Abstract
The emergence of human immunodeficiency virus type 1 (HIV-1) strains resistant to highly active antiretroviral therapy necessitates continued drug discovery for the treatment of HIV-1 infection. Most current drug discovery strategies focus upon a single aspect of HIV-1 replication. A virus-cell-based assay, which can be adapted to high-throughput screening, would allow the screening of multiple targets simultaneously. HIV-1-based vector systems mimic the HIV-1 life cycle without yielding replication-competent virus, making them potentially important tools for the development of safe, wide-ranging, rapid, and cost-effective assays amenable to high-throughput screening. Since replication of vector virus is typically restricted to a single cycle, a crucial question is whether such an assay provides the needed sensitivity to detect potential HIV-1 inhibitors. With a stable, inducible vector virus-producing cell line, the inhibitory effects of four reverse transcriptase inhibitors (zidovudine, stavudine, lamivudine, and didanosine) and one protease inhibitor (indinavir) were assessed. It was found that HIV-1 vector virus titer was inhibited in a single cycle of replication up to 300-fold without affecting cell viability, indicating that the assay provides the necessary sensitivity for identifying antiviral molecules. Thus, it seems likely that HIV-1-derived vector systems can be utilized in a novel fashion to facilitate the development of a safe, efficient method for screening compound libraries for anti-HIV-1 activity.
Currently, the principal regimen for the treatment of individuals infected with human immunodeficiency virus type 1 (HIV-1) involves highly active antiretroviral therapy, which typically includes a combination of a protease inhibitor and a nucleoside and/or nonnucleoside reverse transcriptase (RT) inhibitor. HIV-1 protease inhibitors such as indinavir, ritonavir, and saquinavir prevent proteolytic processing of immature viral polyproteins to produce mature infectious virions (37) Nucleoside RT inhibitors such as zidovudine, didanosine, lamivudine, and stavudine lack the 3′-OH moiety on the ribose sugar and act as chain terminators when incorporated into the elongating DNA chain by the HIV-1 RT, while the nonnucleoside RT inhibitors such as efavirenz, delavirdine, and nevirapine represent a class of diverse polycyclic compounds that bind to a site near the catalytic domain of RT and interfere with polymerase activity (26).
Highly active antiretroviral therapy has greatly decreased morbidity and mortality for millions of HIV-1-infected individuals over the past several years. However, 10 to 50% of patients do not achieve sustained HIV-1 suppression with the current highly active antiretroviral therapy drug cocktails due to difficulties with adherence associated with drug toxicity and viral rebound (17, 37). Long-term use of highly active antiretroviral therapy has been linked to significant adverse side effects, including anemia, neuropathy, pancreatitis, and lipodystrophy syndrome (5, 16, 21, 32). Moreover, the viral genome can evolve very rapidly owing in large part to the lack of proofreading during reverse transcription, which has resulted in the emergence of viral strains resistant to single- and multiple-drug regimens (11).
It has been reported that within 2 years, viral rebound occurred in 20% of patients who were previously treatment naïve and in 36% to 40% who were previously treated patients (18, 28, 31, 36). Additionally, drug-resistant viral strains are archived in latently infected cells, providing HIV-1 with life-long immunity against these forms of treatment (13). Therefore, as the worldwide population of HIV-1-infected individuals continues to grow, the search for additional therapies remains a high priority.
With the advent of high-throughput screening protocols, hundreds of thousands of distinct low-molecular-weight compounds have been assessed for inhibitory effects against homogenously pure recombinant HIV-1 viral enzymes (8, 39). Promising drug candidates from high-throughput screens can be further refined chemically through structural modifications based upon molecular modeling principles in an effort to increase antiviral activity while decreasing toxicity. Such procedures led to the development of the current class of protease inhibitors (19). However, most initial screening focuses upon a single, well-defined aspect of replication with cell-free systems. It would be advantageous to develop a virus-cell-based assay amenable to high-throughput screening because it would allow the screening of a large number of targets, including both less well characterized aspects of replication and well-studied ones. Attempting to develop such an assay with replication-competent HIV-1 would be quite difficult owing to the cumbersome nature of assays for detecting replication-competent virus and the inherent safety concerns which accompany the processing of very large numbers of samples containing replication-competent virus.
Because of their ability to very efficiently introduce genetic material into the genome of a permissive cell, retroviral gene delivery systems based upon the murine leukemia virus for efficient gene transfer into eukaryotic cells have been widely studied (35). Recently, retroviral vector systems derived from HIV-1 have been constructed because of their ability to infect nondividing cells, which might be advantageous for in vivo gene therapy protocols (20, 25, 33).
In this study, we explored the feasibility of using an HIV-1-based producer cell line in an HIV-1 drug screening system. Vector virus produced in this manner can mimic most stages of the HIV-1 life cycle, including viral protein production, assembly, budding, maturation, infection, and integration, without producing replication-competent virus, an obvious safety advantage. Moreover, because marker genes that can be employed in high-throughput screening can be used to monitor infection, retroviral vector systems might be able to serve as the basis for virus-cell-based high-throughput screening assays. However, because the vector virus is restricted to a single round of replication, the assay would have to be sufficiently sensitive to identify potential inhibitors whose effects might otherwise be amplified in assays involving replication-competent virus and multiple rounds of infection. This report details the evaluation of several known HIV-1 inhibitors and indicates that this system could be developed into a powerful screen for identifying novel anti-HIV-1 pharmaceuticals.
MATERIALS AND METHODS
Plasmid construction.
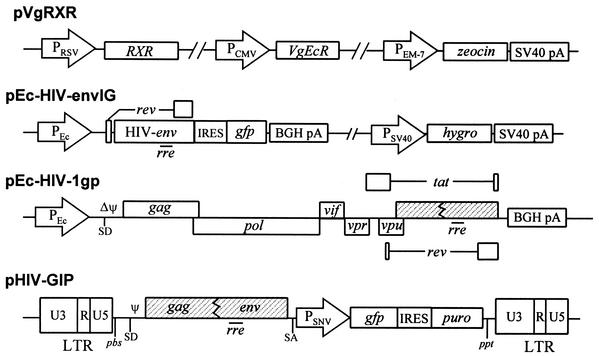
The following plasmids were utilized to create the lentiviral producer cell line: pVgRXR, pEc-HIVenvIG, pEc-HIV-1gp, and pHIV-GIP (Fig. 1). pVgRXR is commercially available (Invitrogen K1001-01) and provides constitutive expression of the transcription factors required for ecdysone inducibility. pEc-HIVenvIG was a modification of vector pEcVIGhyg (34) in which the vesicular stomatitis virus G protein sequence was removed and replaced with a fragment containing the HIV-1 env gene from pTIRevEnv (23). pEc-HIV-1gp was generated by digesting pIND (Invitrogen) with EcoRV and ligating the backbone to the BssHII-SacI fragment of a proviral clone of the HIV-1LAI strain which had encapsidation sequence deletions from nucleotides 241 to 253 and 295 to 328 (24) as well as a deletion from 6630 to 7215, preventing expression of the env gene (23); the numbers correspond to the viral RNA nucleotide sequence. Therefore, pEc-HIV-1gp has open reading frames for Gag, Pol, Tat, Rev, Vpr, Vpu, Vif, and Nef under the control of the ecdysone transcriptional expression system but has mutations, deletions, or disruptions in the packaging signal (ψ), primer binding site, env, and the 5′ and 3′ long terminal repeats. The construction of pHIV-GIP has been described previously; it contains the cis-acting signals for HIV-1 replication and reporter genes which, when transferred to target cells, provide for expression of the enhanced green fluorescent protein and resistance to puromycin (23). The latter reporter gene is expressed in a cap-independent manner due to the presence of an internal ribosomal entry sequence (IRES) (27).
FIG. 1.
Schematic diagrams of expression plasmids and retroviral transducing vector used to establish the #6 producer cell line. The shaded boxes represent nonfunctional HIV-1 genes. Δψ represents a 33-bp deletion within the encapsidation signal downstream of the splice donor site. PRSV, Rous sarcoma virus promoter; PCMV, immediate-early promoter from human cytomegalovirus; PEM-7, synthetic Escherichia coli T7 promoter; PEc, ecdysone promoter; PSNV, spleen necrosis virus U3 promoter; IRES, internal ribosomal entry site from encephalomyocarditis virus (EMCV); gfp, sequence encoding the enhanced green fluorescent protein; PSV40, simian virus 40 early gene promoter; hygro, hygromycin resistance gene; SV40 pA, simian virus 40 late polyadenylation sequence; BGH pA, bovine growth hormone polyadenylation sequence; puro, puromycin resistance gene; rre, cis-acting Rev response element; SD, splice donor site; SA, splice acceptor site; pbs, primer binding site; ppt, polypurine tract; LTR, long terminal repeat.
Cell culture.
293T cells (12) and HeLaT4 cells were maintained in a 5% CO2 incubator at 37°C in minimal essential medium (MEM) containing Earle's salts and l-glutamine (Gibco-BRL 11095-080) supplemented with 10% fetal bovine serum (HyClone), 0.2 mM MEM nonessential amino acid solution (Gibco-BRL 11140-050), 250 U of penicillin per ml, and 250 μg of streptomycin (Gibco-BRL 15070-063) per ml.
Design of HIV-1 packaging cell system.
The #6 packaging cell line was generated by transfection of the pVgRXR and pEc-HIV-1gp vectors into 293T cells, followed by selection with 50 μg of zeocin per ml and screening of individual clones for high levels of inducible RT activity after adding medium containing 10 μM ponasterone A, a synthetic analogue of ecdysone. All transfections were performed following the modified calcium phosphate precipitation procedure (14). Promising clones were then transfected with pEcHIVenvIG. Individual clones were selected for resistance to 150 μg of hygromycin per ml and subsequently screened for inducible expression of green fluorescent protein (GFP) from the HIV-1 env-IRES-GFP cassette. Cell clones that demonstrated a high level of inducibility from the RT and env constructs were finally transfected with pHIV-GIP and selected for resistance to 1 μg of puromycin per ml.
Virus production was assessed upon induction with 10 μM ponasterone A. Vector virus generated from this line (termed #6 HIV-1 producer cells) was, as expected, unable to infect HeLa cells due to the lack of a suitable cell surface receptor for the HIV-1 envelope (data not shown). HeLaT4 cells were utilized for all subsequent experiments as they have been modified to express surface CD4 and are susceptible to HIV-1 infection (30).
RT assay.
RT activity was monitored at day 0 through day 6 postinduction to characterize the induction potential of the ponasterone A-inducible #6 producer cell line. Briefly, 10-μl aliquots were harvested in triplicate on the indicated days and transferred into 96-well plates. Then 50 μl of a cocktail containing 50 mM Tris-HCl (pH 7.8), 7.5 mM KCl, 5 mM MgCl2, 0.05% NP-40, 2 mM dithiothreitol, 5 μg of poly(rA) per ml, 1.57 μg of oligo(dT)12-18 per ml, and 0.5 μCi of [α-32P]TTP was added to each well, followed by incubation for 90 min at 37°C. Samples were replicated onto Whatman DE81 paper, air dried, and washed three times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 10 min and once with 100% ethanol for 5 min. Blots were air dried and analyzed with a Typhoon 8600 variable-mode imager (Amersham Pharmacia Biotech).
HIV-1 p24gag enzyme-linked immunosorbent assay.
The levels of p24gag protein in supernatant samples from uninduced or ponasterone A-induced (days 1 through 4 postinduction) #6 producer cells were assayed in duplicate by a p24gag enzyme-linked immunosorbent assay according to the manufacturer's instructions (NEN Life Science Products).
Western blot for detection of HIV-1 envelope expression.
Western blotting was utilized to assess Env production from uninduced #6 producer cells as well as those induced with ponasterone A for up to 6 days. Cell protein extract preparation, separation via sodium dodecyl sulfate-polyacrylamide, blotting to a polyvinylidene difluoride membrane, and probing with sheep polyclonal anti-gp120 antiserum (provided through the AIDS Research and Reference Reagent Program) were done as described previously (23).
Vector virus production.
The ecdysone-inducible system (Invitrogen) was utilized to control expression of cytotoxic and cytostatic proteins such as HIV-1 Env, protease, and Vpr (2, 22, 38). Twenty-four hours before induction, 2.5 × 106 #6 HIV-1 producer cells were seeded in a 100-mm cell culture plate in 10% MEM. Forty-eight hours later, fresh medium containing 10 μM ponasterone A was added. Forty-eight hours after the first induction, the medium was replaced with fresh 10% MEM-10 μM ponasterone A. Vector virus was harvested 48 h after the second induction. For experiments involving indinavir, 8 × 105 #6 producer cells were seeded in 60-mm plates; 48 and 96 h later, fresh medium containing 10 μM ponasterone A and various concentrations of the protease inhibitor was added.
It should be noted that all manipulations during development of the HIV-1-derived producer cell line and during propagation of vector virus were performed under biosafety level 2/3 as prescribed by the Centers for Disease Control and National Institutes of Health. Although the use of HIV-1 vectors should be significantly safer than handling replication-competent virus, it was felt that it would prudent to continue to utilize this system under biosafety level 2/3 containment. It is anticipated that such practice will be continued when similar systems are used for high-throughput screening.
Infection protocol and drug screening.
The basic infection protocol involved seeding the target HeLaT4 cells at a density of 2 × 105 cells per well in a six-well plate in duplicate. For experiments involving the RT inhibitors, each drug was added at the indicated concentration at the time of the infection and maintained until selection medium containing puromycin was added 24 h later. The RT inhibitors tested were 3′-azido-3′-deoxythymidine (zidovudine) (Sigma), 2′,3′-dideoxyinosine (didanosine) (Sigma), 2′,3′-didehydro-3′-deoxythymidine (stavudine) (Sigma), and 2′,3′-dideoxy-3′-thiacytidine (lamivudine) (Moravek Biochemicals). The protease inhibitor indinavir was added during the induction phase and not during infection or puromycin selection.
Viral vectors from induced #6 producer cells were harvested and passed through a syringe filter outfitted with a 0.45-μm HT Tuffryn membrane (Pall Gelman Laboratory). Serial dilutions of the viral supernatant were prepared and adjusted to contain 8 μg of Polybrene per ml. For the RT inhibitors, the drug concentrations were adjusted to the indicated concentrations after the serial dilution of the viral supernatants. In all cases, 1 ml of each viral supernatant was incubated with the HeLaT4 cells for 6 h at 37°C in 5% CO2. At the conclusion of the infection, the viral supernatant was aspirated and replaced with fresh 10% MEM containing the appropriate RT inhibitor if present during the infection phase. The following day, medium containing 1 μg of puromycin per ml was added (with RT inhibitor again if it was added during the infection phase) and refreshed on days 2 and 4 postinfection. Puromycin-resistant, GFP-positive colonies were counted and used to calculate the viral titer. Each experiment was repeated three times.
Toxicity assays.
A total of 7.5 × 102 HeLaT4 cells were seeded in triplicate on two 96-well plates and incubated in the presence of 0 μM through 2,500 μM zidovudine, didanosine, lamivudine, or stavudine. Seventy-two hours later, cell viability and proliferation were assessed through the cleavage of the tetrazolium salt WST-1 (4-[3-(4-lodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio-]1,3-benzene disulfonate), which utilizes the mitochondrial succinate-tetrazolium reductase system to yield formazan, which can be detected by a scanning multiwell spectrophotometer (Roche). To assess cytotoxicity levels of indinavir on the #6 producer cells, 0 μM through 500 μM indinavir was added to 5 × 103 #6 producer cells, and viability was assessed after 72 h with the WST-1 reagent.
RESULTS
Development and biochemical characterization of an ecdysone-inducible lentiviral producer cell line.
Retroviral vector systems comprise two components, the transducing vector and the packaging cell. The transducing vector contains the marker gene and/or therapeutic gene to be transferred into the target cell during infection. Since the exogenous gene sequences replace some or all of the genes encoding viral proteins, the transducing vectors are defective for replication. The deficient viral proteins are supplied in trans by the packaging cell. The genetic separation of the cis-acting elements contained in the transducing vector and the trans-acting functions supplied by the packaging cell provide a significant level of safety by preventing replication-competent virus from being produced. It should be noted that a producer cell line is a packaging cell that also contains the transducing vector at a different chromosomal location from the trans-acting genes (at least in the case described in this paper). The producer cell yields vector virus that is restricted to a single cycle of replication.
The recombinants employed to create the #6 HIV-1 producer cell line are depicted in Fig. 1. pEc-HIV-gp (gag, pol, vif, vpr, vpu, tat, and rev) and pEc-HIV-envIg (rev and env) express the genes encoding the viral proteins under control of the ecdysone-inducible promoter (Fig. 1). An inducible promoter was used because constitutive expression of some of the viral gene products has cytotoxic and cytostatic effects upon the cells (2, 22, 38). pVgRXR supplies the genes that control activation of the inducible promoter (Fig. 1). pHIV-GIP is the transducing vector containing the marker genes used to monitor vector virus infectivity. The three HIV-1-derived plasmids pEc-HIV-gp, pEc-HIV-envIg, and pHIV-GIP were transfected independently to eliminate the potential for recombination during cotransfection, as detailed in Materials and Methods (design of HIV-1 packaging cell system).
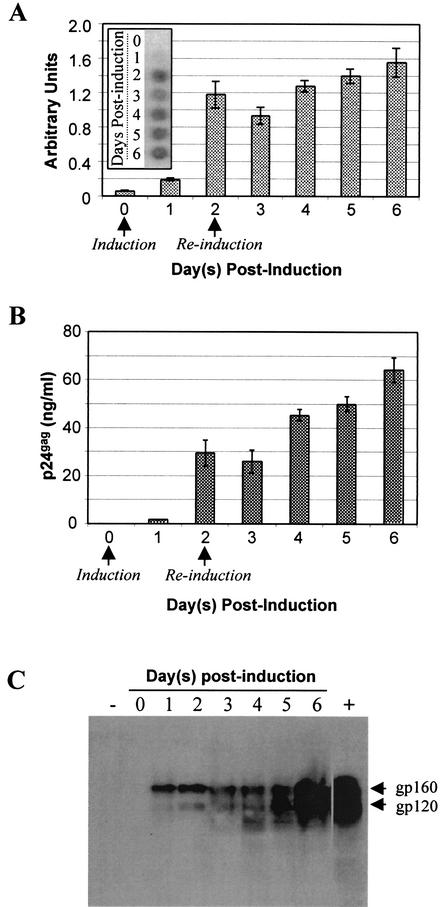
Initial biochemical characterization of the #6 producer cell was performed. The production of three late viral proteins was assessed after induction with the ecdysone homologue ponasterone A. First, RT activity was measured with a standard enzymatic assay. RT is the virus-encoded polymerase required for synthesizing viral DNA with viral RNA as a template and is encoded by the viral pol gene. As can be seen in Fig. 2A, it is undetectable before induction, becoming clearly detectable by day 2. There was at least a 21-fold induction, as measured with the Typhoon 8600 variable-mode imager.
FIG. 2.
Biochemical characterization of #6 producer cell line. (A) Upregulation of reverse transcriptase activity. Supernatant samples from the #6 HIV-1 producer cell line were collected prior to induction and on days 1 through 6 postinduction. RT activity was assayed as described in Materials and Methods. Arrows indicate times at which the medium was replaced with fresh 10% MEM containing 10 μM ponasterone A. The inset depicts the actual phosphorimager induction profile of RT. (B) Induction of p24gag expression. Supernatant was harvested from uninduced and ponasterone A-induced #6 HIV-1 producer cells from day 1 through day 6. The quantity of p24gag was determined by enzyme-linked immunosorbent assay. (C) Western blot analysis of inducible HIV-1 envelope expression. Lane +, cell lysate from wild-type HIV-1-infected HeLaT4 cells; lane −, uninfected HeLaT4 cell lysates.
Second, an enzyme-linked immunosorbent assay was employed to measure expression of the major viral capsid protein, p24gag, and by day 4 postinduction the level of p24gag found in the supernatant was 45.3 ng/ml, while that in supernatant harvested from uninduced #6 HIV-1 producer cells was below the detectable threshold of the assay (1.2 pg/ml) (Fig. 2B). Inducible regulation of both RT and p24gag indicates that appropriate expression is being obtained from pEc-HIV-1gp (Fig. 1).
Third, expression of HIV-1 Env (needed to initiate virus infection) from pEc-HIV-envIG was assessed via Western blotting. Before induction, the envelope glycoproteins were not detectable. Upon induction, a significant quantity of the gp160 precursor Env protein was clearly detectable, followed by an increase in gp120 expression as the gp160 was proteolytically processed to form the mature gp120 and gp41 proteins (37).
Viral vector titers.
Next, vector virus titers were determined with the HIV-GIP transducing vector. HIV-GIP expresses two marker genes, gfp and the puromycin resistance gene (puro), from a single bicistronic mRNA. Expression is driven by the spleen necrosis virus promoter, with gfp being translated in a cap-dependent manner and puro being translated in a cap-independent fashion due to the presence of the encephalomyocarditis internal ribosomal entry site (IRES). After inoculation of the HeLaT4 target cells, titers can then be measured by counting the number of GFP-positive, puromycin-resistant foci and multiplying by the dilution.
In the absence of any drugs, titers from a #6 producer cell line ranged from 1.0 × 104 to 2.5 × 104 IU/ml when supernatant was harvested 4 days after the initial ponasterone A induction. The #6 producer cell line was found to be stable for at least a year and has yielded similar viral titers during that period. It is noteworthy that this is the first report of an ecdysone-inducible HIV-1 packaging cell line that utilizes the HIV-1 envelope. We and other investigators have described the creation of inducible lentiviral packaging cell lines that incorporate the vesicular stomatitis virus G protein envelope for the expanded host range with increased titer afforded by ultracentrifugation (4, 34). However, the system described here utilizes a lentiviral vector system that more closely mimics authentic HIV-1 replication.
Drug screening.
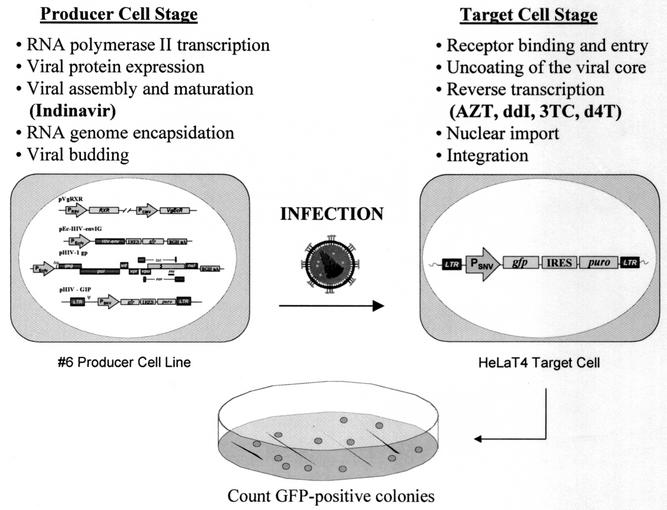
The HIV-1 life cycle reproduced by the #6 HIV-1 producer cell line can be divided into two distinct halves, the producer cell stage and the target cell stage (Fig. 3). The producer cell stage begins with the addition of ponasterone A to the producer cell line and concludes with the harvest of vector virus-containing supernatant from the induced producer cell line. The stages of HIV-1 replication which occur in the producer cell stage include (i) transcription initiation from the HIV-1 long terminal repeat of the integrated HIV-GIP vector to yield viral RNA for encapsidation, (ii) expression of the HIV-1 structural and enzymatic proteins, (iii) virus assembly, including encapsidation of viral RNA, (iv) proteolytic processing of the viral polyproteins during virus maturation, and (v) budding of the virion from the induced producer cell into the extracellular milieu (29).
FIG. 3.
Outline of drug-screening assay system. Vector virus undergoes one round of replication when transmitted from producer cells to target cells. Inhibitory effects of drugs can be evaluated on both producer cells and target cells to distinguish between compounds that act during each stage of replication. See Fig. 1 for an enlargement of the recombinants integrated within the #6 producer cell line. The protease and RT inhibitors are listed in boldface at their stage of action. AZT, zidovudine; ddI, didanosine; 3TC, lamivudine; d4T, stavudine; for other abbreviations, see the legend to Fig. 1.
The target cell stage begins with the harvesting of the conditioned supernatant that contains the vector virus particles and proceeds through infection of the target cells. The following steps occur during the target cell stage: (i) binding of the mature virion to the target cell receptor and coreceptor, (ii) entry of the virus into the cell via membrane fusion, (iii) uncoating of the viral core, (iv) reverse transcription, during which viral DNA is synthesized, (v) import to the nuclear compartment, and (vi) integration of the viral DNA to form a provirus. Drugs that inhibit one or more of these stages during either of the two phases of replication should decrease vector virus titer and be reflected by a reduction in the number of cells expressing gfp and puro.
The ability of this HIV-1-derived vector system to detect the antiviral activity of small compounds was assessed by incubating known HIV-1-inhibitory compounds with vector virus produced by the induced #6 producer cell line. It should be noted that this system provides an additional degree of flexibility allowing determination of at which phase of replication, the producer cell stage and/or the target cell stage, the antiviral compound is acting, ensuring that the compound is working during the appropriate phase of replication. A series of well-characterized HIV-1 inhibitors were incubated with the #6 producer cell line at the time of induction with ponasterone A through supernatant harvest at day 4 during the producer cell stage or during the transduction process of the HeLaT4 cells through the time of puromycin selection during the target cell stage (Fig. 3).
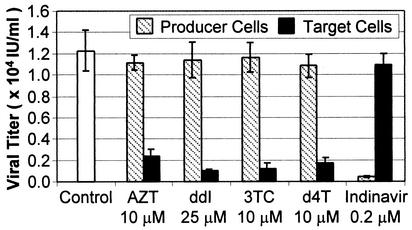
Figure 4 illustrates that when #6 producer cells were incubated with the RT inhibitors during the producer cell stage, no noticeable effect on viral titer was observed. However, a marked decrease in titer was observed when the RT inhibitors were added at the time of infection and maintained throughout the initiation of puromycin selection during the target cell stage. These were the anticipated results because the RT inhibitors should act upon reverse transcription during the target cell stage. In contrast, the addition of the protease inhibitor indinavir during the target cell stage did not result in a significant decrease in viral titer. The effect of indinavir was readily apparent, however, when it was added during the producer cell stage. Again, this was expected because protease inhibitors should act upon virus maturation during the producer cell stage. That both types of inhibitors acted during the appropriate phase of replication provides additional support that the inhibition of titers was due to their direct effects upon virus replication.
FIG. 4.
Effect of inhibitors on HIV-1 vector virus. RT inhibitors and indinavir were incubated with producer cells (at the time of induction with 10 μM ponasterone A until harvest of the supernatant on day 4) or target cells (added at the time of infection along with 8 μg of Polybrene per ml and maintained through the addition of 1 μg of puromycin per ml with 10% MEM during the selection process). Results shown are the means (± standard deviations) of quadruplicate titer determinations. The control consisted of producer or target cells not treated with protease and RT inhibitors. AZT, zidovudine; ddI, didanosine; 3TC, lamivudine; d4T, stavudine.
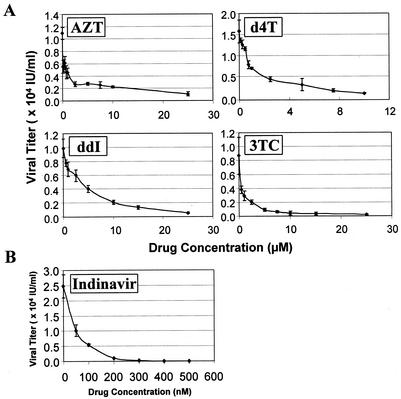
Next, the sensitivity of the system was examined with a wider range of concentrations of RT inhibitors and indinavir. For this series of experiments, the drugs were added only during the stage of the viral replication cycle in which they demonstrated antiviral activity as described above. The resulting plots of drug concentration versus viral vector titer are depicted in Fig. 5. The concentration of each drug required to inhibit vector virus titer by 90% (IC90) was also calculated (Table 1).
FIG. 5.
Effects of increasing concentrations of RT inhibitors and indinavir on viral vector titer. (A) The indicated concentrations of RT inhibitors (zidovudine, didanosine, lamivudine, and stavudine) were added at the time of infection with 8 μg of Polybrene per ml and maintained through the initiation of puromycin selection. AZT, zidovudine; ddI, didanosine; 3TC, lamivudine; d4T, stavudine. (B) Indinavir was added at the time of induction with ponasterone A and maintained through the harvesting of viral supernatant for target cell transduction. Any indinavir remaining from the induction phase would be diluted 10- to 1,000-fold during the serial dilutions to calculate titer. For both panels, the results shown are the means (± standard deviations) of quadruplicate determinations and are representative of three independent experiments.
TABLE 1.
Calculations of IC90 and CC50 values of known antiviral compoundsa
| Compound | Mean IC90 (μM) ± SD | CC50 (μM) |
|---|---|---|
| Zidovudine | 12.5 ± 1.8 | >2,500 (3508.1)b |
| Didanosine | 18.3 ± 2.4 | >>2,500c |
| Stavudine | 9.6 ± 3.2 | 334.8 |
| Lamivudine | 7.7 ± 1.8 | >>2,500c |
| Indinavir | 0.1434 ± 0.0097 | >250 (307.1)b |
The IC90 (amount of compound required to inhibit viral titer by 90%) and CC50 (drug concentration required to inhibit cellular proliferation by 50%) values for each inhibitor are presented. The IC90 and CC50 values for the RT inhibitors were determined on HeLaT4 target cells, while the IC90 and CC50 values for indinavir were determined on the #6 producer cell line. Both sets of values were calculated with the ED50plus worksheet for Microsoft Excel (Mario H. Vargas, Instituto Nacional de Enfermedades Respiratorias, Mexico).
Values were extrapolated based upon tested concentration points and the generation of a best-fit line.
Cell viability was at approximately 100% or greater compared to untreated cells at up to 2,500 μM.
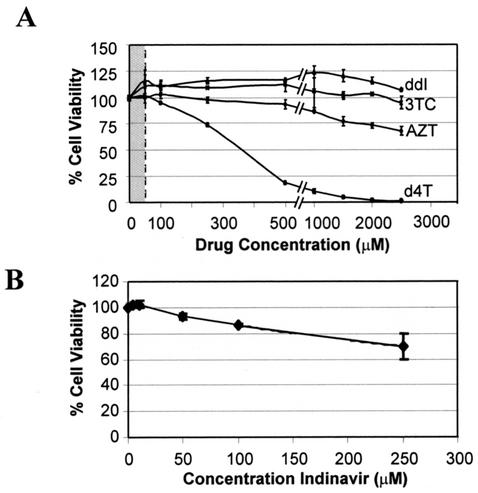
Toxicity assays.
Assays were performed to ensure that the decrease in vector virus titers were not indirect effects due to cellular toxicity. HeLaT4 cells were assayed for evidence of cytotoxicity in the presence of the RT inhibitors up to 2,500 μM with the tetrazolium salt WST-1 method. Figure 6 shows that HeLaT4 cells were greater than 65% viable at concentrations of up to 2,500 μM zidovudine and more than 94% viable at similar concentrations of lamivudine and didanosine. Therefore, the IC90 values of zidovudine, didanosine, and lamivudine were more than 100 times lower than the calculated drug concentration resulting in 50% cytotoxicity (CC50) on the target HeLaT4 cells (Fig. 6, Table 1). The concentration of drug cytotoxic to 50% of the HeLaT4 cells (CC50) treated with stavudine for 72 h was calculated to be 334.8 μM, which exceeded the IC90 concentration in HeLaT4 cells by more than 40-fold.
FIG. 6.
Drug cytotoxicity assays. HeLaT4 cells were assayed for cytotoxicity in the presence of the RT inhibitors zidovudine, didanosine, lamivudine, and stavudine at the indicated concentrations for 72 h (A). AZT, zidovudine; ddI, didanosine; 3TC, lamivudine; d4T, stavudine. Viability of treated cells was assessed with the tetrazolium salt WST-1 reagent as per the manufacturer's instructions (Roche). The cytotoxicity of indinavir was evaluated on the #6 producer cell line (B). Results shown are the means (± standard deviations) of triplicate readings. The shaded region in panel A depicts the effective range of drug inhibitory concentrations towards supernatant harvested from induced #6 HIV-1 producer cells as displayed in Fig. 5. A shaded area is not shown in panel B because it would be too narrow to be seen with the range of coordinates depicted on the abscissa.
The shaded area in Fig. 6A represents the concentration range within which the RT inhibitors were tested for inhibition of titer; this span was significantly lower than the concentrations determined to be cytotoxic to the target HeLaT4 cells. Indinavir was incubated with the #6 HIV-1 producer cell line to determine cytotoxicity because that cell type was the focus of its mode of action (Fig. 6B); the CC50 value was 307.1 μM, which was three logs greater than the corresponding IC90 value. Again, the virus-inhibitory concentration was well below the cytotoxic drug concentration.
DISCUSSION
Traditionally, retroviral packaging cell lines and vectors have been utilized to efficiently transfer exogenous genes into cells. This report outlines the underpinning for a novel use of HIV-1-based vector systems as tools for anti-HIV-1 drug screening. An HIV-1 producer cell line mimics most aspects of HIV-1 replication without producing replication-competent virus because of the genetic separation of the viral cis- and trans-acting sequences. Virus particles that can only undergo a single round of replication are produced, and the target cells into which they integrate cannot support the production of progeny virus.
The lentiviral producer cell line described herein produces all of the key HIV-1 elements required for replication, including all of the regulatory, accessory, and structural viral proteins, under ecdysone-inducible control. In the absence of ponasterone A, a synthetic analogue of ecdysone, the cell line does not produce the requisite viral proteins for virus production, providing an additional degree of safety. The nature of a stable cell line is such that the vector sequences are permanently integrated within the 293T cell background, so production of viral particles is more controlled and consistent compared to production of virus via transient-transfection protocols, which are more prone to batch-to-batch variability as well as to recombination events, increasing the likelihood of generating wild-type HIV-1.
Clearly, an important issue is whether the HIV-1 vector system described here affords the necessary sensitivity for identifying inhibitory compounds, since vector virus replication is confined to a single round of replication. Assays with wild-type HIV-1 encompass multiple rounds of replication over the course of days to weeks, permitting amplification of inhibitory effects. The experimental evidence presented here shows that drug inhibitory effects are readily detectable during a single round of replication (Fig. 5).
Well-characterized RT inhibitors (zidovudine, didanosine, stavudine, and lamivudine) and a protease inhibitor (indinavir) were evaluated in both halves of the system, and corresponding decreases in titer were observed. More specifically, and as expected, the RT inhibitors were effective only during the target cell stage which includes the process of reverse transcription, and the protease inhibitor was only effective at the producer cell stage, where it inhibits maturation. Induction in the presence of the RT inhibitors during the producer cell stage did not decrease viral titers from that of untreated induced #6 controls, and likewise, indinavir was ineffective when added during the target cell stage. Indinavir proved to be a more potent inhibitor of viral titer than the four RT inhibitors, since it was effective in nanomolar concentrations when added during viral production (Table 1). Inclusion of compounds during both the producer cell stage and target cell stage permits examination of their inhibitory potential against essentially all known stages of the HIV-1 life cycle that are reproducible in tissue culture. Selectively retesting potentially promising compounds against both the producer cell stage and the target cell stage could help to narrow its mode of action.
For the compounds tested in this report, the levels of each drug determined to be cytotoxic exceeded the concentrations required for their anti-HIV-1 properties, confirming that the observed reduction in titer was due to the drug's effect on viral replication (Fig. 6). The 72-h incubation period for the cytotoxicity assays described herein was selected to correlate with the time required for the producer and target cell stages (Fig. 3, Materials and Methods). Incubation of some of the inhibitory compounds with 293T or HeLaT4 cells beyond 72 h would be expected to cause a more pronounced cytotoxic effect due to cell overgrowth, which would result in lower CC50 values similar to those reported elsewhere with different cell lines. It should be noted that in multiple reports, the compounds were not associated with toxicity at the highest levels assayed (1, 6,15). This procedure was implemented to ensure that short-term toxicity was not affecting producer or target cell viability, leading to an artificial decrease in viral titer.
The results reported here validate further development of HIV-1 vector systems for high-throughput screening and anti-HIV drug discovery. In its present state, this system should already be useful as a rapid, inexpensive, and safe secondary screen to corroborate the antiviral activity of putative HIV-1 inhibitors. However, for use in high-throughput screening, it would probably be prudent to utilize marker genes that are presently more amenable to high-throughput screening than the gfp gene employed in the HIV-GIP transducing vector (Fig. 1), as high-throughput screening typically employs automated microtiter plate readers. Two such genes are the firefly luciferase gene and the gene encoding the secreted form of alkaline phosphatase (3, 7, 9, 10). Preliminary studies with the secreted alkaline phosphatase gene indicate that it has the requisite sensitivity when treated with known HIV-1 inhibitors (A.L. Pacchia and J. P. Dougherty, unpublished results).
In summary, this report serves as a proof of principle that an HIV-1 producer cell system can be used as the basis of a safe and novel virus-cell-based assay that should be adaptable to a high-throughput screening format to screen for a large number of targets, including both well-understood and less well studied aspects of HIV-1 replication. It is noteworthy that such an assay will also screen for compounds that can disrupt interactions between the virus and the host cell, which might represent the most fruitful targets because interference with such interactions may be the most difficult for the virus to circumvent.
Acknowledgments
This work was supported by grants CA50777, AI43886, and AI51910 from the National Institutes of Health. M.E.A. was supported by National Institutes of Health Fellowship HL10084-02.
We thank Mario H. Vargas (Instituto Nacional de Enfermedades Respiratorias, Mexico) for assistance with the calculation of IC90 values and Sayandip Mukherjee for critically reading the manuscript. Antiserum to HIV-1 gp120 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Michael Phelan.
REFERENCES
- 1.Balzarini, J., A. van Aerschot, P. Herdewijn, and E. De Clercq. 1989. 5-Chloro-substituted derivatives of 2′,3′-didehydro-2′,3′-dideoxyuridine, 3′-fluoro-2′,3′-dideoxyuridine and 3′-azido-2′,3′-dideoxyuridine as anti-HIV agents. Biochem. Pharmacol. 38:869-874. [DOI] [PubMed] [Google Scholar]
- 2.Bartz, S. R., M. E. Rogel, and M. Emerman. 1996. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J. Virol. 70:2324-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, J., J. Hauber, R. Hauber, R. Geiger, and B. R. Cullen. 1988. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene 66:1-10. [DOI] [PubMed] [Google Scholar]
- 4.Burns, J. C., T. Friedmann, W. Driever, M. Burrascano, and J. K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90:8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr, A., J. Miller, M. Law, and D. A. Cooper. 2000. A syndrome of lipoatrophy, lactic acidaemia and liver dysfunction associated with HIV nucleoside analogue therapy: contribution to protease inhibitor-related lipodystrophy syndrome. AIDS 14:F25-F32. [DOI] [PubMed]
- 6.Coates, J. A., N. Cammack, H. J. Jenkinson, I. M. Mutton, B. A. Pearson, R. Storer, J. M. Cameron, and C. R. Penn. 1992. The separated enantiomers of 2′-deoxy-3′-thiacytidine (BCH 189) both inhibit human immunodeficiency virus replication in vitro. Antimicrob. Agents Chemother. 36:202-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen, B. R., and M. H. Malim. 1992. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 216:362-368. [DOI] [PubMed] [Google Scholar]
- 8.Daelemans, D., E. De Clercq, and A. Vandamme. 2001. A quantitative GFP-based bioassay for the detection of HIV-1 Tat transactivation inhibitors. J. Virol. Methods 96:183-188. [DOI] [PubMed] [Google Scholar]
- 9.de Wet, J. R., K. V. Wood, M. DeLuca, D. R. Helinski, and S. Subramani. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:725-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wet, J. R., K. V. Wood, D. R. Helinski, and M. DeLuca. 1986. Cloning firefly luciferase. Methods Enzymol. 133:3-14. [DOI] [PubMed] [Google Scholar]
- 11.Drosopoulos, W. C., L. F. Rezende, M. A. Wainberg, and V. R. Prasad. 1998. Virtues of being faithful: can we limit the genetic variation in human immunodeficiency virus? J. Mol. Med. 76:604-612. [DOI] [PubMed] [Google Scholar]
- 12.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by with an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 14.Gorman, C. 1985. High efficiency gene transfer into mammalian cells, p. 143-190. In D. M. Glover (ed.), DNA cloning. IRL Press, Oxford, UK.
- 15.Gu, Z., M. A. Wainberg, N. Nguyen-Ba, L. L'Heureux, J. M. de Muys, T. L. Bowlin, and R. F. Rando. 1999. Mechanism of action and in vitro activity of 1′,3′-dioxolanylpurine nucleoside analogues against sensitive and drug-resistant human immunodeficiency virus type 1 variants. Antimicrob. Agents Chemother. 43:2376-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman, J. S., and P. J. Easterbrook. 2001. The metabolic toxicities of antiretroviral therapy. Int. J. STD AIDS 12:555-562. [DOI] [PubMed] [Google Scholar]
- 17.Isada, C. M. 2001. New developments in long-term treatment of HIV: the honeymoon is over. Cleveland Clin. J. Med. 68:804-807. [DOI] [PubMed] [Google Scholar]
- 18.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, P. S. 1998. Strategies for antiviral drug discovery. Antivir. Chem. Chemother. 9:283-302. [PubMed] [Google Scholar]
- 20.Kafri, T., U. Blomer, D. A. Peterson, F. H. Gage, and I. M. Verma. 1997. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat. Genet. 17:314-317. [DOI] [PubMed] [Google Scholar]
- 21.Kakuda, T. N. 2000. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin. Ther. 22:685-708. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan, A. H., and R. Swanstrom. 1991. The HIV-1 gag precursor is processed via two pathways: implications for cytotoxicity. Biomed. Biochim. Acta 50:647-653. [PubMed] [Google Scholar]
- 23.Kaul, M., H. Yu, Y. Ron, and J. P. Dougherty. 1998. Regulated lentiviral packaging cell line devoid of most viral cis-acting sequences. Virology 249:167-174. [DOI] [PubMed] [Google Scholar]
- 24.Kim, V. N., K. Mitrophanous, S. M. Kingsman, and A. J. Kingsman. 1998. Minimal requirement for a lentivirus vector based on human immunodeficiency virus type 1. J. Virol. 72:811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klages, N., R. Zufferey, and D. Trono. 2000. A stable system for the high-titer production of multiply attenuated lentiviral vectors. Mol. Ther. 2:170-176. [DOI] [PubMed] [Google Scholar]
- 26.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 27.Koo, H. M., A. M. Brown, R. J. Kaufman, C. M. Prorock, Y. Ron, and J. P. Dougherty. 1992. A spleen necrosis virus-based retroviral vector which expresses two genes from a dicistronic mRNA. Virology 186:669-675. [DOI] [PubMed] [Google Scholar]
- 28.Ledergerber, B., M. Egger, M. Opravil, A. Telenti, B. Hirschel, M. Battegay, P. Vernazza, P. Sudre, M. Flepp, H. Furrer, P. Francioli, and R. Weber. 1999. Clinical progression and virological failure on highly active antiretroviral therapy in HIV-1 patients: a prospective cohort study. Swiss HIV Cohort Study. Lancet 353:863-868. [DOI] [PubMed] [Google Scholar]
- 29.Levy, J. A. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57:183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333-348. [DOI] [PubMed] [Google Scholar]
- 31.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris, A. A., and A. Carr. 1999. HIV nucleoside analogues: new adverse effects on mitochondria? Lancet 354:1046-1047. [DOI] [PubMed] [Google Scholar]
- 33.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 34.Pacchia, A. L., M. E. Adelson, M. Kaul, Y. Ron, and J. P. Dougherty. 2001. An inducible packaging cell system for safe, efficient lentiviral vector production in the absence of HIV-1 accessory proteins. Virology 282:77-86. [DOI] [PubMed] [Google Scholar]
- 35.Parkman, R., K. Weinberg, G. Crooks, J. Nolta, N. Kapoor, and D. Kohn. 2000. Gene therapy for adenosine deaminase deficiency. Annu. Rev. Med. 51:33-47. [DOI] [PubMed] [Google Scholar]
- 36.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 37.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 38.Sodroski, J., W. C. Goh, C. Rosen, K. Campbell, and W. A. Haseltine. 1986. Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. Nature 322:470-474. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, N., B. G. Stewart, J. C. Moore, R. L. Greasham, D. K. Robinson, B. C. Buckland, and C. Lee. 2000. Directed evolution of toluene dioxygenase from Pseudomonas putida for improved selectivity toward cis-indandiol during indene bioconversion. Metab. Eng. 2:339-348. [DOI] [PubMed] [Google Scholar]