage, 100 μl of each passage vial was added to corresponding wells on a 96-well microtiter plate for observation of inclusions. A laboratory reference strain, serovar D/uw-3/cx, which is antimicrobial sensitive, was used in all comparisons and in experiments comparing the MIC, MCC, and MCC3 (the MCC after three passages in shell vials; see below).
The transition point MIC (MICTP) was defined as the concentration of drug in which 90% or more of the inclusions were altered in size and morphology. The MIC was defined as the concentration of drug that is one twofold dilution more concentrated than the MICTP. The MCC was defined as the lowest concentration of drug that produced no morphologically normal inclusions by one freeze-thaw passage in microtiter plates (microtiter plate method) or by one passage in shell vials (shell vial method) in antimicrobial-free medium. The MCC3 was determined to be the MCC after three passages in shell vials (shell vial method) in antimicrobial-free medium.
To determine the effect of cell line variation on MCC3 levels, several representative Chlamydia strains were passed three times in McCoy, BGMK, HeLa and HEp-2 cells. To test for selection of antimicrobial-resistant organisms, chlamydiae surviving the highest MCC3 were retested against the respective antimicrobial and the MIC was determined.
A twofold dilution scheme with C. trachomatis strain UW-3 (serovar D) inocula ranging from ∼320,000 to 300 IFU/well (48-well microtiter plate) was used to determine the effect of inoculum size on the MIC, MCC, and MCC3. The effect on MICs of the time that elapsed between infection and the addition of a given antimicrobial was tested by adding the drug at 2-h intervals up to 24 h after infection.
RESULTS
Antimicrobial susceptibility testing of Chlamydia species in different cell lines.
The MICs of tetracycline, doxycycline, and ofloxacin were comparable in all cell lines for serovar D (Table 1). Interestingly, there was considerable MIC variation when C. trachomatis was tested against azithromycin and erythromycin, with the lowest MICs being those for isolates tested in HeLa and HL cell lines and with the MICs for those tested in the BGMK cell line being significantly higher (Table 1). The MCC and MCC3 for C. trachomatis (Table 1) were highest in McCoy and BGMK cells.
TABLE 1.
Effect of cell line on MIC, MCC, and MCC3 for C. trachomatis serovar Da
| Cell type (origin) | Doxycycline
|
Azithromycin
|
Erythromycin
|
Ofloxacin
|
Tetracycline
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MCC | MCC3 | MIC | MCC | MCC3 | MIC | MCC | MCC3 | MIC | MCC | MCC3 | MIC | MCC | MCC3 | |
| McCoy (human) | 0.064 | >8 | 64 | 0.125 | >8 | 64 | 0.25 | >8 | 128 | 1 | >8 | 128 | 0.25 | >8 | 64 |
| HeLa (human) | 0.064 | 1 | 4 | 0.016 | 1 | 8 | 0.032 | 2 | 4 | 1 | 4 | 16 | 0.25 | 2 | 16 |
| BGMK (simian) | 0.064 | >8 | 64 | 1.0 | >8 | 64 | 2 | >8 | 128 | 1 | >8 | 128 | 0.25 | >8 | 64 |
| HEp-2 (human) | 0.064 | 1 | 4 | 0.064 | 4 | 32 | 0.125 | 4 | 8 | 1 | 4 | 64 | 0.25 | 4 | 32 |
| HL (human) | 0.064 | 2 | 32 | 0.008 | 4 | 32 | 0.016 | 4 | 0.5 | 1 | 8 | 32 | 0.25 | 4 | 32 |
| Vero (simian) | 0.064 | 2 | 8 | 0.25 | 2 | 4 | 0.5 | 4 | 16 | 1 | 8 | 32 | 0.25 | 2 | 16 |
All values are in micrograms per milliliter. The MIC was defined as one twofold dilution more than the MICTP; MCC, minimal chlamydicidal concentration following one freeze-thaw passage in microtiter plates; MCC, minimal chlamydicidal concentration following one passage in shell vials; MCC3, minimal chlamydicidal concentration following three passages in shell vials.
MICs for C. pneumoniae strain TW-183 and C. psittaci strain 6BC were comparable for each antimicrobial tested (not shown). The MCC and MCC3 for C. pneumoniae were highest in HEp-2 and HL cells (not shown), while the corresponding values for C. psittaci were highest in McCoy and BGMK cells (not shown).
Effect of inoculum size on antimicrobial susceptibility testing in C. trachomatis.
There was no observed difference in MICs when the inoculum size of viable chlamydia was varied over the range of 300 to 300,000 IFU/well. However, when an inoculum of less than 300 IFU/well was used, it was difficult to find a sufficient number of inclusions to read accurately. Variation of inoculum size did influence MCCs by shell vial passage, with elevation of MCCs and MCC3s above the MIC (Table 2). However, when the inoculum fell below 5,000 IFU/well, there was no observed difference between the MIC and MCC. This suggests that the survival rate of Chlamydia exposed to an effective antimicrobial is approximately one bacterium per 5,000 infected cells. Inclusions produced from surviving bacterium by definition of the chlamydial developmental cycle must have been produced from viable elementary bodies generated from aberrant inclusions, since no normal inclusions are observed. Independent incubation death studies done in our laboratory showed that elementary bodies could not survive in cell-free growth medium at an incubation temperature of 37°C for periods of longer than 24 h (unpublished data).
TABLE 2.
Effect of inoculum size on doxycycline MIC, MCC, and MCC3 for C. trachomatis serovar Da
| Inoculum (103 IFU/well) | MIC | MCC | MCC3 |
|---|---|---|---|
| 320 | 0.064 | >8 | 64 |
| 160 | 0.064 | >8 | 64 |
| 80 | 0.064 | >8 | 64 |
| 40 | 0.064 | >8 | 64 |
| 20 | 0.064 | >8 | 64 |
| 10 | 0.064 | >8 | 64 |
| 5 | 0.064 | >8 | 64 |
| 2.5 | 0.064 | 0.064 | 0.064 |
| 1.25 | 0.064 | 0.064 | 0.064 |
| 0.625 | 0.064 | 0.064 | 0.064 |
| 0.3125 | 0.064 | 0.064 | 0.064 |
All values are in micrograms per milliliter. The MIC was defined as one twofold dilution more than the MICTP; MCC, minimal chlamydicidal concentration following one freeze-thaw passage in microtiter plates; MCC, minimal chlamydicidal concentration following one passage in shell vials; MCC3, minimal chlamydicidal concentration following three passages in shell vials.
Effect of the time between infection and the addition of antimicrobial agents on antimicrobial susceptibility testing in C. trachomatis.
There were no observable differences in MICs when an antimicrobial was added at 2-h intervals from 0 to 8 h after infection, but the MIC increased steadily after 8 h, suggesting that the drug had less of an effect on chlamydial development after 8 h postinfection (data not shown).
Antimicrobial susceptibility testing of reference strains of Chlamydia species and clinical isolates of C. trachomatis.
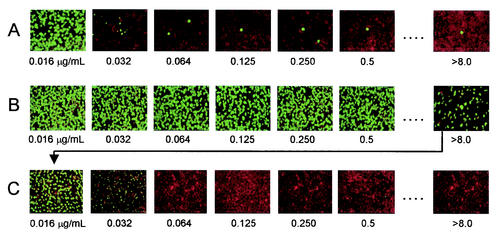
Table 3 shows the results of in vitro susceptibility testing of the C. trachomatis, C. pneumoniae, and C. psittaci reference strains against doxycycline, azithromycin, and ofloxacin. For the tetracycline-resistant C. suis strain, the doxycycline MICs were considerably higher than those for the other reference strains tested, but the azithromycin and ofloxacin MICs were comparable to those for the other reference strains. Figure 1 illustrates the elevated doxycycline MIC (homotypic resistance) demonstrated by the C. suis strain R-19 relative to that for the sensitive C. trachomatis strain serovar D. The MCCs determined by microtiter plate passage were consistently lower than those determined by shell vial passage for all of the Chlamydia reference strains. Further, the MCCs determined by shell vial passage were consistently lower than the MCC3s. The MCC3s were consistently >100 times the MICs, demonstrating the survival of some organisms at high drug concentrations. Normal inclusions formed from surviving chlamydia at higher MICs after one passage (MCC) are illustrated in Fig. 2A. When Chlamydia strains reflecting the highest MCC3s were retested, there was no homotypic elevation of MICs, but rather a repeat of the initial MIC pattern with the heterotypic survival of a small number of organisms at higher MICs of a given antimicrobial (Fig. 2B and C). For some strains, the retesting of surviving organisms reflecting the highest MCC3s was repeated up to five times, still with no apparent elevation of the MIC for this select group of organisms. When MCC3s were determined for isolates in different cell lines, those for isolates cultured in the McCoy and BGMK cell lines were comparable. For Chlamydia in HeLa cell lines, the MCC3s were elevated (>10 times the MIC), but never to the higher degree seen in the McCoy and BGMK cell lines, exemplifying the various degrees of infectivity that different cell lines have for Chlamydia. For C. pneumoniae strains, the MCC3s were found to reach those seen in McCoy cells when HEp-2 cells were used.
TABLE 3.
Antimicrobial susceptibilities of reference strains of Chlamydia speciesa
| Chlamydia reference strain(s) | Doxycycline
|
Azithromycin
|
Ofloxacin
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microtiter
|
Shell vial
|
Microtiter
|
Shell vial
|
Microtiter
|
Shell vial
|
|||||||
| MIC | MCC | MCC | MCC3 | MIC | MCC | MCC | MCC3 | MIC | MCC | MCC | MCC3 | |
| C. trachomatis B, D, E, F, G, H, I, Ia, J, and K (human genital serovars) | 0.064 | 0.125 | >8 | 16-32 | 0.125 | 0.25 | >8 | 64 | 0.5 | 1.0 | >8 | >128 |
| C. trachomatis L2 (LGV serovar) | 0.064 | 0.125 | >8 | 32 | 0.125 | 0.25 | >8 | 64 | 0.5 | 1.0 | >8 | >128 |
| C. trachomatis MoPn (mouse pneumonitis strain) | 0.064 | 0.5 | >8 | 64 | 0.125 | 0.5 | >8 | 128 | 0.5 | 1.0 | >8 | >128 |
| C. suis R-19 (porcine strain)b | 1.0 | 2.0 | >8 | >128 | 0.125 | 0.25 | >8 | >128 | 0.5 | 1.0 | >8 | >128 |
| C. pneumoniae TW-183, CWL-029c | 0.064 | 0.125 | 4-8 | 32-64 | 0.125 | 0.25 | 4-8 | 64 | 0.5 | 1.0 | >8 | >128 |
| C. psittaci 6BC, GPIC | 0.064 | 0.250 | >8 | >128 | 0.125 | 0.50 | >8 | >128 | 0.5 | 2.0 | >8 | >128 |
All values are in micrograms per milliliter. The MIC was defined as one twofold dilution more than the MICTP; MCC, minimal chlamydicidal concentration following one freeze-thaw passage in microtiter plates; MCC, minimal chlamydicidal concentration following one passage in shell vials; MCC3, minimal chlamydicidal concentration following three passages in shell vials.
Tetracycline-resistant (MIC, >4 μg/ml) porcine strain provided by Arthur A. Andersen, National Animal Disease Center, Ames, Iowa.
C. pneumoniae strains grown in HEp-2 cells.
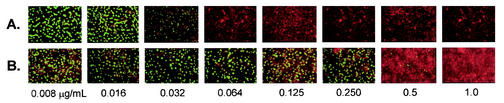
FIG. 1.
Sequential photomicrographs of MIC growth patterns of C. trachomatis serovar D (A) and C. suis strain R-19 (B) at various concentrations of doxycycline. The C. suis strain demonstrates homotypic resistance, in which most of the organisms survive at concentrations well above MIC for the C. trachomatis strain. Also shown is an example of the MICTP, the transition point where the concentration of antimicrobial first noticeably influences chlamydial inclusion growth (MICTP is 0.032 μg/ml for C. trachomatis and 0.5 μg/ml for C. suis).
FIG. 2.
Sequential photomicrographs of growth C. trachomatis serovar D at three different passage levels after initial exposure to given concentrations of doxycycline. (A) Illustration of MCC with heterotypic survival at concentrations well above the MICs after one passage in antimicrobial-free medium. (B) Illustration of MCC3 with complete growth of surviving chlamydiae after three passages in antimicrobial-free medium. (C) Illustration of the MIC pattern seen after retesting chlamydiae from the highest drug concentration in which organisms survived (arrow).
Antimicrobial susceptibility testing of clinical isolates of C. trachomatis.
Susceptibility testing of the different clinical isolates of C. trachomatis showed only slight variation (by one twofold dilution) of the doxycycline, azithromycin, and ofloxacin MICs (Table 4). There was no apparent difference in the MIC, MCC, or MCC3 when strains from individuals with treatment failure or persistence were compared with those from persons with single-episode isolates.
TABLE 4.
Antimicrobial susceptibilities of clinical isolates of Chlamydia trachomatisa
| C. trachomatis clinical isolate(s) | Doxycycline
|
Azithromycin
|
Ofloxacin
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microtiter
|
Shell vial
|
Microtiter
|
Shell vial
|
Microtiter
|
Shell vial
|
|||||||
| MIC | MCC | MCC | MCC3 | MIC | MCC | MCC | MCC3 | MIC | MCC | MCC | MCC3 | |
| Single isolates (n = 30) | 0.064-0.125 | 0.125-0.250 | >8 | 16-64 | 0.125-0.250 | 0.25-0.50 | >8 | 64 | 0.5-1.0 | 1-2 | >8 | >128 |
| Treatment failure isolates (n = 6) | 0.064-0.125 | 0.125-0.250 | >8 | 16-64 | 0.125-0.250 | 0.25-0.50 | >8 | 64 | 0.5-1.0 | 1-2 | >8 | >128 |
| Recurrent same-serovar isolates (n = 6) | 0.064-0.125 | 0.125-0.250 | >8 | 16-64 | 0.125-0.250 | 0.25-0.50 | >8 | 64 | 0.5-1.0 | 1-2 | >8 | >128 |
| CDC strains | ||||||||||||
| Treatment failures (n = 2)b | 0.064-0.125 | 0.125-0.250 | >8 | 16-64 | 0.125-0.250 | 0.25-0.50 | >8 | 64 | 0.5-1.0 | 1-2 | >8 | >128 |
| Sensitive controls (n = 2)c | 0.064-0.125 | 0.125-0.250 | >8 | 16-64 | 0.125-0.250 | 0.25-0.50 | >8 | 64 | 0.5-1.0 | 1-2 | >8 | >128 |
All values are in micrograms per milliliter. The MIC was defined as one twofold dilution more than the MICTP; MCC, minimal chlamydicidal concentration following one freeze-thaw passage in microtiter plates; MCC, minimal chlamydicidal concentration following one passage in shell vials; MCC3, minimal chlamydicidal concentration following three passages in shell vials.
Strains from two treatment failures provided by the CDC and determined at CDC laboratories to be multidrug resistant.
Strains from two controls provided by the CDC and determined at CDC laboratories to be sensitive isolates.
DISCUSSION
Cell culture systems using immunofluorescence staining to identify chlamydial inclusions are the most common methodology employed in antimicrobial susceptibility testing of Chlamydia. Such systems are not completely physiologic in that during natural infection, chlamydiae are usually exposed to antimicrobials long after the establishment of intracellular infection and the induction of a host inflammatory response. In contrast, with the in vitro systems, many investigators add antimicrobials simultaneously with or shortly after the infection (28). The failure to detect infectious chlamydiae in cell cultures in vitro does not exclude a viable state that could recur after antibiotics are removed (3). Dreses-Werringloer et al. used a reverse transcriptase PCR (RT-PCR) assay to demonstrate the presence of primary 16S rRNA transcripts in chlamydia-infected cells treated with 0.5 μg of azithromycin per ml over a period of 16 days (12). These results support our finding of survival of small numbers of organisms at concentrations well above the MIC for all antimicrobials tested. Hence, it is possible that in vivo, chlamydial organisms could survive in patients treated with a single dose of azithromycin despite being susceptible in vitro.
However, in vitro culture systems do not allow for the role of immune clearance mechanisms in a host. For example, Rasmussen et al. demonstrated that despite the effect of antimicrobials on chlamydia growth and development, the infected cells continued to be recognized and killed by cytotoxic T cells (24). The importance of the host response is further supported by findings by Parks et al. demonstrating possible spontaneous clearance of chlamydial infection in a minority of untreated patients. Resolution of infection was associated with increasing duration of clinical follow-up (22). Hence, heterotypic survival of small numbers of infectious elementary bodies may occur in vitro, but in natural infections, host factors may eradicate the remaining surviving chlamydial organisms.
We found that when Chlamydia isolates were tested in the presence of tetracycline, doxycycline, and ofloxacin, antimicrobial susceptibilities were comparable with all cell lines utilized. However, the MICs and MCCs varied significantly by cell line when tested with azithromycin and erythromycin, particularly being increased in BGMK cells. Many laboratory conditions, such as pH, temperature, nutrients present in the media, polarity of the cell type infected, and secretion of cytokines by infected cells, likely differ from natural infection and may affect the ability of a particular antimicrobial to penetrate intracellularly and exert its action (13, 16, 25, 26, 29, 30). Rota demonstrated that McCoy cells in media with a high glucose concentration, a pH closer to neutrality, and a higher temperature during centrifugation (33 to 35°C being more effective than 20 to 25°C) produced higher numbers of C. trachomatis inclusions upon infection (26). These conditions could be more variable in vivo, and therefore the in vitro MIC may not necessarily correlate with that in vivo. Wyrick et al. reported that use of polarized host cells, rather than nonpolarized cells such as laboratory-adapted HeLa or McCoy cell monolayers, resulted in more efficient transport and internal concentration of azithromycin within host cells and that C. trachomatis appeared more susceptible to the lethal action of azithromycin in polarized cells (29). They postulated that basolateral uptake of antibiotics was compromised when cells were cultured in vitro on nonporous surfaces, compared with internalization of antibiotics in vivo by mucosal epithelial cells via basolateral membranes. The polarity of the various cell lines tested with azithromycin may have influenced its uptake, which may explain the differences in azithromycin MICs in different cell lines. Thus, the type of cell line used influences the MIC. In addition, extensive quality control and monitoring of a given cell line is necessary to achieve optimal sensitivity.
Additional variables that may influence results of susceptibility testing in Chlamydia, but which have been incompletely studied, include the inoculum size, the interval between the establishment of infection and the administration of an antimicrobial, and the timing of antimicrobial removal (21). We observed no differences in MICs when antimicrobials were added from 0 to 8 h after infection, while the MIC increased steadily after 8 h. Notomi et al. reported that the addition of ofloxacin or sparfloxacin even at concentrations up to 64 μg/ml could not inhibit the formation of C. trachomatis inclusions when added 20 h after the start of infection (21). These findings suggest that addition of an antimicrobial after a certain time interval following infection may lessen the effect of the antimicrobial on chlamydial development, but more studies are needed to clarify this time interval for various antimicrobials.
A critical methodological aspect of susceptibility testing in Chlamydia is the endpoint utilized for defining the MIC, since distinguishing aberrant from normal inclusions can be tedious, and subjective interpretation may lead to severalfold variation in the MICs. At the MICTP, there is a dramatic alteration in the morphology and size of the inclusions, leading to nearly every inclusion being morphologically aberrant. At further serial dilutions, occasional inclusions are seen and it becomes difficult to determine what can be considered an inclusion of typical morphology. Hence, the MICTP provides a consistent end point where almost all chlamydial inclusions are inhibited, and one twofold concentration above the MICTP would seem to provide a reasonable standardized endpoint to define the MIC. By using this definition, different groups performing antimicrobial susceptibility testing of Chlamydia organisms would achieve reproducible and comparable MIC results. While small numbers of chlamydiae survive at concentrations well above the MIC, the use of these organisms to define the MIC is problematic. It is likely that differences in interpretation of inclusion morphology at higher antimicrobial concentrations may explain the differences in MICs that can arise when the same isolates are tested by different laboratories. This, for example, may explain why our MIC results on the C. trachomatis strains obtained from the Centers for Disease Control differ from the Centers' findings. Many other research groups have demonstrated their reproducibility of determining the MICTP (5, 8, 11, 18, 27, 29).
Recently, an RT-PCR-based method for antimicrobial susceptibility testing in C. trachomatis has been described which may be more sensitive and specific than standard cell culture methods, since it has the ability to detect viable chlamydiae that otherwise may be unrecognizable upon immunofluorescence staining of chlamydial inclusions (8). Cross et al. reported RT-PCR to be consistently more sensitive than conventional cell culture methods and demonstrated MICs that were 1.6-fold higher for erythromycin by this method than by conventional methods (8). RT-PCR may certainly be a useful method to validate the results of Chlamydia susceptibility tests that utilize cell culture methods; further studies of this type are needed.
Relatively few reports have described antimicrobial resistance in clinical isolates of C. trachomatis. One explanation for the apparent lack of resistance in C. trachomatis may be the organism's unique developmental cycle. Since gene replication occurs isolated within an intracellular inclusion in an infected epithelial cell, acquisition of antibiotic resistance genes from other organisms would be difficult (28). However, it is plausible that point mutations leading to antimicrobial resistance may occur under selective pressure from exposure to subinhibitory concentrations of antimicrobials. Selection for antimicrobial resistance has been clearly demonstrated in the laboratory by serial passage of C. trachomatis strains in subinhibitory concentrations of sulfonamides, penicillins, rifampin, and fluoroquinolones (10, 17). Dessus-Babus et al. exposed L2 strains of C. trachomatis to subinhibitory concentrations of ofloxacin (0.5 μg/ml) and sparfloxacin (0.015 μg/ml) and after four passages isolated two mutant strains that demonstrated high-level resistance to various fluoroquinolones, particularly sparfloxacin (1,000-fold increase in the MIC) (10). The MICs of doxycycline and erythromycin for the mutant strains were identical to those for the reference strain. A point mutation was found in the gyrA quinolone resistance-determining region of both mutant strains. Similar results were recently reported by Morrisey and coworkers (20). The ability to select for antimicrobial resistance in the laboratory by exposure to subinhibitory antimicrobial concentrations suggests that a similar mechanism could lead to the development of resistant chlamydial organisms in humans (or animals) exposed to antimicrobial therapy.
Despite selection for antimicrobial resistance to C. trachomatis in the laboratory, it is unclear whether continuous exposure to antimicrobials promotes resistance in vivo. The finding of antimicrobial resistance in swine C. suis isolates supports this possibility. Andersen and Rogers reported stable high-level resistance to tetracycline (MIC, ≥5 μg/ml) in eight swine C. suis isolates that had undergone 10 to 15 passages in tetracycline-free medium before testing (1). Since tetracycline congeners were used as swine feed additives, it seems plausible that the selective pressure of continuous exposure to this antimicrobial may have selected for more resistant swine C. trachomatis strains.
Relatively few clinical isolates of C. trachomatis with antimicrobial resistance have been described. In 1990, Jones et al. reported five patients who were infected with C. trachomatis isolates that were resistant (MIC, ≥8 μg/ml) to tetracycline, doxycycline, and erythromycin but were sensitive to ofloxacin (15). Resistance was demonstrated only when a relatively large inoculum was used, and <1% of a population of organisms showed high-level resistance. Further, the fully resistant phenotypes either died or lost their resistance on serial passage in antibiotic-free medium. Treatment failure was suspected in four of the five patients. In 1997, Lefèvre et al. reported a single C. trachomatis strain that was isolated from a symptomatic patient following tetracycline therapy as having resistance to tetracycline (MIC and MCC, >64 μg/ml) (18). Similar to the report by Jones et al. (15), fewer than 1% of organisms demonstrated high-level resistance. In 2000, Somani et al. reported three patients who were infected with strains of C. trachomatis that exhibited resistance (MCC, >4 μg/ml) to doxycycline, ofloxacin, and azithromycin (27). A heterotypic pattern of resistance, with only a small population of organisms demonstrating resistance, was also noted, similar to the reports of Jones et al. (15) and Lefevre et al. (18). Two of the three patients had clinical evidence of treatment failure, and the third patient was the wife of one of the treatment failures. In the 44 clinical isolates tested in our study, survivability of chlamydial organisms in high concentrations of antimicrobials was seen in all isolates tested and hence did not correlate with their apparent clinical outcome (i.e., success or failure of therapy). It is possible that this phenomenon, i.e., survivability of small numbers of chlamydial organisms in the presence of high levels of antimicrobials, has evolved due to selective pressure from frequent exposure to antimicrobials, or this may be an innate characteristic that is only now being detected due to heightened sensitivity of tissue culture methods.
In conclusion, susceptibility testing for Chlamydia requires careful attention to conditions which may affect both the ability of chlamydial organisms to infect cells in culture and the efficacy of a given antimicrobial through mechanisms such as intracellular uptake. Since MICs and MCCs varied by cell line when tested against azithromycin and erythromycin, one cell line should be consistently used for Chlamydia susceptibility testing. Our study suggests that using McCoy cells for susceptibility testing for C. trachomatis and HEp-2 cells for C. pneumoniae testing offers the most reliable and consistent results. Since a common endpoint is needed for defining the MIC, we suggest defining the MIC as the concentration of antimicrobial that is one twofold concentration more than the MICTP. Survivability of chlamydial organisms at antimicrobial concentrations well above the MIC, heterotypic survival, appears to be a common phenomenon, but studies with larger numbers of patients with treatment failures or same-serovar recurrences are needed to meaningfully assess the correlation of heterotypic survival with clinical outcome. To our knowledge, homotypic resistance, as demonstrated by the tetracycline-resistant C. suis strain, has not been detected in any human Chlamydia species.
Acknowledgments
This work was supported by grant AI-48769 from the National Institutes of Health and by a grant from ActivBiotics.
REFERENCES
- 1.Andersen, A. A., and K. G. Rogers. 1998. Resistance to tetracycline and sulfadiazine in swine C. trachomatis isolates, p. 313-316. In R. S. Stephens, G. I. Byrne, G. Christiansen, I. N. Clarke, J. T. Grayston, R. G. Rank, G. L. Ridgeway, P. Saikku, J. Schachter, and W. E. Stamm (ed.), Chlamydial infections. Proceedings of the 9th International Symposium on Human Chlamydial Infection, Napa, Calif. Berkeley University Press, Berkeley, Calif.
- 2.Barnes, R. C., R. E. Roddy, and W. E. Stamm. 1986. Serovars of Chlamydia trachomatis causing repeated genital infection, p. 503-507. In D. Oriel, G. Ridgway, J. Schachter, D. Taylor-Robinson, and M. Ward (ed.), Chlamydial infections. Cambridge University Press, Cambridge, United Kingdom.
- 3.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1993. Morphologic and antigenic characterization of interferon γ-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. USA 90:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blythe, M. J., B. P. Katz, B. E. Batteiger, J. A. Ganser, and R. B. Jones. 1992. Recurrent genitourinary chlamydial infections in sexually active female adolescents. J. Pediatr. 121:487-493. [DOI] [PubMed] [Google Scholar]
- 5.Catalan, F., A. Milovanovic, C. Prouteau, and M. Soulignac. 1998. Evaluation of in vitro activity of ofloxacin against 73 strains of Chlamydia trachomatis isolated form gynecologic infections. Pathol. Biol. (Paris) 46:144-146. [PubMed] [Google Scholar]
- 6.Cates, W. 1999. Estimates of the incidence and prevalence of sexually transmitted diseases in the United States. Sex. Transm. Dis. 26(Suppl.):S2-S7. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1995. Chlamydia trachomatis genital infections-United States. Morb. Mortal. Wkly. Rep. 46:193-198. [PubMed] [Google Scholar]
- 8.Cross, N. A., D. J. Kellock, G. R. Kinghorn, M. Taraktchoglou, E. Bataki, K. M. Oxley, P. M. Hawkey, and A. Eley. 1999. Antimicrobial susceptibility testing of Chlamydia trachomatis using a reverse transcriptase PCR-based method. Antimicrob. Agents Chemother. 43:2311-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean, D., R. J. Suchland, and W. E. Stamm. 2000. Evidence of long-term cervical persistence of C. trachomatis by omp1 genotyping. J. Infect. Dis. 182:909-916. [DOI] [PubMed] [Google Scholar]
- 10.Dessus-Babus, S., C. M. Bébéar, A. Charron, C. Bébéar, and B. De Barbeyrac. 1998. Sequencing of gyrase and topoisomerase IV quinolone-resistance-determining regions of Chlamydia trachomatis and characterization of quinolone-resistant mutants obtained in vitro. Antimicrob. Agents Chemother. 42:2474-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donati, M., F. M. Rodriguez, A. Olmo, L. D'Apote, and R. Cevenini. 1999. Comparative in-vitro activity of moxifloxacin, minocycline, and azithromycin against Chlamydia spp. J. Antimicrob. Chemother. 43:825-827. [DOI] [PubMed] [Google Scholar]
- 12.Dreses-Werringloer, U., I. Padubrin, H. Zeidler, and L. Köhler. 2000. Effects of azithromycin and rifampin on Chlamydia trachomatis infection in vitro. Antimicrob. Agents Chemother. 45:3001-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gladue, R. P., and M. E. Snider. 1990. Intracellular accumulation of azithromycin by cultured human fibroblasts. Antimicrob. Agents Chemother. 34:1056-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Institute of Medicine. 1997. The hidden epidemic: confronting sexually transmitted diseases. National Academy Press, Washington, D.C.
- 15.Jones, R. B., B. Vander Der Pol, D. H. Martin, and M. K. Shepard. 1990. Partial characterization of Chlamydia trachomatis isolates resistant to multiple antibiotics. J. Infect. Dis. 162:1309-1315. [DOI] [PubMed] [Google Scholar]
- 16.Kagnoff, M. F., and L. Eckmann. 1997. Epithelial cells as sensors for microbial infection. J. Clin. Investig. 100:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keshishyan, K., L. Hanna, and E. Jawefz. 1973. Emergence of rifampin resistance in Chlamydia trachomatis. Nature 244:173-174. [DOI] [PubMed] [Google Scholar]
- 18.Lefèvre, J. C., J. P. Lepargneur, D. Guion, and S. Bei. 1997. Tetracycline-resistant Chlamydia trachomatis in Toulouse, France. Pathol. Biol. 45:376-378. [PubMed] [Google Scholar]
- 19.Lenart, J., A. A. Andersen, and D. D. Rockey. 2000. Growth and development of tetracycline-resistant Chlamydia suis. Antimicrob. Agents Chemother. 45:2198-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrisey, C., I. H. Salman, S. Bakker, D. Farrell, C. M. Bebear, and G. Ridgway. 2002. Serial passage of chlamydia spp. in subinhibitory fluoroquinolone concentrations. J. Antimicrob. Chemother. 49:757-761. [DOI] [PubMed] [Google Scholar]
- 21.Notomi, T., Y. Ikeda, and A. Nagayama. 1999. Minimum inhibitory and minimal lethal concentration against Chlamydia trachomatis dependent on the time of addition and duration of the presence of antibiotics. Chemotherapy 45:242-248. [DOI] [PubMed] [Google Scholar]
- 22.Parks, K. S., P. B. Dixon, C. M. Richey, and E. W. Hook III. 1997. Spontaneous clearance of Chlamydia trachomatis infection in untreated patients. 24:229-235. [DOI] [PubMed]
- 23.Pedersen, L. N., H. O. Kjær, J. K. Møller, T. F. Ørntoft, and L. Ostergaard. 2000. High-resolution genotyping of Chlamydia trachomatis from recurrent urogenital infections. J. Clin. Microbiol. 38:3068-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen, S. J., P. Timms, P. R. Beatty, and R. S. Stephens. 1996. Cytotoxic-T-lymphocyte-mediated cytolysis of L cells persistently infected with Chlamydia spp. Infect. Immun. 64:1944-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridgway, G. L., J. M. Owen, and J. D. Oriel. 1976. A method for testing the antibiotic susceptibility of Chlamydia trachomatis in a cell culture system. J. Antimicrob. Chemother. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 26.Rota, T. R. 1980. Techniques for culturing and determining antimicrobial susceptibility of Chlamydia trachomatis. Arch. Androl. 4:63-69. [DOI] [PubMed] [Google Scholar]
- 27.Somani, J., V. B. Bhullar, K. A. Workowski, C. E. Farshy, and C. M. Black. 2000. Multiple drug-resistant Chlamydia trachomatis associated with clinical treatment failure. J. Infect. Dis. 181:1421-1427. [DOI] [PubMed] [Google Scholar]
- 28.Stamm, W. E. 2000. Potential for antimicrobial resistance in Chlamydia pneumoniae. J. Infect. Dis. 181(Suppl.):S456-S459. [DOI] [PubMed] [Google Scholar]
- 29.Wyrick, P. B., C. H. Davis, S. T. Knight, and J. Choong. 1993. In-vitro activity of azithromycin on Chlamydia trachomatis infected, polarized human endometrial epithelial cells. J. Antimicrob. Chemother. 31:139-150. [DOI] [PubMed] [Google Scholar]
- 30.Wyrick, P. B., C. H. Davis, J. E. Raulston, S. T. Knight, and J. Choong. 1994. Effect of clinically relevant culture conditions on antimicrobial susceptibility of Chlamydia trachomatis. Clin. Infect Dis. 19:931-936. [DOI] [PubMed] [Google Scholar]