Abstract
The genetic basis of antibiotic resistance in 113 unrelated group B streptococci was studied by PCR. Ninety-four strains were resistant to tetracycline-minocycline, and tet(M) was detected in 85% of these isolates. Seventeen erythromycin-resistant strains contained the erm(B), erm(TR), or mef(A) gene. Eleven strains exhibited high-level resistance to kanamycin due to the presence of the aphA3 gene; eight of these strains were also highly resistant to streptomycin; aad-6-related sequences were detected in seven strains.
Group B streptococci (GBS) are the main cause of neonatal infections. Intrapartum antibiotic prophylaxis is recommended for colonized women to prevent neonatal GBS disease, and aminopenicillin is recommended as the first-line antibiotic (1). Penicillin remains highly active, and resistance to this agent has not been reported. In the case of β-lactam allergy, clindamycin (CM) or erythromycin (EM) is a therapeutic alternative (1). Most isolates are resistant to tetracycline (TC), and resistance to EM and related antibiotics has emerged in recent years in several countries (2, 9, 10, 13, 19).
The first aim of this work was to assess the prevalence of antibiotic resistance in GBS strains isolated in a 700-bed Parisian general hospital (Necker-Enfants Malades) between January 1998 and December 1999. Beta-hemolytic colonies and suspected nonhemolytic colonies were identified as GBS by using a commercial latex agglutination test (bio-Mérieux, Marcy l'Etoile, France). They were serotyped by agglutination with commercial kits purchased from Bio-Rad (Marnes la Coquette, France) and Dako Corporation (Carpinteria, Calif.). All strains were stored at −80°C in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) containing 20% glycerol until being tested. Screening for antibiotic resistance was first carried out by the disk diffusion method with Mueller-Hinton plates containing 5% sheep blood (Bio-Rad) in accordance with the guidelines of the Comité de l'Antibiogramme de la Société Française de Microbiologie (8).
Between January 1998 and December 1999, 664 GBS strains were isolated from different patients. These strains were recovered from urine samples (n = 295), from genital specimens of pregnant women (n = 75) and nonpregnant adults (n = 111), from gastric fluid or ear specimens of colonized or infected newborns (n = 89), from pus of different origins (n = 74), from blood cultures (n = 17), and from cerebrospinal fluid (n = 3). During this 2-year period, the rates of antibiotic resistance remained stable. The percentages of strains resistant to TC, macrolides-lincosamides-streptogramin B (MLS), and high levels of kanamycin (KM) were 85, 15, and 10, respectively. All strains were susceptible to penicillin, ampicillin, cefotaxime, vancomycin, teicoplanin, and rifampin and exhibited low-level resistance to gentamicin. The distribution of GBS capsular serotypes was also stable during our study: Ia, 15%; Ib, 6%; II, 8%; III, 39%; V, 20%; nongroupable, 12%. It has been shown that the distribution of GBS capsular serotypes is influenced by the type of infection (noninvasive versus invasive) and the patient type (neonate versus adult) (4). Accordingly, the 20 strains of our collection isolated from blood cultures (n = 17) or cerebrospinal fluid (n = 3) belong to either serotype III (75%) or V (25%). Since 1990, serotypes III, I, and V have been the most prevalent serotypes recovered from invasive infections (4). Serotype V was the most common serotype recovered from nonpregnant adults with invasive GBS disease and the second and third most common serotype recovered from pregnant women and neonates with early-onset disease (4). Therefore, our results reflect these contemporary trends. No association between sample origin and susceptibility to antibiotics was found. However, 45% of the MLS-resistant strains belonged to type V. This finding confirms previous data (12) suggesting that antibiotic resistance is more frequently encountered in strains of serotype V than in strains of other serotypes.
The second aim of our study was to determine, by PCR amplification, the genetic basis of antibiotic resistance in GBS. This was done with a sampling of 113 unrelated isolates representative of the 664 GBS strains isolated during the 2-year survey period. These 113 GBS strains were all isolates excluding duplicates obtained during a 4-month period from urine samples (n = 50), genital specimens from pregnant women and nonpregnant adults (n = 33), gastric fluid or ear specimens from colonized or infected newborns (n = 15), pus from different origins (n = 12), blood cultures (n = 2), and cerebrospinal fluid (n = 1). The antibiotic resistance rate of this sampling was similar to that observed during the 2-year survey, and the serotype distribution was almost identical to that of the 664 strains isolated during the 2-year period (Ia, 14%; Ib, 8%; II, 9%; III, 37%; V, 18%; nongroupable, 14%).
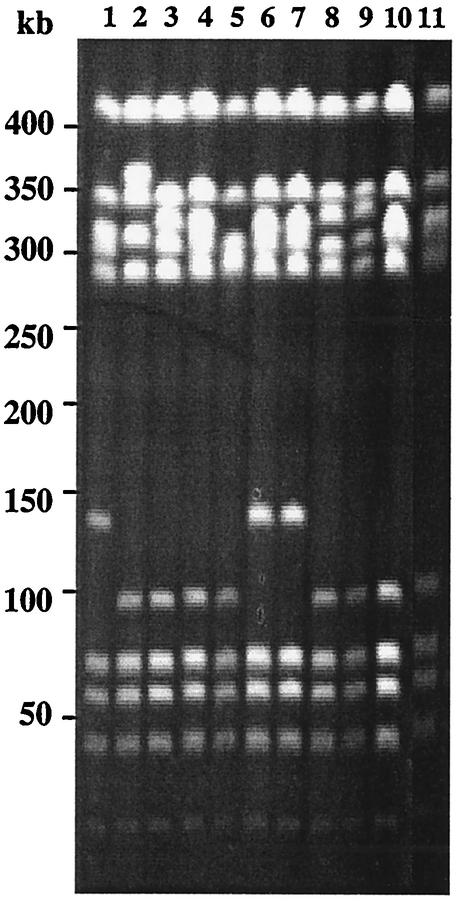
We first examined the genetic diversity of the 113 selected strains by pulsed-field gel electrophoresis (PFGE) as previously described (22). Chromosomal DNAs were digested with restriction enzyme BssHII or SmaI and separated with a Bio-Rad contour-clamped homogeneous electric field mapper with a switch time of 5 to 35 s for 22 h at a 120° angle with a voltage gradient of 6 V/cm at 14°C. PFGE banding patterns were compared by using a computer system (Biocapt; Vilmer Lourmat). Cluster analysis (unweighted pair group average) was used to calculate similarity and dissimilarity among GBS isolates. A difference was considered significant if the similarity coefficient was <60%. PFGE typing of the 113 strains yielded 19 distinct patterns and a total of 90 pulsotypes (data not shown). Fifty percent of the type V strains were highly related and clustered within the same pattern (Fig. 1 shows part of this analysis). This pattern was almost identical to that of the predominant French type V clone, which is clonally related to the predominant U.S. serotype V GBS strain (Fig. 1) (11, 18, 19).
FIG. 1.
Analysis of SmaI-digested genomic DNAs of 11 serotype V GBS isolates by PFGE Lanes: 1 to 10, strains isolated during this work; 11, predominant French serotype V strain clonally related to the predominant U.S. type V strain.
Screening for antibiotic susceptibility was performed as described above. The resistance phenotypes of EM-resistant GBS strains were determined by the double-disk test with EM and CM disks as previously described (25). Resistance to both EM and CM indicated a constitutive MLS resistance phenotype, and blunting of the CM inhibition zone proximal to the EM disk indicated an inducible MLS resistance phenotype. Resistance to EM and susceptibility to CM with no blunting suggested an M phenotype. The MICs were determined by the agar dilution method in Mueller-Hinton broth, and the plates were incubated under air at 37°C. Categorization was done on the basis of the following Comité de l'Antibiogramme de la Société Française de Microbiologie breakpoints: EM and CM, ≥4 μg/ml, TC and minocycline (MN), >8 μg/ml, penicillin, ≥8 μg/ml, KM and streptomycin (SM), >250 μg/ml (8). All of the strains were susceptible to penicillin, vancomycin, teicoplanin, and rifampin and exhibited low-level resistance to gentamicin (data not shown). The MICs of the antibiotics tested for the 113 strains tested are shown in Table 1. Ninety-three strains (81%) were resistant to TC and MN (TC MIC90 [MIC for 90% of the strains tested], 64 μg/ml; MN MIC90, 32 μg/ml). Seventeen strains (16%) had decreased susceptibility to EM, and 12 were Emr Cmr, indicating an MLS constitutive phenotype (MIC90, >256 μg/ml). Four Emr strains were Emr Cms with blunting, indicating an MLS inducible phenotype (EM MIC90, >4 μg/ml; CM MIC90, 0.125 μg/ml), and one was Emr Cms without blunting, suggesting an M phenotype (EM MIC, 2 μg/ml; CM MIC, 0.064 μg/ml). Eleven strains (9.7%) exhibited high-level resistance to KM (HLR-KM) and SM (HLR-SM) (KM MIC90, >1,024 μg/ml; SM MIC90, >1,024 μg/ml). Six combinations of resistance could be distinguished (Table 2). MLS and aminoglycoside resistances were always associated with TC-MN resistance.
TABLE 1.
Antibiotic susceptibilities of 113 GBS strains
| Antibiotic | MIC50a (μg/ml) | MIC90 (μg/ml) | MIC range |
|---|---|---|---|
| Penicillin | 0.032 | 0.064 | <0.032-0.064 |
| EM | 0.032 | 256 | 0.016->256 |
| CM | 0.016 | 256 | 0.016->256 |
| TC | 32 | 64 | 0.064-256 |
| MN | 16 | 32 | 0.064-64 |
| Gentamicin | 8 | 16 | 8-16 |
| KM | 32 | 32 | 16->1,024 |
| SM | 32 | 32 | 16->1,024 |
| Rifampin | 0.25 | 0.5 | 0.125-1 |
| Vancomycin | 0.25 | 0.5 | 0.25-0.5 |
MIC50, MIC for 50% of the strains tested.
TABLE 2.
Distribution of S. agalactiae strains according to their resistance phenotypes and genotypes
| Resistance phenotype | No. of strains (n = 113) | No. of positive PCR amplifications with specific primers for following resistance gene:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| tet(M) | tet(O) | tet(T) | tet(L) | erm(A/TR) | erm(B) | mef(A) | aphA3 | aad-6 | int-Tn | ||
| Susceptible | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Tcr Mnr | 74 | 67 | 6 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 64 |
| Tcr Mnr Emr | 4 | 3 | 1 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 3 |
| Tcr Mnr Emr Cmr | 5 | 5 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 4 |
| Tcr Mnr Kmr Smr | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 3 |
| Tcr Mnr Emr Kmr Smr | 2 | 2 | 1 | 0 | 0 | 1 | 1 | 0 | 2 | 2 | 2 |
| Tcr Mnr Emr Cmr Kmr Smr | 6 | 2 | 4 | 0 | 0 | 0 | 6 | 0 | 6 | 3 | 5 |
We searched for the presence of antibiotic resistance genes by PCR. Total DNAs were extracted by using the Instagene matrix (Bio-Rad), and PCR amplifications were performed as previously described (22). The primers used to amplify the various TC, EM, KM, and SM resistance determinants and the sizes of the amplicons are listed in Table 3. Amplification of DNA from positive controls with the corresponding primers yielded PCR products of the expected size (data not shown). Two known mechanisms of resistance to TC have been reported in streptococci and enterococci: efflux by proton antiporters [Tet(L) and Tet(K)] and ribosome protection [Tet(M), Tet(O), Tet(S), and Tet(T)] (24). The tet(M) resistance gene was detected in 82 TC-MN-resistant strains (83%), tet(O) was detected alone in 10 strains and in association with tet(M) in 2 strains, and tet(T) was detected alone in 1 strain. The tet(L) determinant, which confers resistance to TC but not MN, was detected in one strain in association with tet(M) and tet(O) (Table 2). The tet(K) and tet(S) determinants were not detected. The tet(T) determinant, originally detected in Streptococcus pyogenes (7), was found for the first time in S. agalactiae. Combination of TC resistance determinants has been reported previously in various gram-positive bacteria and in S. agalactiae (24). tet(M) is the most prevalent resistance determinant, accounting for TC resistance in gram-positive bacteria (24), and is often associated with conjugative elements of the Tn916 family (24). Consistently, the int-Tn gene, encoding the integrase of Tn916, was found in 88% of the strains harboring tet(M). These results demonstrate that TC resistance in GBS is due to the acquisition of Tn916-related transposons. The reason why Tn916-mediated TC resistance is so widely disseminated in GBS, compared to other streptococcal or enterococcal species, remains unclear since these bacteria are exposed to similar antibiotic selective pressures.
TABLE 3.
Primers and control strains used in PCR experiments
| Target gene | Forward and reverse primers (5′-3′) | Amplimer size (bp) | Control strain | Reference |
|---|---|---|---|---|
| tet(M) | GTGGAGTACTACATTTACGAG | 359 | Enterococcus faecalis BM4110::Tn1545 | 21 |
| GAAGCGGATCACTATCTGAG | ||||
| tet(O) | GCGGAACATTGCATTTGAGGG | 538 | Streptococcus anginosus MG23 | 7 |
| CTCTATGGACAACCCGACAGAAG | ||||
| tet(S) | CGCTACATTTGCGAGACTCAG | 569 | Listeria monocytogenes BM4210/pIP811 | 23 |
| GGCTCTCATACTGAATGCCAC | ||||
| tet(T) | CAGTGGGAATATAAGGACACGTC | 644 | Streptococcus pyogenes A498 | 7 |
| CAAGCCTTCTCTACAGCATC | ||||
| tet(K) | GTAGGATCTGCTGCATTCCC | 552 | Staphylococcus aureus RN4220/pT181 | 16 |
| CACTATTACCTATTGTCGC | ||||
| tet(L) | GGATCGATAGTAGCCATGGG | 516 | Listeria monocytogenes BM4212/pIP812 | 23 |
| GTATCCCACCAATGTAGCCG | ||||
| int-Tn | GATGGTATTGATGTTGTAGG | 528 | Enterococcus faecalis BM4110::Tn1545 | 21 |
| GGTCTATATTGACAAGACG | ||||
| erm(B) | GGTAAAGGGCATTTAACGAC | 454 | Enterococcus faecalis BM4110::Tn1545 | 21 |
| CGATATTCTCGATTGACCCA | ||||
| erm(A/TR) | TCAGGAAAAGGACATTTTACC | 423 | Streptococcus agalactiae NEMLJ12 | This work |
| ATACTTTTTGTAGTCCTTCTT | ||||
| erm(C) | TCAAAACATAATATAGATAAA | 649 | Staphylococcus aureus RN4220/pE194 | 14 |
| GCTAATATTGTTTAAATCGTCAAT | ||||
| mef(A) | AGTATCATTAATCACTAGTGC | 328 | Streptococcus agalactiae NEMLJ20 | This work |
| TTCTTCTGGTACTAAAAGTGG | ||||
| mreA | AGACACCTCGTCTAACCTTC | 498 | Streptococcus agalactiae NEM316 | 5 |
| TCTGCAGGTAAGTAAGTGCG | ||||
| aphA3 | GGGGTACCTTTAAATACTGTAG | 848 | Enterococcus faecalis BM4110::Tn1545 | 21 |
| TCTGGATCCTAAAACAATTCATCC | ||||
| aad-6 | AGAAGATGTAATAATATAG | 978 | Listeria monocytogenes BM4210/pIP811 | 23 |
| CTGTAATCACTGTTCCCGCCT |
Fifteen percent of the strains studied exhibited decreased susceptibility to EM. In streptococci, two major mechanisms accounting for resistance to MLS antibiotics are recognized. Cross-resistance to all MLS antibiotics is due to methylation of the 23S rRNA by a methyltransferase encoded by an erm (EM resistance methylase) gene (17). Resistance to 14- and 15-member macrolides is mediated by a proton-dependent active drug efflux system encoded by the mef (macrolide efflux) gene (6). We searched for the presence of sequences related to erm(B), erm(A/TR), erm(C), and mef(A) by PCR. The merA gene, initially described as a macrolide efflux determinant but now known to encode a riboflavin kinase in all GBS strains, was used as a PCR positive control (5). The MLS resistance phenotype was due to the presence of the erm(B) and erm(TR) genes, which were distributed differently in strains expressing resistance constitutively (90.5 and 9.5%, respectively) or inducibly (10 and 90%, respectively). The strain exhibiting an M phenotype harbored the mef(A) gene (Table 2). The erm(C) determinant was not detected. Combinations of EM resistance determinants were not found in our study as previously reported in S. pneumoniae and S. pyogenes and more recently in S. agalactiae (3, 9, 10, 13). Our results confirm that the erm(B) gene and, to a lesser extent, erm(TR) are widely distributed among GBS strains. In contrast, the mef(A) gene is rare among GBS compared to S. pyogenes or other beta-hemolytic streptococci (15). Finally, our study shows that the presence of erm(TR) was encountered significantly (P < 0.02) more frequently in strains belonging to serotype V than in strains of other serotypes (data not shown). This observation may simply reflect the clonality of the type V strains in our sampling.
Eleven strains show HLR-KM, and as previously shown in streptococci and enterococci, this resistance phenotype is due to the presence of aphA3 (20). Among these 11 strains, 8 were also highly resistant to SM and sequences related to the aad-6 gene were detected in 7 strains (Table 2). The SM resistance of the remaining strain was probably due to a chromosomal mutation(s).
In conclusion, our results show that TC resistance due to the presence of tet(M) and conjugative transposons such as Tn916 was detected in at least 80% of the GBS strains studied. Most importantly, EM and CM resistance was detected in 15% of the strains. These results emphasize the need to monitor the epidemiology and genetic basis of antibiotic resistance in GBS.
Acknowledgments
We thank E. Bingen for the gift of clinical isolates and C. Tinsley for critical reading of the manuscript.
This work was supported by the Institut Pasteur (Programme Transversal de Recherche 17), INSERM, and the University Paris V.
REFERENCES
- 1.American Academy of Pediatrics Committee on Infectious Diseases and Committee on Fetus and Newborn. 1997. Revised guidelines for prevention of early-onset group B streptococcal (GBS) infection. Pediatrics 99:489-496. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, J. I., D. J. Diekema, S. K. Hunter, P. R. Rhomberg, M. A. Pfaller, R. N. Jones, and G. V. Doern. 2000. Group B streptococci causing neonatal bloodstream infection: antimicrobial susceptibility and serotyping results from SENTRY centers in the Western Hemisphere. Am. J. Obstet. Gynecol. 183:859-862. [DOI] [PubMed] [Google Scholar]
- 3.Bingen, E., F. Fitoussi, C. Doit, R. Cohen, A. Tanna, R. George, C. Loukil, N. Brahimi, I. Le Thomas, and D. Deforche. 2000. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob. Agents Chemother. 44:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173:365-373. [DOI] [PubMed] [Google Scholar]
- 5.Clancy, J., F. Dib-Hajj, J. W. Petitpas, and W. Yuan. 1997. Cloning and characterization of a novel macrolide efflux gene, merA, from Streptococcus agalactiae. Antimicrob. Agents Chemother. 41:2719-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clancy, J., J. W. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 7.Clermont, D., O. Chesneau, G. De Cespédès, and T. Horaud. 1997. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob. Agents Chemother. 41:112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comité de l'Antibiogramme de la Société Française de Microbiologie. 1999. Communiqué 1999. Société Française de Microbiologie, Paris, France.
- 9.de Azavedo, J. C., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Mouy, D., J. D. Cavallo, R. Leclercq, and R. Fabre. 2001. Antibiotic susceptibility and mechanisms of erythromycin resistance in clinical isolates of Streptococcus agalactiae: French multicenter study. Antimicrob. Agents Chemother. 45:2400-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, J. A., K. D. Farmer, and R. R. Facklam. 1998. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J. Clin. Microbiol. 36:2115-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez, M., M. E. Hickman, and C. J. Baker. 1998. Antimicrobial susceptibilities of group B streptococci isolated between 1992 and 1996 from patients with bacteremia or meningitis. Antimicrob. Agents Chemother. 42:1517-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitoussi, F., C. Doit, P. Geslin, N. Brahimi, and E. Bingen. 2001. Mechanisms of macrolide resistance in clinical pneumococcal isolates in France. Antimicrob. Agents Chemother. 45:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horinouchi, S., and R. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 150:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataja, J., P. Huovinen, M. Skurnik, The Finnish Study Group for Antimicrobial Resistance, and H. Seppälä. 1999. Erythromycin resistance genes in group A streptococci in Finland. Antimicrob. Agents Chemother. 43:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan, S. A., and R. P. Novick. 1983. Complete nucleotide sequence of pT181, a tetracycline resistance plasmid from Staphylococcus aureus. Plasmid 10:251-259. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Thomas-Bories, I., F. Fitoussi, P. Mariani-Kurkdjian, J. Raymond, N. Brahimi, P. Bidet, V. Lefranc, and E. Bingen. 2001. Clonal relationship between U.S. and French serotype V group B streptococcus isolates. J. Clin. Microbiol. 39:4526-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, F. Y., P. H. Azimi, L. E. Weisman, J. B. Philips III, J. Regan, P. Clark, G. G. Rhoads, J. Clemens, J. Troendle, E. Pratt, R. A. Brenner, and V. Gill. 2000. Antibiotic susceptibility profiles for group B streptococci isolated from neonates, 1995-1998. Clin. Infect. Dis. 31:76-79. [DOI] [PubMed] [Google Scholar]
- 20.Ounissi, H., E. Derlot, C. Carlier, and P. Courvalin. 1990. Gene homogeneity for aminoglycoside-modifying enzymes in gram-positive bacteria. Antimicrob. Agents Chemother. 34:2164-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poyart, C., J. Celli, and P. Trieu-Cuot. 1995. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrob. Agents Chemother. 39:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poyart, C., C. Pierre, G. Quesne, B. Pron, P. Berche, and P. Trieu-Cuot. 1997. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother. 41:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poyart-Salmeron, C., C. Carlier, P. Trieu-Cuot, A. L. Courtieu, and P. Courvalin. 1990. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 335:1422-1426. [DOI] [PubMed] [Google Scholar]
- 24.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 25.Seppälä, H., A. Nissinen, Q. Yu, and P. Huovinen. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J. Antimicrob. Chemother. 32:885-891. [DOI] [PubMed] [Google Scholar]