Abstract
We previously reported a new marine bacterium, Pseudoalteromonas phenolica sp. nov. O-BC30T, which produced a bactericidal antibiotic against methicillin-resistant Staphylococcus aureus (MRSA). In the present study, we purified an anti-MRSA substance (MC21-A) from the methanol extract of the cells of P. phenolica O-BC30T and analyzed its chemical structure. MC21-A was determined to be 3,3′,5,5′-tetrabromo-2,2′-biphenyldiol by spectrometric analyses. Its anti-MRSA activity against 10 clinical isolates of MRSA was comparable to that of vancomycin (MC21-A MICs, 1 to 2 μg/ml; vancomycin MICs, <0.25 to 2 μg/ml). This substance was also high active against Enterococcus serolicida, Enterococcus faecium, and Enterococcus faecalis but was less active against Streptococcus spp. A time-kill study also demonstrated that MC21-A was bactericidal and that its killing rate was much higher than that of vancomycin. The postantibiotic effect (PAE) of MC21-A against a clinical MRSA isolate, strain E 31243, was also comparable to that of vancomycin (MC21-A PAEs, 1.46 to 1.65 h; vancomycin PAEs, 0.84 to 1.43 h). However, a lysis experiment demonstrated that this substance failed to lyse MRSA cells. This substance also did not lyse human erythrocytes. A SYTOX Green staining experiment implied that this substance permeabilized the cell membrane of MRSA as its mode of action. When its activities against a hypersensitive Escherichia coli mutant (KO 1489) and wild-type strains were tested, MC21-A exhibited higher levels of activity against the former. Furthermore, MC21-A was not cytotoxic to human normal fibroblast, rat pheochromocytoma, and Vero cells at concentrations up to 50 μg/ml. These results suggest that MC21-A might be useful as a lead compound in the development of new types of anti-MRSA substances with modes of action different from those of vancomycin and teicoplanin.
Methicillin-resistant Staphylococcus aureus (MRSA) is the most problematic gram-positive bacterium in public health not only because it is highly prevalent but also because it has become resistant to almost all available antibiotics except vancomycin and teicoplanin (44). Recently, its susceptibility to vancomycin has decreased, and vancomycin-intermediate and vancomycin-resistant S. aureus have increasingly been found (16, 21, 35) in several countries. Furthermore, a decrease in the susceptibility of MRSA to teicoplanin has also been reported in several hospitals around the world (28, 43). The evidence of MRSA resistance to vancomycin and teicoplanin, which are antibiotics of last resort, makes the need for alternative antibiotics and chemotherapeutics after vancomycin and teicoplanin treatments have failed particularly urgent.
Although the chemical compounds of marine microorganisms are less well known than those of their terrestrial counterparts, in the last decade several bioactive substances have been isolated from marine bacteria and are new resources for the development of medically useful compounds. Antibiotics from marine microorganisms have been reported, including loloatins from Bacillus (14), agrochelin and sesbanimides from agrobacterium (1, 2), pelagiomicins from Pelagiobacter variabilis (23), δ-indomycinone from a Streptomyces sp. (6), and dihydrophencomycin methyl ester from Streptomyces (36). In particular, some species of the genus Pseudoalteromonas (formerly Alteromonas) (11) produce both antibiotics and several bioactive substances (12, 13, 18, 25, 29, 38, 39, 45). For example, Pseudoalteromonas rubra (12) and Pseudoalteromonas aurantia (13) have been reported to be antibiotic-producing bacteria. The several biologically active substances, antibacterial and algicidal toxins, as well as extracellular enzymes, produced by Pseudoalteromonas spp. have been reviewed by Holmström and Kjelleberg (22).
With this background, we conducted a screening program for anti-MRSA substance-producing marine bacteria; one of the isolates, strain O-BC30T, showed high levels of anti-MRSA activity on a coculture plate with MRSA. Phenotypic characterization, 16S rRNA gene sequence analysis, and DNA-DNA hybridization suggested that strain O-BC30T is a new bacterial species in the genus of Pseudoalteromonas (24); Pseudoalteromonas phenolica sp. nov. is its proposed name. In this study we purified the anti-MRSA substance produced by strain O-BC30T, determined its chemical structure, and evaluated its antibacterial and bactericidal activities, especially against clinical isolates of MRSA, compared to those of vancomycin.
This paper describes the purification, chemical structure elucidation, and in vitro antibacterial activity of the newly discovered anti-MRSA substance, MC21-A, as well as our investigation of its mechanism of action against MRSA.
MATERIALS AND METHODS
Bacterial strains and media.
The following strains were used to evaluate the antibacterial activities of MC21-A: 10 clinical isolates of MRSA (strains E 31224, E 31237, E 31243, E 31256, E 31271, E 31280, and E 31283, kindly provided by S. Araki, Eisai Co. Ltd., Tokyo, Japan), strain 7B29 (kindly provided by T. Someya, Saga University, Saga, Japan), and strains GIFU 12361 and GIFU 12364 (kindly provided by H. Yamamoto, Gifu University, Gifu, Japan); a reference strain of MRSA (ATCC 33591); two strains of methicillin-sensitive S. aureus (MSSA; IFO 15035 and ATCC 25923); one strain of Bacillus subtillis (IFO 14419); three strains of Enterococcus serolicida (E 9053, E 9568, and NG 8206, kindly provided by S. Araki, Eisai Co. Ltd.); three strains of E. faecium (NBRC 3128, NBRC 3535, and NBRC 3826); three strains of E. faecalis (NBRC 3971, NBRC 3989, and NBRC 12964); and Streptococcus mutans (NBRC 13955), Streptococcus pneumoniae (GTC 261), and Streptococcus pyogenes (GTC 262) (kindly provided by the Gifu Type Culture Collection, Department of Microbiology, Gifu University). Vibrio alginolyticus V-7 (kindly provided by T. Takeda, Hokkaido Kushiro Fisheries Experimental Station, Hokkaido, Japan), Pseudomonas aeruginosa IFO 13736, hyperpermeable Escherichia coli mutant strains JARV15 and B1LK0 (kind gifts of T. Palmer, John Innes Centre, Norwich Research Park, Norwich, United Kingdom), and E. coli KO 1489 (kind gift of A. Wright, Tufts Medical School, Boston, Mass.) were also used to evaluate the antibacterial activity of MC21-A. Bacterial strains IFO 15035, IFO 14419, and IFO 13736 were purchased from the Institute for Fermentation Osaka (IFO), Osaka, Japan. Bacterial strains with an NBRC designation were purchased from the NITE Biological Resource Center, Chiba, Japan. All of the bacterial strains except V. alginolyticus and P. phenolica O-BC30T were stocked in Trypticase soy broth (TSB) medium (Difco Laboratories, Detroit, Mich.) containing 20% (vol/vol) glycerol at −80°C; V. alginolyticus and P. phenolica O-BC30T (IAM 14989T, KCTC 12086T) were stocked in ZoBell 2216E broth medium containing polypeptone (5 g/liter; Katayama Chemical, Osaka, Japan), yeast extract (1 g/liter; Nihon Seiyaku, Osaka, Japan), and 75% artificial seawater (pH 7.5; Jamarin Laboratory, Osaka, Japan) containing the same concentration of glycerol at −80°C. The bacteria were slanted on the respective media before being used in the experiments. For MRSA in particular, the Trypticase soy agar (TSA) medium was supplemented with 6 μg of oxacillin (Sigma Chemical Co., St. Louis, Mo.) per ml. The agar medium was supplemented with 3% defibrinated blood sheep (Cedarlane Laboratories Ltd., Hornby, Ontario, Canada) for Streptococcus spp.
Antibiotics and reagents.
Vancomycin and amoxicillin were purchased from Sigma Chemical Co. Chloramphenicol and amphotericin B were purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan). Gramicidin D and SYTOX Green nucleic acid stain were purchased from ICN Biomedical Inc. (Aurora, Ohio) and Molecular Probes (Eugene, Oreg.), respectively.
Fermentation and isolation of MC21-A.
A seed culture was prepared by inoculation of 80 ml of ZoBell 2216E broth medium in a 100-ml Erlenmeyer flask and incubation at 25°C for 24 h with stirring. The seed culture (10 ml) was inoculated onto ZoBell 2216E agar medium on plates 29 cm in diameter, and the plates were incubated at 25°C for 5 days, since the highest anti-MRSA activity of the bacterial crude extract was reached after 5 days of culture (data not shown). The culture was extracted with methanol (MeOH) from the agar surface, and the extracts were centrifuged at 3,000 × g for 15 min. The supernatant of the MeOH extract was partitioned with CHCl3 and water, and the CHCl3 layer was concentrated to dryness.
Purification of MC21-A.
MC21-A was purified by silica gel 60 (Merck, Darmstadt, Germany) and cosmosil (Nacalai Tesque Inc., Kyoto, Japan) column chromatographies and high-pressure liquid chromatography (HPLC) on a reversed-phase column (4.6 mm [diameter] by 250 mm; Mightysil RP-8 GP; Kanto Chemical Co., Inc., Tokyo, Japan). All of the solvents used in this purification process were purchased from Sigma-Aldrich Japan (Tokyo, Japan). The active CHCl3 extract (780 mg) was chromatographed on a silica gel column, with elution with n-hexane-ethyl acetate (8/1) and n-hexane-ethyl acetate-ethanol (8/1/0.5). The active fractions found in the n-hexane-ethyl acetate (8/1) eluate were further subjected to cosmosil column chromatography, with elution with 60 to 80% MeOH. To purify the anti-MRSA substance, the active fraction was finally subjected to HPLC with a preparative Mightysil RP-8 GP column, with the gradients eluted with acetonitrile and water containing 0.1% trifluoroacetic acid (Sigma-Aldrich Japan).
Spectrometric analyses of MC21-A.
The spectrogram obtained by electron impact (EI)-mass spectrometry was recorded with an EI-MS spectrometer (JMS-DX303; Jeol). The 13C and 1H nuclear magnetic resonance (NMR), one-dimensional nuclear overhouser effect, 1H decoupling, and heteronuclear multiple bond coherence spectra were recorded in CDCl3 with a Unity Plus 500 Varian NMR spectrometer at 500.2 and 125.8 MHz for 13C and 1H NMR, respectively. The UV-visible spectrum was determined in MeOH with a UV-visible spectrophotometer (V-550; Jasco). The infrared (IR) spectrum was recorded in KBr with an IR spectrometer (FT/IR-610; Jasco).
Test for antibacterial activity.
Comparison of the MICs of MC21-A and vancomycin was conducted by the standard microdilution method described by the National Committee for Clinical Laboratory Standards (32) with ZoBell 2216E broth medium for V. alginolyticus and cation-adjusted Mueller-Hinton broth (CAMHB) medium (Difco Laboratories) for all other organisms. For Streptococcus spp., CAMHB was supplemented with lysed horse blood (Cedarlane Laboratories Ltd.). The final volume of CAMHB or ZoBell 2216E broth medium containing MC21-A or vancomycin was 100 μl per well, to give a starting inoculum density of 5 × 105 cells/ml.
Time-kill experiment.
The time-kill experiment was conducted by the method described by Aeschlimann and Rybak (4) and Entenza et al. (10). The experiments were conducted in 50-ml Erlenmeyer flasks containing 25 ml of fresh TSB medium (Difco Laboratories) inoculated with an overnight culture of a clinical isolate of MRSA (E 31243) or a reference strain of MRSA (ATCC 33591) to give an initial bacterial density of 106 cells/ml. The inoculation was carried out immediately after addition of MC21-A or vancomycin at final concentrations that consisted of the MIC and two, four, and eight times the MIC. The flasks were further incubated at 37°C with stirring with a magnetic stirrer at 200 rpm. The viable cell counts of MRSA were estimated at various incubation times by the plating method. To minimize the effect of antibiotic carryover, the samples were centrifuged at 1,600 × g for 15 min. Then, the medium was replaced with fresh TSB medium, serially diluted 10-fold, and plated on TSA medium. The plates were incubated at 37°C for 24 h, and then the colonies were counted.
PAE.
Postantibiotic effects (PAEs) were determined by the method described by Aeschlimann et al. (3) and Craig and Gudmundsson (7). MC21-A or vancomycin was added to the test tube containing TSB medium to give final concentrations that consisted of the MIC, two times the MIC, and four times the MIC; and the test tubes were inoculated with an overnight culture of MRSA at 106 cells/ml. Test tubes without antibiotics were used as controls. The test tubes were incubated at 37°C for 1 h and then centrifuged at 1,600 × g for 15 min. The medium was replaced with fresh TSB medium, diluted to 1:1,000 in Erlenmeyer flasks, and incubated at 37°C with stirring. Samples were taken at several time intervals; the numbers of viable cells were counted on TSA medium. The PAE was calculated by the following equation: PAE = T − C, where T represents the time required for the bacterial cell counts in the test cultures to increase 1 log10 CFU/ml above the bacterial count observed immediately after drug removal, and C represents the time required for the bacterial cell count in the untreated control culture to increase 1 log10 CFU/ml. The values of T and C were determined either by linear regression (if R was ≥0.95) or by visual inspection of the regrowth curve. Each PAE experiment was performed with seven replications to ensure reproducibility.
Bacteriolytic assay.
The seed cultures of MRSA (E 31243 and ATCC 33591) in TSB medium were washed twice with sterile 0.9% NaCl, and the absorbance was adjusted to 0.1 at 660 nm. The bacterial cell suspension was divided into aliquots of 5 ml each, placed into sterile test tubes, and exposed to MC21-A at various concentrations or to lysostaphin (Wako Pure Chemical Industries Ltd.) at 1 μg/ml as the positive control. Untreated bacterial suspensions were used as the negative control. The concentration of MeOH (the solvent for MC21-A) in each tube was less than 0.1% (vol/vol). These test tubes were incubated at 37°C by shaking at 120 rpm. The absorbance at 660 nm was measured 0, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, 6, 8, and 12 h after incubation; and the relative absorbance was calculated by dividing each absorbance by that for the negative control. Each treatment was conducted in triplicate.
Bacterial cell membrane permeabilization test.
The permeabilization of the MRSA cell membrane induced by MC21-A was compared to that induced by antibiotics with different modes of action. The experiment conducted was based on a previously described method (30, 37) and was done in a sterile 0.9% NaCl solution containing 5% TSB. This medium was filtered through a 0.2-μm-pore-size filter before autoclaving of the medium. The MICs of chloramphenicol and amoxicillin for MRSA (ATCC 33591) were determined by the standard method described above. Bacterial cells (108 cells/ml), antibiotic, and SYTOX Green (5 μM) were combined and dispensed into a black microplate (Costar, Cambridge, Mass.) at 100 μl/well. After various periods of incubation at 37°C, the fluorescence intensities were measured with a fluorescence multiwell plate reader (Cytoflour II; PerSeptive Biosystems, Foster City, Calif.) at an excitation wavelength of 485 nm and an emission wavelength of 530 nm.
Fluorescence microscopy.
The bacterial cells stained with SYTOX Green were observed under a fluorescence microscope (BX50; Olympus, Tokyo, Japan) equipped with 100-W mercury arc lamp. A green fluorescence filter was used to determine the fluorescence of bacterial cells stained with SYTOX Green.
Erythrocyte hemolysis.
The hemolytic activity of MC21-A compared to those of known membrane-active substances, gramicidin D and amphotericin B, were determined by spectrometric analysis as described previously (20, 40) with human erythrocytes. Blood from a male human was drawn with heparin as the anticoagulant. The erythrocytes were centrifuged (1,000 × g for 5 min), washed three times with 10 volumes of 10 mM Tris-HCl buffer (pH 7.4) containing 0.9% NaCl, and resuspended to 10% in the same buffer. The erythrocytes were treated with MC21-A, gramicidin D, or amphotericin B at final concentrations ranging from 3.1 to 50 μg/ml and incubated at 37°C for 2 h. Triton X-100 (0.1%) was used to obtain a positive control hemolysis value, and 1% dimethyl sulfoxide (DMSO) or MeOH was used as a negative control, as MC21-A was dissolved in MeOH and the other two substances were dissolved in DMSO. The final concentration of DMSO or MeOH in each test sample was 1%. After incubation, the test samples were centrifuged at 1,000 × g for 5 min. The amount of hemoglobin released from the cells was determined by measuring the absorbance of the supernatants at 540 nm (V-550 UV-visible spectrophotometer; Jasco) after dilution 10-fold with 10 mM Tris-HCl buffer.
Cytotoxicity test.
The cytotoxic activity of MC21-A was evaluated by a previously described method (19). Human normal dermal fibroblasts (HDFs; Morinaga Institute of Biological Science, Yokohama, Japan), human leukemic cells (MOLT-4 cells; JCRB9031; Japanese Cancer Research Resources Bank), Madin-Darby canine kidney (MDCK) cells, African green monkey kidney cells (Vero cells; ATCC CCL 81), and rat pheochromocytoma cells (PC12D cells; kind gift of M. Sano, Aichi Colony Developmental Disorder Research Center, Kasugai, Aichi, Japan) were used to evaluate the cytotoxicity of MC21-A. HDF, MOLT-4, and Vero cells were cultured and maintained in enhanced RPMI-DMEM-F12 medium (Kyokuto Pharmaceutical Industrial Inc., Tokyo, Japan) containing 10% fetal bovine serum (Sigma Chemical Co.) at 37°C for 3 days in a humidified 5% CO2 incubator (19). PC12D and MDCK cells were cultured and maintained in Dulbecco's modified Eagle's medium (Gibco BRL, Rockville, Md.) as described previously (33, 42). After incubation, the medium was replaced with the respective fresh medium. Then, the cells were harvested and seeded into a 96-well plate at 5 × 103 cells/100 μl/well. Then, serially twofold diluted MC21-A was added to give final concentrations ranging from 0.4 to 50 μg/ml. The cells were incubated under the same conditions described above. The viability and proliferation of cells were determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. The growth rate relative to that of the control treatment was calculated.
RESULTS
Fermentation, isolation, and purification of MC21-A.
Strain O-BC30T grew well when it was cultured to produce MC21-A on ZoBell 2216E medium and was confluent on day 2. The antibacterial activity was detected in the MeOH extract of the bacterial cells but not in the agar of the plates, indicating that the antibacterial substance(s) might be bound on the cell surface. The remaining bacterial cells in the MeOH extracts were further sonicated to explore the intracellular products for the presence of the antibacterial substance, but antibacterial activity was not noted. Further isolation was performed by partition of the MeOH extract with CHCl3 and water. A total of 6.4 g of bacterial cells was harvested from five large petri dishes (diameter, 25 cm) and dried to give 780 mg of extract. An antibacterial substance, MC21-A (2.1 mg), was purified by silica gel and cosmosil column chromatographies and finally by HPLC on a reversed-phase column.
Chemical structure of MC21-A.
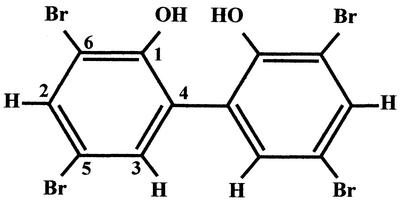
EI-MS indicated that the molecular weight of MC21-A was 501.7, and the fragmentation pattern suggested the presence of bromine (Table 1). The typical chemical shifts of aromatic benzene at 7.68 and 7.35 ppm were noted from the 1H NMR spectrum (Table 2). The presence of benzene and bromine was also confirmed by the IR spectrum, which showed absorptions at 1,455, 1,388, 1,220, 856, and 684 cm−1 (Table 1). High-resolution MS indicated that the molecular formula of MC21-A is C12H6Br4O2. This molecular formula along with the 1H NMR and 13C NMR data implied that MC21-A is a symmetrical aromatic benzene. Further analyses by one-dimensional NOE, 1H-decoupling, and HMBC finally determined that the structure of MC21-A is 3,3′,5,5′-tetrabromo-2,2′-biphenyldiol (Fig. 1).
TABLE 1.
Physicochemical properties of MC21-A
| Property | Result |
|---|---|
| Appearance | White powder |
| Molecular formula | C12H6Br4O2 |
| EI-MS (m/z) | 503.7, 501.7, 343.9, 341.9, 339.9 |
| FDa-MS (m/z) | 504, 502, 500 |
| UV λmax in MeOH (nm) | 211.5, 301 |
| IR νmax (cm−1) | 3,366, 1,455, 1,388, 1,220, 856, 684 |
| Solubility | |
| Soluble in: | MeOH, CHCl3 |
| Insoluble in: | H2O, hexane |
FD, field desorption.
TABLE 2.
1H and 13C NMR data for MC21-Aa
| Position | 13C NMR (ppm) | 1H NMR (ppm) | J value (Hz) |
|---|---|---|---|
| 1 | 148.9 | ||
| 2 | 134.6 | 7.68 d | |
| 3 | 133.5 | 7.35 d | 2.3 |
| 4 | 125.8 | ||
| 5 | 112.9 | ||
| 6 | 112.0 | ||
| 5.82 (OH) |
The 1H and 13C NMR spectra were recorded in CDCl3.
FIG. 1.
Chemical structure of MC21-A.
MIC.
MC21-A showed antibacterial activity comparable to that of vancomycin. The MICs of MC21-A for MSSA, MRSA, E. serolicida, E. faecalis, E. faecium, and Bacillus subtilis were 1, 1 to 2, <0.25 to 1, <0.25 to 1, 0.5 to 1, and 0.25 μg/ml, respectively. The MICs of vancomycin for the same bacterial strains were 1, 0.25 to 2, <0.25 to 0.5, 2, 1 to 2, and <0.25 μg/ml, respectively (Table 3). However, MC21-A was less active against S. pneumoniae, S. pyogenes, and S. mutans, for which MICs were 4, 8, and 16 μg/ml, respectively. This substance was slightly active against wild-type E. coli strains, for which MICs were 64 μg/ml; was moderately active against hyperpermeable E. coli mutant strain KO 1489, for which the MIC was 16 μg/ml; but was not active against P. aeruginosa and V. alginolyticus at up to 64 μg/ml.
TABLE 3.
Comparative antibacterial activities of MC21-A produced by P. phenolica sp. nov. O-BC30T and vancomycin
| Bacterial strain | MIC (μg/ml)
|
|
|---|---|---|
| MC21-A | vancomycin | |
| MSSA ATCC 25923 | 1 | 1 |
| MSSA IFO 15035 | 1 | 1 |
| MRSA ATCC 33591 | 1 | 2 |
| MRSA E 31224 | 1 | 1 |
| MRSA E 31237 | 1 | 1 |
| MRSA E 31243 | 1 | 1 |
| MRSA E 31256 | 1 | 0.5 |
| MRSA E 31271 | 1 | 0.5 |
| MRSA E 31280 | 1 | 1 |
| MRSA E 31283 | 2 | <0.25 |
| MRSA 7B29 | 2 | 1 |
| MRSA GIFU 12361 | 1 | 1 |
| MRSA GIFU 12364 | 2 | 2 |
| B. subtilis IFO 14419 | 0.25 | <0.25 |
| E. serolicida E 9053 | <0.25 | 0.5 |
| E. serolicida E 9568 | 1 | <0.25 |
| E. serolicida NG 8206 | 1 | 0.5 |
| E. faecalis NBRC 3971 | <0.25 | 2 |
| E. faecalis NBRC 12964 | <0.25 | 2 |
| E. faecalis NBRC 3989 | 1 | 2 |
| E. faecium NBRC 3826 | 1 | 2 |
| E. faecium NBRC 3128 | 1 | 2 |
| E. faecium NBRC 3535 | 0.5 | 1 |
| S. pneumoniae GTC 261 | 4 | 0.5 |
| S. pyogenes GTC 262 | 8 | 0.25 |
| S. mutants NBRC 13955 | 16 | 16 |
| E. coli NBRC 12734a | 64 | NDe |
| E. coli NBRC 14360a | 64 | ND |
| E. coli NBRC 13540a | 64 | ND |
| E. coli B1LK0b | 64 | ND |
| E. coli JARV15c | 64 | ND |
| E. coli KO 1489d | 16 | ND |
| P. aeruginosa IFO 13736 | >64 | >64 |
| V. alginolyticus V-7 | >64 | ND |
Bactericidal activity of MC21-A.
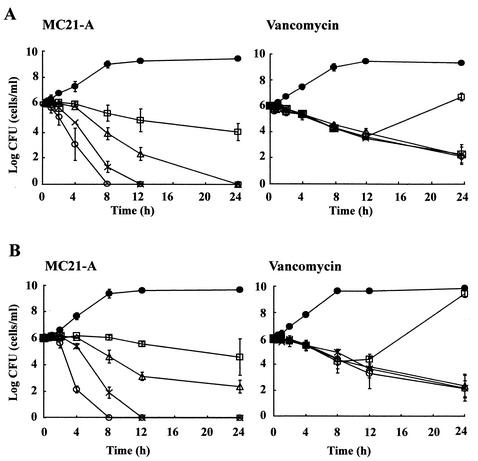
The time-kill study showed that MC21-A was able to decrease the counts of both a clinical isolate and a reference strain of MRSA (Fig. 2). At two times the MIC (2 μg/ml), this substance decreased the numbers of viable bacterial cells of these MRSA strains after 8 h of exposure. A decrease in bacterial cell counts was more readily found when the strains were exposed to MC21-A at higher concentrations. At four times the MIC (4 μg/ml) and eight times the MIC (8 μg/ml), the viable cell counts decreased drastically after 4 and 2 h of exposure, respectively. This substance effectively killed both a clinical isolate and a reference strain of MRSA after 12 h of exposure at 4 μg/ml and after 8 h of exposure at 8 μg/ml. There were no differences in the time-kill patterns and the killing rates between these two MRSA strains exposed to MC21-A.
FIG. 2.
Comparative bactericidal activities of MC21-A and vancomycin against a reference strain of MRSA ATCC 33591 (A) and a clinical isolate of MRSA, E 31243 (B). •, growth control; □, MIC; ▵, two times the MIC; ×, four times the MIC; ○, eight times the MIC. The values with standard error bars are mean values from duplicate experiments.
Vancomycin at two, four, and eight times the MIC gradually decreased the bacterial cell counts. However, MC21-A decreased the bacterial cells counts more significantly than vancomycin at the same concentrations and after the same lengths of exposure, indicating that the killing rates of MC21-A for MRSA were much higher than those of vancomycin. Unlike MC21-A, vancomycin failed to kill MRSA completely even at eight times the MIC after 24 h of exposure. The regrowth of the bacterium was observed at the MIC of vancomycin but not at the MIC of MC21-A.
PAE.
The PAE of MC21-A at the MIC was significantly longer than that of vancomycin at the MIC. However, the PAEs were relatively similar at two and four times the MIC of each compound (Table 4). As the concentrations of MC21-A increased from the MIC to four times the MIC, the PAEs showed only moderate increases. The PAEs of vancomycin increased more significantly with increases in the concentration.
TABLE 4.
PAEs of MC21-A and vancomycin against clinical isolate MRSA E 31243
| Concn (multiple of MIC) | PAE (h)a
|
|
|---|---|---|
| MC21-A | Vancomycin | |
| 1 | 1.46 ± 0.31 | 0.84 ± 0.17 |
| 2 | 1.54 ± 0.25 | 1.33 ± 0.21 |
| 4 | 1.65 ± 0.35 | 1.43 ± 0.31 |
PAE values are mean values from seven replications of independent experiments.
Bacteriolytic activity.
Reduction of the absorbance of the MRSA cell suspensions was not observed in the presence of MC21-A at up to four times the MIC (4 μg/ml) until the end of the incubation period (Fig. 3). In contrast, the absorbance of MRSA cell suspensions treated with 1 μg of lysostaphin per ml were reduced drastically early in the incubation period. These results indicate that MC21-A does not lyse MRSA cells.
FIG. 3.
Bacteriolytic activities of MC21-A against a reference strain of MRSA (ATCC 33591) and a clinical isolate of MRSA (E 31243). •, positive control (1 μg of lysostaphin per ml); ○, the MIC; ▵, two times the MIC; □, four times the MIC. Relative absorbance was calculated by dividing the absorbance for the treated tube by that for the negative control tube. The values with standard error bars are mean values from triplicate experiments.
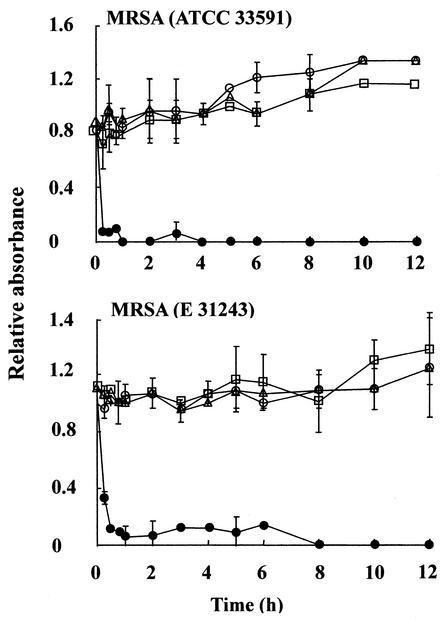
Bacterial cell membrane permeabilization.
SYTOX Green staining indicated that MC21-A rapidly permeabilized the cell membranes of MRSA. The fluorescence intensities of SYTOX Green in the bacterial cells treated with MC21-A at the MIC and four times the MIC increased drastically during a 30-min incubation period (Fig. 4). However, no increase in fluorescence intensity was observed in cells treated with MC21-A at 1/4 or 1/16 the MIC, even after 4 h of incubation. Treatment of the bacterial cells with amoxicillin at the MIC and four times the MIC also increased the fluorescence intensities, although not as much as the treatments with MC21-A at comparable concentrations did. The other two antibiotics tested, chloramphenicol and vancomycin, did not permeabilize the bacterial cell membranes, as indicated by the low SYTOX Green fluorescence intensity after 4 h of incubation.
FIG. 4.
Cells membrane permeabilization of MRSA induced by MC21-A (A), chloramphenicol (B), amoxicillin (C), and vancomycin (D). ▵, untreated control; □, 1/16 the MIC; ○, 1/4 the MIC; ▪, the MIC; •, 4 times the MIC. The MICs of chloramphenicol and amoxicillin were each 64 μg/ml. The fluorescence values with standard error bars are the corrected mean values from quadruplet measurements. The corrected fluorescence values are each fluorescence value subtracted from the background value.
Fluorescence microscopy.
A bright green fluorescence of MRSA bacterial cells stained with SYTOX Green in the presence of MC21-A was observed under a fluorescence microscope. In contrast, such fluorescence intensity was not observed when the bacterial cells were stained with SYTOX Green in the absence of MC21-A.
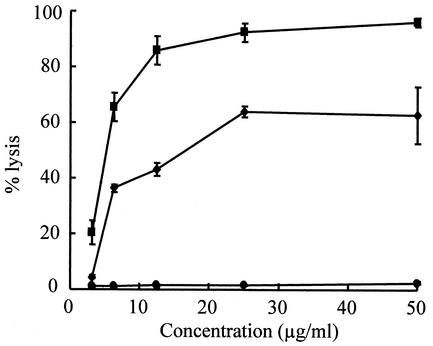
Hemolytic activity.
MC21-A did not lyse human erythrocytes (Fig. 5). In contrast, the other two membrane-active antibiotics, amphotericin B and gramicidin D, readily lysed the erythrocytes at low concentrations. The same results were obtained when mouse erythrocytes were used in this experiment (data not shown).
FIG. 5.
Hemolytic activity of MC21-A (•) compared to those of amphotericin B (▪) and gramicidin D (♦) against human red blood cells. The values are represented as the percentage of total lysis compared to the lysis caused by 0.1% Triton X-100. Each value with a standard error bar is the mean value from triplicate experiments.
Cytotoxicity.
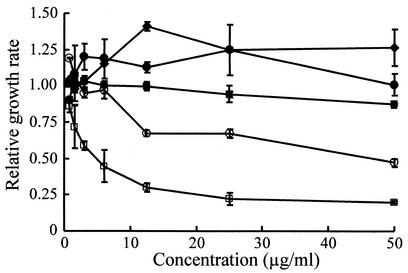
MC21-A was not cytotoxic to HDF cells at up to 50 μg/ml (Fig. 6). The same results were observed against a rat adrenal medullary pheochromocytoma cells (PC12D cells) and African green monkey kidney cells (Vero cells). This substance exhibited moderately cytotoxic activity against human leukemic cells (MOLT-4 cells) but significant cytotoxic activity against MDCK cells.
FIG. 6.
Cytotoxicity of MC21-A to HDFs (♦), human leukemic cells (MOLT-4 cells) (○), MDCK cells (□), African green monkey kidney cells (Vero cells) (•), and rat pheochromocytoma cells (PC12D cells) (▪). The relative growth rates with standard error bars are mean values from triplicate experiments.
DISCUSSION
This paper describes the fermentation, purification, and elucidation of the chemical structure of an anti-MRSA substance (MC21-A) produced by a novel marine bacterium, P. phenolica sp. nov. O-BC30T, as well as its in vitro antibacterial activity and mechanism of action. P. phenolica O-BC30T is a new marine bacterial species in the genus Pseudoalteromonas that produces phenolic anti-MRSA substances (24). The main product of this strain is an anti-MRSA substance, designated MC21-A, derived from the chloroform extract of the bacterial cells. In this study, MC21-A was determined to be 3,3′,5,5′-tetrabromo-2,2′-biphenyldiol. This substance is a natural newly discovered substance, and this is the first report of a marine bacterium which produces this antibiotic. To the best of our knowledge, the anti-MRSA activity and the mechanism of action of 3,3′,5,5′-tetrabromo-2,2′-biphenyldiol have not yet been reported, although polybrominated compounds have recently been isolated from several marine organisms. Polybrominated biphenyl ethers from an Indonesian sponge, Dysidea herbacea, have been shown to have activities against B. subtilis but not against gram-negative bacteria. However, their anti-MRSA activities have not been reported (17).
MC21-A demonstrates activity specifically against gram-positive bacteria, but it is less active against Streptococcus spp. This substance is not active against two gram-negative bacteria, P. aeruginosa and V. alginolyticus. In this way it differs from the antibiotics produced by Pseudoalteromonas luteoviolacea, the Pseudoalteromonas species most closely related to P. phenolica sp. nov. O-BC30T. P. luteoviolacea produces pentabromopseudilin and violacein, which are active against both gram-positive and gram-negative bacteria (18, 29).
The time-kill experiment indicated that MC21-A is rapidly bactericidal against both a reference strain and a clinical isolate of MRSA (Fig. 2). The killing rate of MC21-A is much higher than that of vancomycin. MC21-A exhibits concentration-dependent bactericidal activity. Its killing rate was significantly higher with an increase in the concentration. At four and eight times the MIC, MC21-A was able to kill MRSA completely after 8 and 12 h of incubation. A slower killing rate was observed at lower concentrations. MC21-A at 2 μg/ml was able to kill MRSA ATCC 33591 completely, while vancomycin acted slowly even at eight times the MIC. The slow killing rate of vancomycin against MRSA and MSSA has been reported also by Löwdin et al. (27). The time-kill pattern of MC21-A against MRSA is similar to that of phenethylguanidine (RWJ-49815), an antibacterial substance that inhibits the two-component signal transduction system of bacteria (5). This agent kills MRSA rapidly, and a further investigation has demonstrated that it also effectively permeabilizes the S. aureus cell membrane after a short exposure (20). Speculating that a similar mechanism may be at work for MC21-A, we decided to investigate MC21-A-induced membrane permeabilization of MRSA by the SYTOX Green straining method (26, 34, 37). Our results indicated that MC21-A at the MIC rapidly permeabilizes bacterial cell membranes. These results could be confirmed by observation by fluorescence microscopy, which demonstrated a green fluorescence of a high intensity for bacterial cells treated with MC21-A. This observation revealed that SYTOX Green penetrates the cells and binds to the nucleic acid.
The cell membrane permeabilization activity of MC21-A is more intense and rapid than that of amoxicillin, an antibiotic known to damage membranes. Amoxicillin at four times the MIC permeabilized the bacterial cell membranes after 30 min of incubation. Our result agreed with those from the study of Novo et al. (34), in which amoxicillin permeabilizes the S. aureus cell membrane within 45 min of incubation. Chloramphenicol does not permeabilize the MRSA cell membrane even at four times the MIC because it binds to the 50S subunit of the ribosome and inhibits interaction between the aminocyl-tRNA and the ribosome. De novo protein synthesis in S. aureus is not necessary for the maintenance of membrane permeability (15, 34). A similar result is also obtained when the bacterial cells are treated with vancomycin for a short period because it inhibits the synthesis of peptidoglycan in bacterial cell walls. It does not alter the permeabilities of bacterial cell membranes.
When the activity of MC21-A was evaluated against three strains of wild type E. coli and three strains of hypersensitive E. coli mutant, the substance exhibited fourfold higher levels of activity against strain KO 1489, a sodium dodecyl sulfate-sensitive E. coli mutant (8, 9). However, its activities against two E. coli mutants (B1LK0 and JARV15) and wild-type E. coli were identical, although strains B1LK0 and JARV15 have compromised outer membrane permeability barriers and, specifically, defects in the lipopolysaccharide component of the outer membrane. Strain B1LK0 is sensitive to erythromycin, rifampin, and ampicillin at relatively high concentrations: 300, 800, and 25 μg/disk, respectively (41).
The PAE of MC21-A is significantly longer than that of vancomycin at the MIC but is relatively no different from that of vancomycin at two and four times the MIC. The fast regrowth of MRSA after treatment with vancomycin at the MIC might be due to the presence of a concentration that is inadequate to inhibit the synthesis of new peptidoglycan; hence, higher concentrations of vancomycin are required to improve its antibacterial effect. The PAE of vancomycin reflects the length of time that the amount of peptidoglycan needed for bacterial growth can be kept at a critical level (27).
Although MC21-A permeabilizes the MRSA cell membrane, bacteriolytic activity was not observed against the same bacterial strain, as reflected by the constant density of bacterial cells treated with various concentrations of MC21-A during the incubation period (Fig. 3). This phenomenon indicates that MC21-A does not give rise to the disintegration of cells or cell organelles by rupture of the outer membranes but alters the permeability of the cell membrane as its mode of action. However, bacterial cell membrane permeabilization by MC21-A without lysis of the cells indicates that the specific mode of action and the target-based mode of action of this substance are still unclear. The effect of MC21-A on macromolecular synthesis in bacterial cells must be evaluated by measuring the incorporation of labeled precursors to determine any target-specific activity of this substance.
To evaluate the eukaryotic membrane damage caused by MC21-A, its hemolytic activity against human red blood cells was determined. The result indicated that this substance does not hemolyze human erythrocytes (Fig. 5). These data support the results reported by Hilliard et al. (20), who concluded that the hemolytic activity does not correlate with the bacterial membrane damage and permeabilization. In contrast, amphotericin B and gramicidin D readily lyse human erythrocytes. The erythrocytes are almost completely lysed by treatment with amphotericin B at 50 μg/ml for 2 h. The hemolytic activity of gramicidin D is somewhat lower than that of amphotericin B.
The cytotoxicity of MC21-A and the growth alteration caused by MC21-A were evaluated with five mammalian cell lines by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. This assay is widely used to determine cell proliferation and viability as well as to evaluate cytotoxicity based on the reduction of the dye by the mitochondrial dehydrogenase of viable cells to yield a dark blue formazan (31). MC21-A is not cytotoxic to HDF, PC12D, and Vero cells but is moderately cytotoxic to MOLT-4 cells. This substance is significantly cytotoxic to MDCK cells, as determined by drastic decreases in the growth of treated cells. MC21-A does not alter the growth of PC12D, HDF, and Vero cells.
In conclusion, MC21-A, a substance produced by a newly identified marine bacterium, P. phenolica sp. nov. O-BC30T, showed aggressive activity against gram-positive bacteria, especially MRSA. Its structure was determined to be 3,3′,5,5′-tetrabromo-2,2′-biphenyldiol. This substance rapidly permeabilized the cell membranes of MRSA as its mode of action, but it did not lyse bacterial cells or human erythrocytes. At concentrations up to 50 μg/ml, this substance also demonstrated selective cytotoxicity to HDF, Vero, and PC12D cells.
Although further studies on the pharmacokinetics and pharmacological properties of MC21-A are necessary, our findings suggest that this antibiotic, with a mode of action that differs from those of vancomycin and teicoplanin, may be useful as a lead compound in the development of new anti-MRSA substances to anticipate the rapid increment of new resistance in MRSA.
Acknowledgments
We are grateful to S. Araki (Eisai Co. Ltd.), T. Sumeya (Saga University), H. Yamamoto (Gifu University), T. Palmer (John Innes Centre), A. Wright (Tufts Medical School), the Gifu Type Culture Collection (Department of Microbiology, Gifu University), and T. Takeda (Hokkaido Kushiro Fisheries Experimental Station) for kindly donating the bacterial strains tested in this study. We also thank M. Sano for kindly providing the PC12D cell line.
REFERENCES
- 1.Acebal, C., L. M. Cañedo, J. L. F. Puentes, J. P. Baz, F. Romero, F. De La Calle, M. D. G. Grávalos, and P. Rodrigues. 1999. Agrochelin, a new cytotoxic antibiotic from a marine agrobacterium. Taxonomy, fermentation, isolation, physicochemical properties and biological activity. J. Antibiot. 52:983-987. [DOI] [PubMed] [Google Scholar]
- 2.Acebal, C., R. Alcazar, L. M. Cañedo, F. De La Calle, P. Rodriguez, F. Romero, and J. L. F Puentes. 1998. Two marine agrobacterium producers of sesbanimide antibiotics. J. Antibiot. 51:64-67. [DOI] [PubMed] [Google Scholar]
- 3.Aeschlimann, J. R., E. Hershberger, and M. J. Rybak. 1999. Analysis of vancomycin population susceptibility profiles, killing activity, and postantibiotic effect against vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aeschlimann, J. R., and M. J. Rybak. 1998. Pharmacodynamic analysis of the activity of quinopristin-dalfopristin against vancomycin-resistant Enterococcus faecium with differing MBCs via time-kill-curve and postantibiotic effect methods. Antimicrob. Agents Chemother. 42:2188-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett, J. F., R. M. Goldschmidt, L. E. Lawrence, B. Foleno, R. Chen, J. P. Demers, S. Johnson, R. Konojia, J. Fernandez, J. Bernstein, L. Licata, A. Donetz, S. Huang, D. J. Hlasta, M. J. Macielag, K. Ohemeng, R. Frechette, M. B. Frosco, D. H. Klaubert, J. M. Whiteley, L. Wang, and J. A. Hoch. 1998. Antibacterial agents that inhibit two-component signal transduction systems. Proc. Natl. Acad. Sci. USA 95:5317-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biabani, M. A. F., H. Laatsch, E. Helmke, and H. Weyland. 1997. δ-Indomycinone: a new member of pluramycin class of antibiotics isolated from marine Streptomyces sp. J. Antibiot. 50:874-877. [DOI] [PubMed] [Google Scholar]
- 7.Craig, W. A., and S. Gudmundsson. 1991. Postantibiotic effect, p. 403-431. In V. Lorian (ed.), Antibiotics in laboratory medicine, 3rd ed. The Williams & Wilkins Co., Baltimore, Md.
- 8.Daugelavic̆ius, R., J. K. H. Bamford, A. M. Grahn, E. Lanka, and D. H. Bamford. 1997. The IncP plasmid-encoded cell envelope-associated DNA transfer complex increases cell permeability. J. Bacteriol. 179:5195-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daugelavic̆ius, R., E. Bakien, and D. H. Bamford. 2000. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob. Agents Chemother. 44:2969-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Entenza, J. M., O. Marchetti, M. P. Glauser, and P. Moreillon. 1998. Y-688, a new quinolone active against quinolone-resistant Staphylococcus aureus: lack of in vivo efficacy in experimental endocarditis. Antimicrob. Agents Chemother. 42:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauthier, G., M. Gauthier, and R. Christen. 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequence and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combination. Int. J. Syst. Bacteriol. 45:755-761. [DOI] [PubMed] [Google Scholar]
- 12.Gauthier, M. J. 1976. Alteromonas rubra sp. nov., a new marine antibiotic-producing bacterium. Int. J. Syst. Bacteriol. 26:459-466. [Google Scholar]
- 13.Gauthier, M. J., and V. A. Breittmayer. 1979. A new antibiotic-producing bacterium from seawater: Alteromonas aurentia sp. nov. Int. J. Syst. Bacteriol. 29:366-372. [Google Scholar]
- 14.Gerard, J. M., P. Haden, M. T. Kelly, and R. J. Andersen. 1999. Loloatins A-D, cyclic decapeptide antibiotics produced in culture by a tropical marine bacterium. J. Nat. Prod. 62:80-85. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, D. 1995. Tetracycline and chloramphenicol, p. 306-317. In G. Mandell, J. Bennet, and R. Dolin (ed.), Principles and practice of infectious disease. Churchill Livingstone, New York, N.Y.
- 16.Hanaki, H., H. Labischinski, Y. Inaba, N. Kondo, H. Murakami, and K. Hiramatsu. 1998. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 42:315-320. [DOI] [PubMed] [Google Scholar]
- 17.Handayani, D., R. A. Edrada, P. Proksch, V. Wray, L. Witte, R. W. M. Van Soest, A. Kunzmann, and Soedarsono. 1997. Four new bioactive polybrominated diphenyl ethers of the sponge Dysidea herbacea from West Sumatra, Indonesia. J. Nat. Prod. 60:1313-1316. [DOI] [PubMed] [Google Scholar]
- 18.Hanefeld, U., H. G. Floss, and H. Laatsch. 1994. Biosynthesis of the marine antibiotic pentabromopseudilin. 1. The benzene ring. J. Org. Chem. 59:3604-3608. [Google Scholar]
- 19.Harada, H., and Y. Kamei. 1997. Selective cytotoxicity of marine algae extracts to several human leukemic cell lines. Cytotechnology 25:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilliard, J. J., R. M. Goldschmidt, L. Licata, E. Z. Baum, and K. Bush. 1999. Multiple mechanism of action for inhibitors of histidine protein kinase from bacterial two-component systems. Antimicrob. Agents Chemother. 43:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 22.Holmström, C., and S. Kjelleberg. 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 23.Imamura, N., M. Nishijima, T. Takadera, K. Adachi, M. Sakai, and H. Sano. 1997. New anticancer antibiotics, pelagiomicins produced by a new marine bacterium Pelagiobacter variabilis. J. Antibiot. 50:8-12. [DOI] [PubMed] [Google Scholar]
- 24.Isnansetyo, A., and Y. Kamei. Pseudoalteromonas phenolica sp. nov., a novel marine bacterium that produces phenolic anti-methicillin-resistant Staphylococcus aureus (MRSA) substances. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 25.James, S., C. Holsmström, and S. Kjelleberg. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. App. Environ. Microbiol. 62:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebaron, P., P. Catala, and N. Parthuisot. 1998. Effectiveness of SYTOX Green stain for bacterial viability assessment. Appl. Environ. Microbiol. 64:2697-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Löwdin, E., I. Odenholt, and O. Cars. 1998. In vitro studies of pharmacodynamic properties of vancomycin against Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 42:2739-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mainardi, J. L., D. M. Shlaes, R. V. Goering, J. H. Shlaes, J. F. Acar, and F. W. Goldstein. 1995. Decreased teicoplanin susceptibility of methicillin-resistant strains of Staphylococcus aureus. J. Infect. Dis. 171:1646-1650. [DOI] [PubMed] [Google Scholar]
- 29.McCarthy, S. A., R. M. Johnson, D. Kakimoto, and T. Sakata. 1985. Effects of various agents on the pigment (violacein) and antibiotic production of Alteromonas luteoviolacea. Bull. Jpn. Soc. Sci. Fish. 51:1115-1121. [Google Scholar]
- 30.Mortimer, F. C., D. V. Mason, and V. A. Gant. 2000. Flow cytometric monitoring of antibiotic-induced injury in Escherichia coli using cell-impermeant fluorescence probes. Antimicrob. Agents Chemother. 44:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. 1997. Method for dilution antimicrobial susceptibility test for bacteria that grow aerobically, 4th ed., p, 1-29. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 33.Nishikawa, J., Y. Takeyama, T. Ueda, Y. Hori, N. Ueno, M. Yamamoto, and Y. Saitoh. 1995. Induction of apoptotic cell death by pancreatitis-associated ascitic fluid in Mardin-Darby canine kidney cells. FEBS Lett. 373:19-22. [DOI] [PubMed] [Google Scholar]
- 34.Novo, D. J., N. G. Perlmutter, R. H. Hunt, and H. M. Shapiro. 2000. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeabilization, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 44:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patersen, D. L. 1999. Reduced susceptibility of Staphylococcus aureus to vancomycin- a review of current knowledge. Commun. Dis. Intell. 24:69-73. [DOI] [PubMed] [Google Scholar]
- 36.Pusecker, K., H. Laatsch, E. Helmke, and H. Weyland. 1997. Dihydrophencomycin methyl ester, a new phenazine derivative from a marine Streptomycete. J. Antibiot. 50:479-483. [DOI] [PubMed] [Google Scholar]
- 37.Roth, B. L., M. Poot, S. T. Yue, and P. J. Millard. 1997. Bacterial viability and antibiotic susceptibility testing with SYTOX Green nucleic acid stain. App. Environ. Microbiol. 63:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiozawa, A., T. Kagasaki, A. Torikata, N. Tanaka, K. Fujimoto, T. Hata, Y. Furukawa, and S. Takahashi. 1995. Thiomarinol B and C, new antimicrobial antibiotics produced by a marine bacterium. J. Antibiot. 48:907-909. [DOI] [PubMed] [Google Scholar]
- 39.Shiozawa, H., A. Shimada, and S. Takahashi. 1997. Thiomarinol D, E, F and G, new hybrid antimicrobial antibiotics produced by a marine bacterium: isolation, structure and antimicrobial activity. J. Antibiot. 50:449-452. [DOI] [PubMed] [Google Scholar]
- 40.Singh, M. P., P. J. Petersen, W. J. Weiss, F. Kong, and M. Greenstein. 2000. Saccharomicins, novel heptadecaglycoside antibiotics produced by Saccharothrix espanaenses: antibacterial and mechanistic activities. Antimicrob. Agents Chemother. 44:2154-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanley, N. R., K. Findlay, B. C. Berks, and T. Palmer. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsang, C. K., A. Sagara, and Y. Kamei. 2001. Structure-activity relationship of a neurite outgrowth-promoting substance purified from the brown alga, Sargassum macrocarpum, and its analogues on PC12D cells. J. Appl. Phycol. 13:349-357. [Google Scholar]
- 43.Vaudaux, P., P. Francois, B. Berger-Bächi, and D. P. Lew. 2001. In vivo emergence of subpopulations expressing teicoplanin or vancomycin resistance phenotypes in a glycopeptide-susceptible, methicillin-resistant strain of Staphyloccus aureus. J. Antimicrob. Chemother. 47:163-170. [DOI] [PubMed] [Google Scholar]
- 44.Witte, W. 1999. Antibiotic resistance in gram-positive bacteria: epidemiological aspects. J. Antimicrob. Chemother. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 45.Yoshikawa, K., T. Takadera, K. Adachi, M. Nishijima, and H. Sano. 1997. Karormicin, a novel antibiotic specifically active against marine gram-negative bacteria, produced by a marine bacterium. J. Antibiot. 50:949-953. [DOI] [PubMed] [Google Scholar]