Abstract
Actinoplanes friuliensis produces the lipopeptide antibiotic friulimicin. This antibiotic is active against gram-positive bacteria such as multiresistant Enterococcus and Staphylococcus strains. It consists of 10 amino acids that form a ring structure and 1 exocyclic amino acid to which an acyl residue is attached. By a reverse genetic approach, biosynthetic genes were identified that are required for the nonribosomal synthesis of the antibiotic. In close proximity two genes (glmA and glmB) were found which are involved in the production of methylaspartate, one of the amino acids of the peptide core. Methylaspartate is synthesized by a glutamate mutase mechanism, which was up to now only described for glutamate fermentation in Clostridium sp. or members of the family Enterobacteriaceae. The active enzyme consists of two subunits, and the corresponding genes overlap each other. To demonstrate enzyme activity in a heterologous host, it was necessary to genetically fuse glmA and glmB. The resulting gene was overexpressed in Streptomyces lividans, and the fusion protein was purified in an active form. For gene disruption mutagenesis, a host-vector system was established which enables genetic manipulation of Actinoplanes spp. for the first time. Thus, targeted inactivation of biosynthetic genes was possible, and their involvement in friulimicin biosynthesis was demonstrated.
In recent years an increasing number of multiresistant pathogens have been observed in chemotherapy and in the medical treatment of bacterial infections. Strains of methicillin resistant gram-positive Enterococcus and Staphylococcus strains that are also resistant or intermediately resistant to the glycopeptide antibiotic vancomycin have been described (29). Therefore, the development of new and potent antibiotics is a real necessity. In a screen for new antibiotics with activities against multiresistant gram-positive bacteria, a group of eight lipopeptides was isolated from the rare actinomycete Actinoplanes friuliensis (2). The target of the antibiotics is bacterial cell wall synthesis, which is probably inhibited by an antibiotic-mediated complexation of the carrier bactoprenylphosphate (H. Decker, personal communication).
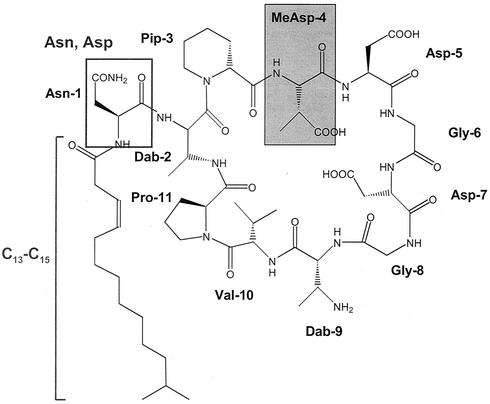
Elucidation of the structure of the antibiotic revealed that all lipopeptides possess an identical macrocyclic peptide as the central element. It is N-terminally linked via diamino butyric acid either to an acylated asparagine or aspartic acid residue (Fig. 1) (56). Whereas the structures of four lipopeptides (A 1437A, A 1437B, A1437E, and A 1437G) are identical to those of known peptide antibiotics such as amphomycin, tsushimycin, and aspartomycin, the other four lipopeptides represent a new class of antibiotics called friulimicins A to D (56).
FIG. 1.
Chemical structure of the lipopeptide antibiotic friulimicin B from A. friuliensis. The positions of the amino acid residues are marked by numbers. Dab, diamon butyrate; Pip, pipecolinic acid; Asn, asparagine; Asp, aspartate; Gly, glycine; Val, valine, Pro, proline. The unusual amino acid methylaspartate (MeAsp) is marked by a grey box.
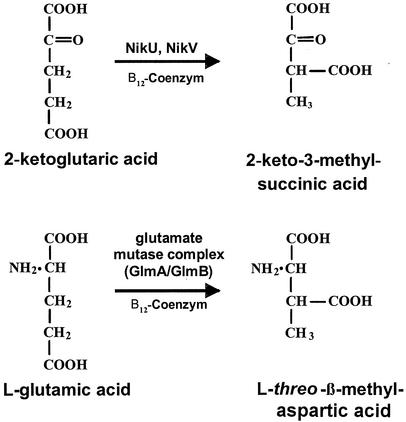
In addition to proteinogenic amino acids, the peptide core of friulimicin is characterized by the existence of the unusual amino acids diamino butyric acid, pipecolinic acid, and methylaspartic acid (L-threo-β-methylaspartic acid) (56). Methylaspartic acid is normally found as an intermediate of the mesaconate pathway for (S)-glutamate fermentation in Clostridium spp. (7) and in members of the family Enterobacteriaceae (19). It is produced by the reversible rearrangement of l-glutamate, which is catalyzed by the adenosylcobamide (coenzyme B12)-dependent enzyme glutamate mutase. The investigation of the glutamate mutase in Clostridium cochlearium (16) has revealed that the active enzyme consists of two subunits (GlmS and GlmE) that form a GlmS2GlmE2 tetramer whose assembly is mediated by coenzyme B12. The smaller protein component, GlmS, has been identified to be the coenzyme B12-binding subunit (16).
In addition to glutamate fermentation, a glutamate mutase-like reaction was found in the biosynthesis of the peptidyl nucleoside antibiotic nikkomycin in Streptomyces tendae Tü901 (24). It was speculated that the corresponding glutamate mutase-like proteins NikU and NikV catalyze the isomerization of 2-ketoglutaric acid to 2-keto-3-methylsuccinic acid (Fig. 2) (24).
FIG. 2.
Comparison of the glutamate mutase reaction to the NikU-NikV-catalyzed reaction in nikkomycin biosynthesis from S. tendae.
In this paper, we describe the isolation of the glutamate mutase genes glmA and glmB as part of the biosynthetic gene cluster of friulimicin. The involvement of these genes in antibiotic biosynthesis was verified by gene disruption mutagenesis and by heterologous expression studies in Streptomyces lividans.
MATERIALS AND METHODS
Cloning, restriction mapping, and in vitro manipulation of DNA.
Methods for isolation and manipulation of DNA were as described by Sambrook et al. (44) and Hopwood et al. (17). Restriction endonucleases were purchased from various suppliers and used according to the instructions of those suppliers.
Gene disruption mutagenesis and transformation.
The gene disruption mutant GM18 was generated by intergeneric conjugation between A. friuliensis and Escherichia coli ET12567 (pUB307) with plasmid pMOGM, as described in this paper. Plasmids were introduced in S. lividans by polyethylene glycol (PEG)-mediated transformation of protoplasts, as described by Hopwood et al. (17). Transformation of E. coli was performed by the CaCl2 method described by Sambrook et al. (44). E. coli XL1 Blue was used for standard cloning experiments.
Intergeneric conjugation between E. coli and A. friuliensis.
The vector pDS401 was used for intergeneric conjugation between E. coli and A. friuliensis (Table 1). Methylase-negative strain E. coli ET12567 (pUB307) (14) was used as the E. coli donor strain.
TABLE 1.
Bacterial strains, cosmids, and plasmids
| Strains, phages, and plasmids | Relevant genotype and phenotype | Source or reference(s) |
|---|---|---|
| A. friuliensis | ||
| HAG010964 | Friulimicin-producing wild type | 2 |
| GM18 | Non-friulimicin-producing mutant, Aprar | This study |
| S. lividans T7 | tsr, T7 RNA polymerase gene | Altenbuchner, personal communication |
| E. coli | ||
| BL21 (DE3)pLysS | F−ompT hsdFB (rB−mB−) gal dcm (DE3) pLysS (Camr) | 12, 52 |
| ET12567(pUB307) | F− dam-13::Tn9 dcm-6 hsdM hsdR lacYT, pUB307 | 14 |
| XL1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1, lac [F′ proAB lacIqZΔ, M15Tn10 (Tetr)] | 8 |
| Cosmids | ||
| pOJ446 | Streptomyces-E. coli cosmid shuttle vector, Aprar | 22 |
| 18M80 | pOJ446 carrying approx. 20 kb of the friulimicin biosynthetic gene cluster | This study |
| pKC505 | Streptomyces-E. coli cosmid shuttle vector, Aprar | 40 |
| Plasmids | ||
| pK18mob | pK18 derivative carrying the oriT region of plasmid RP4, Kanr | 46 |
| pDS401 | pK18mob derivative, a PstI-EcoRI fragment of pKC505 containing the apramycin resistance gene cloned in the MscI site | This study |
| pMOGM | pDS401 carrying an internal 1.3-kb HincII fragment of the glmA-glmB region of the friulimicin biosynthetic gene cluster | This study |
| pOP7 | pK19 carrying a 4.9-kb BamHI fragment of the friulimicin biosynthetic gene cluster | This study |
| pRSETB | T7 promoter expression vector, six-His tag, bla | Invitrogen, Karlsruhe, Germany |
| pEM4 | Streptomyces-E. coli shuttle vector, tsr, PermE | 39 |
| pGM9 | aphII ble tsr, temperature-sensitive Streptomyces vector | 31 |
| pIJ4026 | pUC18 carrying ermE gene from Saccharopolyspora erythraea | 22 |
| pJOE2775cat | E. coli expression vector, rhaP rrnB bla cat | 58 (modified) |
| pWHM3 | Streptomyces-E. coli shuttle vector, trs bla | 55 |
| pEH15 | pK19 carrying the ermE promoter (PermE) as a 0.3-kb BamHI-KpnI fragment | 15 |
| pEHGA1 | pUC18 carrying glmA as a PCR-generated fragment | This study |
| pEHGB1 | pUC18 carrying glmB as a PCR-generated fragment | This study |
| pEHG1 | pUC18 carrying glmA and glmB as a PCR-generated fragment | This study |
| pEHGA2 | pJOE2775cat carrying glmA as NdeI-BamHI fragment of pEHGA1 | This study |
| pEHGB2 | pRSETB carrying glmB as BamHI-HindIII fragment of | This study |
| pEHGB1 | ||
| pEHG2 | pRSETB carrying glm as NdeI-HindIII fragment of pEHG1 | This study |
| pEHGA3 | pRSETB carrying glmA as NdeI-HindIII fragment of pEHGA2 | This study |
| pEHGB3 | pGM9 carrying pEHGB2 as HindIII fragment | This study |
| pEHGA4 | pGM9 carrying pEHGA3 as HindIII fragment | This study |
| pEHG∗1 | pUC18 carrying glmBA∗ as PCR-generated fragment | This study |
| pEHG∗2 | pEHGB2 carrying glmBA∗ as MluI-HindIII fragment | This study |
| pEHG∗3 | pGM9 carrying pEHG∗2 as HindIII fragment | This study |
| pEHEG | pEH15 carrying glm as XbaI-HindIII fragment of pEHG2 | This study |
| pEHK | pGM9 carrying pRSETB as HindIII fragment | This study |
| pCH93 | pUC18 carrying nikU/nikV as a 1.6-kb BamHI-StyI fragment | 24 |
The A. friuliensis recipient strain was cultivated in 100 ml of tryptic soy broth (TSB) medium (Bacto tryptic soy broth; Becton, Dickinson & Co, Sparks, Md.) in a 500-ml baffled Erlenmeyer flask which was inoculated with 1 ml of a homogenized stationary-phase culture. The incubation was carried out at 30°C for 5 days on a rotary shaker at 180 rpm. The culture was diluted 1 to 10 and further incubated for 16 h under the same conditions. Then, the mycelium was homogenized and diluted 1 to 5 (10 ml TSB plus 2.5 ml of mycelium), and incubation was carried out for from 1 h to a maximum of 5 h under the conditions described above. For the conjugation approach, the cells were pelleted by centrifugation (5,000 × g, 10 min) and resuspended in 2 ml of TSB. A total of 200 μl of A. friuliensis cells (approximately 108 cells/ml) and 200 μl of the E. coli donor strain (a washed overnight culture; approximately 108 cells/ml) were mixed and then spread onto three medium 65 plates (4 g of glucose, 4 g of yeast extract, 10 g of malt extract, 2 g of CaCO3, 12 g of agar [pH 7.2]; according to the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). After 24 to 30 h, the plates were overlaid with 3 ml of R2L soft agar (60) containing spectinomycin (A. friuliensis is intrinsically resistant to spectinomycin) and apramycin (final concentrations, 50 and 100 μg/ml, respectively). After 7 to 10 days, the number of transconjugants was determined.
Southern hybridization.
Southern hybridization was carried out with the nonradioactive DIG DNA Labeling and Detection kit from Roche (Mannheim, Germany). Hybridization experiments with the nikU and nikV genes as a probe were performed at 68°C and a stringent washing step with 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS).
DNA sequencing and analysis.
DNA fragments containing friulimicin biosynthetic genes were subcloned in the sequencing vectors pK19, pUC18, and pBluesript SK(+). The DNA sequences were determined by standard techniques (45). The DNA fragments were examined for open reading frames with the codon usage program described elsewhere (6, 51). The programs BLAST (1), CLUSTAL W (53), Genedoc (33), and Treeview (36) were used for homology searches, multiple-sequence alignments, and construction of a phylogenetic tree.
Isolation of peptide synthetase genes by PCR.
In order to identify the peptide synthetase genes involved in the synthesis of friulimicin, a PCR approach was carried out with primers derived from conserved core motifs of peptide synthetases (26): primers oligo2 (TTC ACS TCS GGS TCS ACS GGS; core motif 2, FTSGSTG), oligo5 (ATC GAG CTS GGS GAG ATC GAG; core motif 5, IELGEIE), and oligo6 (SGA GTG SCC SCC SAG CTC GAA; core motif 6, FELGGHS). The following reaction mixture was used: 1 μg of chromosomal DNA from A. friuliensis, 1.0 μM each primer (oligo2-oligo6 and oligo5-oligo6), 10 μl of 10× reaction buffer (with 15 mM MgCl2), 5% dimethyl sulfoxide (DMSO), 0.2 mM deoxynucleoside triphosphates, and 0.5 μl of Taq polymerase (Qiagen, Hilden, Germany). After denaturation (3 min, 94°C), 25 cycles of amplification (1 min at 94°C for denaturation, 1.5 min at 55°C for annealing, 2 min at 72°C for amplification) were performed in a PTC100 thermocycler from MJ Research, Inc. (Watertown, Mass.). The PCR products were electrophoretically separated in a 1% agarose gel, isolated by gel elution (Qiaquick; Qiagen), and directly used for cloning.
To identify peptide synthetase genes in cosmid pools or cosmid clones, the same PCR approach was carried out but with 0.1 μg of DNA used as the template.
Isolation of glmA and glmB by PCR.
glmA and glmB were separately isolated by PCR with pOP7 as the template. pOP7 contains a 4.9-kb BamHI fragment of the cluster carrying glmA and glmB. The following reaction mixture was used: 0.5 μg of pOP7 as the template, 1.0 μM primer 1A (5′-AGA ATT CCA TAT GAA TCT CAC GTA CGC-3′) and primer 2A (5′-AAA GAT CTC GAC GCG ACT GCC GCG C-3′) for glmA amplification and primer 1B (5′-AAG GAT CCG TGA CCG CCG CGG CGC CCT TC-3′) and primer 2B (5′-AAT AAG CTT TCA TGG TGC TCC TTC GTC GTA-3′) for glmB amplification (the sequences of the restriction sites used for cloning are underlined), 10 μl of 10× reaction buffer (containing 20 mM MgCl2), 5% DMSO, 0.2 mM deoxynucleoside triphosphates, and 1 μl of Pwo polymerase (Roche). After denaturation (5 min, 95°C), 25 cycles of amplification (5 min at 95°C, 2 min at 61°C [glmA] and 64°C [glmB], 1.5 min at 72°C) were performed. The PCR products were separated electrophoretically in a 1% agarose gel, isolated by gel elution, and cloned into the sequencing vector pUC18.
Heterologous expression of glmA and glmB.
Luria-Bertani medium (50 ml) with 150 μg of ampicillin per ml in a 100-ml Erlenmeyer flask was inoculated with 0.5 ml of an overnight culture of E. coli XL1 Blue(pEHGA2) and incubated at 37°C and 180 rpm until an optical density at 600 nm of 0.3 was attained. Synthesis of GlmA (the glmA gene was cloned downstream of rhamnose-inducible promoter rhaP of pJOE2775cat) was induced by the addition of 0.2% rhamnose, and the culture was allowed to grow for 5 h. The cells were then harvested by centrifugation at 5,000 × g and 4°C for 10 min. For purification under denaturing conditions, the cells were resuspended and incubated in buffer B (1 h at room temperature) (Ni-NTA Spin kit; Qiagen). For purification under native conditions, the cells were resuspended in lysis buffer (Ni-NTA Spin kit; Qiagen) and were broken twice with a French press (10,000 lb/in2). The insoluble protein fraction was harvested by centrifugation at 13,000 × g for 30 min.
In both cases, soluble proteins were purified with Ni-NTA Spin Columns (Qiagen) according to the protocol of the manufacturer. Expression of glmB and large-scale purification of the His-tagged protein were performed by affinity chromatography by a protocol described by Heinzelmann et al. (15).
Assembly of glmBA by recombinant PCR.
The 5′ terminus of glmA was genetically fused to the 3′ terminus of glmB via a sequence encoding an 11-amino-acid (Gly-Gln)5-Gly linker segment, as described by Chen and Marsh (11). For this, four oligonucleotides were designed: oligo AL1 [5′-(GGACAA)5GGAAATCTCACGTACGCCATTCCGGGC-3′], oligo A2 (5′-AAGCTTTCACGCGACTCGGGCGCT-3′), oligo B1 (5′-AGTGGCACATCGTGCTCTACGGCGTACA-3′), and oligo BL2 [5′-(TCCTTG)5TCCTGGTGCTCCTTCGTCGTACCG-3′]. Oligo A2 primes from the 3′ terminus of glmA (including the stop codon), and it also carries the sequence of an additional HindIII site; oligo AL1 and oligo BL2 were designed to introduce the linker segment at the 3′ terminus of glmB and the 5′ terminus of glmA, respectively; and oligo B1 primes toward the 3′ terminus of glmB upstream of an internal MluI site. A 572-bp DNA fragment of the 3′ region of glmB was amplified with 1 μM oligo B1 and oligo BL2 as the primers and 0.5 μg of plasmid pOP7 as the template, 10 μl of 10× reaction buffer (containing 30 mM MgSO4), 5% DMSO, 0.2 mM deoxynucleoside triphosphates, and 1 μl of Pwo polymerase (Roche). After denaturation (5 min at 95°C), 30 cycles of amplification (1 min at 94°C, 1.5 min at 70°C, and 2 min at 72°C) were performed. The PCR product was designated glmB*.
To amplify glmA, 1 μM each oligo AL1 and oligo A2 were used in the same reaction mixture and by use of the PCR protocol described above for glmB*, but with an annealing temperature of 64°C. The resulting 476-bp glmA fragment was designated glmA*.
In a third PCR, glmA* and glmB* were assembled by using 1 μM each oligo A2 and oligo B1 as primers, 5% DMSO, 10 μl of 10× reaction buffer for herculase, and 1 μl of herculase (Stratagene, La Jolla, Calif.). A 1,010-bp fusion fragment (glmBA*) was generated by using the same PCR protocol described above for the amplification of glmB*. glmBA* was separated by gel electrophoresis and isolated by gel elution. Plasmid pEHGB2 and glmBA* were both restricted with MluI and HindIII, and a 521-bp glmB fragment was exchanged for the 1,010-bp glmBA* fragment, resulting in pEHG*2. Cloning of pEHG*2 as a HindIII fragment in pGM9 resulted in the Streptomyces-E. coli shuttle vector pEHG*3.
Purification of the His-tagged protein.
The purification of the His-tagged proteins from S. lividans T7, which possesses a thiostrepton-inducible T7 RNA polymerase gene (J. Altenbuchner, personal communication), was performed by a procedure described by Heinzelmann et al. (15).
Determination of glutamate mutase activity.
A coupled enzyme assay described by Barker et al. (4) was used to assay for glutamate mutase activity spectroscopically. This test is based on the glutamate mutase-catalyzed formation of l-threo-β-methyl aspartic acid. In a second enzyme reaction that is catalyzed by methylaspartase from Clostridium tetanomorphum (kindly provided by W. Buckel), methylaspartate is converted to mesaconate. The formation of mesaconate is linked to the increase in the extinction coefficient at 240 nm. Because it was shown that the glutamate mutase from A. friuliensis is not active in the recommended buffer (50 mM Tris-HCl [pH 8.3], 10 mM KCl, 1 mM MgCl2), the reaction buffer had to be modified (buffer 2 consisted of 37.5 mM Tris-HCl, 15 mM KCl, 1.25 mM MgCl2, 12.5 mM KH2PO4, 0.0125% β-mercaptoethanol, 0.25 mM dithiothreitol).
Nucleotide sequence accession numbers.
The nucleotide sequence data for the friulimicin biosynthetic genes reported here have been assigned EMBL accession no. AJ488769. The genes used for construction of the alignments and of the phylogenetic tree are deposited in GenBank under accession numbers AF008569, U67612, AJ246005, AJ250581, AP002553, X80997.
RESULTS
Development of a host-vector system for A. friuliensis.
As a prerequisite for molecular genetic experiments with A. friulienesis, it was necessary to develop a host-vector system. To identify selectable markers for the introduction of DNA into A. friuliensis, we first determined the susceptibility of A. friuliensis to different antibiotics at concentrations normally used for streptomycetes. A. friuliensis was resistant to thiostrepton (25 μg/ml), streptomycin (50 μg/ml), and spectinomycin (50 μg/ml) but was sensitive to gentamicin (25 μg/ml), erythromycin (50 μg/ml), kanamycin (50 μg/ml), neomycin (10 μg/ml), hygromycin (50 μg/ml), and apramycin (50 μg/ml). In subsequent transformation experiments, apramycin was used as the most convenient antibiotic.
Because no DNA-uptake system has been described for Actinoplanes spp., we first tested standard streptomycete transformation methods like electroporation (38) and PEG-mediated transformation (22). In these experiments, replicative derivatives of the Streptomyces vectors pIJ101 (21), SCP2 (47), and pSG5 (30) and Amycolatopsis plasmids such as pMEA derivatives (59), as well as nonreplicative plasmids carrying a chromosomal fragment of A. friuliensis, were used. In order to achieve transformation, the following parameters were varied: the regeneration medium (yeast-malt medium, R2YE medium, MS medium [22]), the brand of PEG (PEG 1000, 1500, and 4000 from Serva [Heidelberg, Germany] and Koch & Light [Haverhill, England]), the PEG concentration (20 to 35%), growth conditions, and the DNA preparation (from S. lividans and methylase-negative E. coli strain ET12567) (25). Despite these efforts, no Actinoplanes recombinants were obtained.
An alternative method of DNA uptake, intergeneric conjugation between E. coli and Streptomyces and other actinomycetes, has been described by Mazodier et al. (28) and Wohlleben and Pielsticker (61); and this method can be adapted for many other actinomycetes (14, 27, 57). To increase the conjugation efficiency, methylase-negative E. coli strain ET12567 carrying the transfer genes of plasmid RP4 on plasmid pUB307 (14) was used. In our experiments, fragments of friulimicin biosynthetic genes were subcloned in pDS401 (Table 1), and the strain was transformed with the resulting plasmids. By variation of the incubation times and the regeneration media, an optimized protocol (described in Material and Methods) was developed. The frequency of transconjugant formation per recipient strain was determined to be 10−6 to 10−7.
Identification of putative peptide synthetase genes involved in friulimicin biosynthesis.
In order to identify the peptide synthetase genes that are involved in the nonribosomal synthesis of friulimicin, a PCR approach was performed with primers that were derived from conserved core motifs of peptide synthetases (26). Several internal peptide synthetase gene fragments were found by these experiments. By subsequent screening of a cosmid library from A. friuliensis, the cosmids carrying peptide synthetase genes were identified. By comparison of the restriction patterns obtained with different restriction enzymes, the cosmids could be arranged into two different groups (group 1, six overlapping cosmids; group 2, one cosmid), indicating that A. friuliensis contains at least two different biosynthetic gene clusters of compounds synthesized by nonribosomal peptide synthesis.
Identification of a glutamate mutase gene.
The peptide core of friulimicin contains the unusual amino acid methylaspartate, which is known to be synthesized by glutamate mutases in the mesaconate pathway of glutamate fermentation in Clostridium spp. (7) and members of the family Enterobacteriaceae (19). Therefore, we intended to identify the friulimicin biosynthetic gene cluster by screening further for putative glutamate mutase genes using the cosmids carrying peptide synthetase genes. In a first approach, PCR experiments were performed with primers that were derived from conserved regions of the Clostridium enzymes (primer 1, amino acid positions 172 to 179 of the MutE protein from C. cochlearium; primer 2, amino acid positions 415 to 422 of the MutE protein from C. cochlearium). With both chromosomal DNA of A. friuliensis and DNA of the cosmids, no amplification was observed (data not shown). Therefore, a second approach was carried out by Southern hybridization experiments with the glutamate mutase-like genes nikU-nikV from S. tendae as a probe (kindly provided by C. Bormann). With a 1.6-kb BamHI-StyI fragment of pCH93 carrying the corresponding genes, cosmid clones of both groups (see above) were screened. Whereas all cosmids of group 1 showed no signal, a 4.9-kb BamHI DNA fragment of the cosmid clone of group 2 (named 18 M80) was found to hybridize. In further subcloning and hybridization experiments, a 1.4-kb BamHI-SacI fragment and a 1.9-kb SacI fragment carrying putative glutamate mutase genes were identified (data not shown).
Characterization of glutamate mutase genes glmA and glmB.
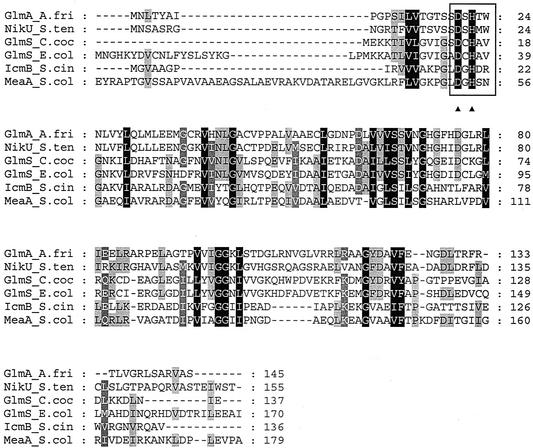
By sequence analysis of the hybridizing fragments, two complete open reading frames (glmA and glmB) encoding proteins of 145 and 415 amino acids were identified. The deduced GlmA and GlmB proteins showed similarities to the subunits of glutamate mutase complex from C. cochlearium, GlmS and GlmE (for GlmA, 17% identity to GlmS; for GlmB, 21% identity to GlmE). GlmA and GlmB were found to have the highest degrees of similarity to the NikU and NikV proteins from S. tendae, with identities of 50 and 52%, respectively. Conserved residues were identified in the deduced amino acid sequence of GlmA. These residues are known to be involved in the binding of cofactor B12 (Fig. 3) (54). The 3′-terminal end of glmA and the 5′-terminal end of glmB overlap by 4 bp, indicating that both genes probably form a single transcriptional unit. Thus, this potential translational coupling, which is characteristic of many bacterial genes whose products are required in equimolar quantities (34), suggests a possible connection between the functions of the products of these two open reading frames.
FIG. 3.
Alignment of protein sequences of the small subunits of cofactor B12-dependent mutases including the glutamate mutase subunit GlmA from A. friuliensis. Conserved amino acid residues are marked by letters in reverse type. First level of shading (black), 100% minimum degree of conservation; second level of shading, 80% minimum degree of conservation; third level of shading, 60% minimum degree of conservation. The characteristic aspartic acid and histidine residues involved in the binding of cofactor B12 are indicated by arrowheads. The cofactor B12-binding region is marked by a box. GlmA_A.fri, glutamate mutase subunit GlmA from A. friuliensis; NikU_S.ten, NikU protein from S. tendae; GlmS_C.coc, glutamate mutase subunit GlmS from C. cochlearium; IcmB_S.cin, isobuytryl-CoA mutase subunit IcmB from Streptomyces cinnamonensis; MeaA_S.col, cofactor B12-binding domain of the MeaA protein from Streptomyces collinus.
Partial characterization of the friulimicin biosynthetic gene cluster.
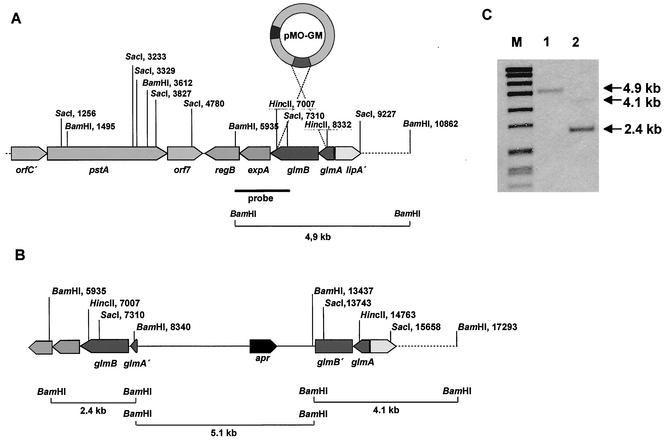
In further experiments, adjacent DNA fragments of glmA-glmB were subcloned, and a DNA sequence of approximately 9.2 kb was determined (Fig. 4A). Two open reading frames of 837 bp (expA) and 903 bp (regB) were found downstream of glmB. These open reading frames are transcribed in the same direction as the glutamate mutase genes. The deduced expA gene product showed similarity to the ATP binding component of ABC transporters (e.g., 41% identity to an ABC transporter component from Streptomyces griseus [9]) (Table 2). The deduced RegB protein is similar to antibiotic biosynthesis regulatory proteins like syrP of the syringomycin biosynthesis from Pseudomonas syringae (51% identity) (62). Next to regB, the complete genes orf7 (906 bp) and pstA (3,186 bp) and the incomplete gene orfC′ were identified downstream of regB. Whereas Orf7 and OrfC′ are similar only to hypothetical proteins of unknown function from Rhizobium sp. strain NGR234 and Mesorhizobium loti, the pstA gene product shows a structure typical of that of a peptide synthetase involved in the nonribosomal synthesis of peptides. The highest similarity was found to the pyoverdine synthetase B from Pseudomonas fluorescens (36% identity). Since PstA lacks a C-terminal condensation domain, it can be speculated that PstA activates the first amino acid (asparagine or aspartic acid) of the peptide antibiotic. By comparing the sequence of the amino acid residues within the substrate binding pocket of the adenylation domain with the specificity-conferring code (as described elsewhere [10, 50]), it was not possible to predict a significant specificity of the deduced PstA protein for asparagine or aspartate or for any other amino acid.
FIG. 4.
Partial characterization of the friulimicin biosynthetic gene cluster and inactivation of the glutamate mutase genes glmA and glmB by gene disruption mutagenesis. (A) Arrangement of genes on a 9.2-kb DNA fragment carrying a part of the friulimicin biosynthetic gene cluster. orfC′, 5′-terminal end of the orfC gene; pstA, peptide synthetase gene; orf7, orf7 gene; regB, transcription regulator gene; expA, gene encoding the membrane part of an ABC transporter; glmB, glutamate mutase gene; glmA, glutamate mutase gene; lipA, acyl coenzyme A synthase gene. (B) Inactivation of glmA-glmB by gene disruption mutagenesis with plasmid pMOGM. The restriction sites used in the subcloning experiments are shown. The hybridizing BamHI DNA fragments are marked by brackets. (C) The correct gene disruption in mutant GM18 shown by Southern hybridization experiments with the 1.4-kb BamHI-SacI fragment as a probe. Lane 1, BamHI digested chromosomal DNA of A. friuliensis wildtype; lane 2, BamHI digested chromosomal DNA of GM18; lane M, molecular weight marker VII (Roche).
TABLE 2.
Genes identified on approximately 9.2 kb of the friulimicin biosynthetic gene cluster from A. friuliensis
| Open reading frame | Match in database | % Identity | Accession no. of homologous proteina | Proposed function |
|---|---|---|---|---|
| orfC | Hypothetical Y4RH protein from Rhizobium sp. strain NGR234 | 28 | NP444046.1 | Unknown |
| pstA | Pyoverdine synthetase B from P. fluorescens | 36 | AF237701 | Nonribosomal peptide synthesis |
| orf7 | Hypothetical protein from M. loti | 41 | NP085770.1 | Unknown |
| regB | SyrP-like protein from Streptomyces avermitilis | 51 | AB070953 | Transcriptional regulator |
| expA | ABC transporter protein, transmembrane component from S. griseus | 41 | AJ300302 | ABC transporter |
| glmB | NikV protein from S. tendae | 52 | CAC11144.1 | 2-Keto-3-methyl succinic acid synthesis |
| glmA | NikU protein from S. tendae | 50 | CAC11143.1 | 2-Keto-3-methyl succinic acid synthesis |
| lipA′ | Putative acyl coenzyme A synthase from Nostoc sp. strain PCC 7120 | 39 | NP 486718.1 | Acyl coenzyme A synthesis |
The amino acid sequences are deposited in Genbank
The incomplete gene lipA was identified upstream of glmA in the direction opposite that of the characterized glmA-glmB region,. The deduced LipA gene product is similar to acyl-CoA synthases from different microorganisms. It can be speculated, that the gene is involved in the synthesis and linkage of the fatty acid part of the antibiotic.
Gene disruption mutagenesis of the glutamate mutase gene.
In order to prove the involvement of the glutamate mutase genes in friulimicin biosynthesis, gene disruption mutagenesis was performed. A 1.3-kb HincII fragment of the glmA-glmB region (Fig. 4A) was subcloned into pDS401, resulting in plasmid pMOGM. By using the host-vector system described above, the glutamate mutase genes were inactivated by integration of the plasmid via homologous recombination of the cloned fragment and the chromosomal copy to generate mutant strain GM18 (Fig. 4B). The correct integration was proved by Southern hybridization (Fig. 4C). Whereas mutant GM18 produced remarkably less friulimicin than the wild type did (the level of production was approximately 40% of that by the wild type), inactivation of the peptide synthetase gene pstA led to the complete loss of friulimicin production (data not shown), indicating that the characterized region represents a part of the friulimicin biosynthetic gene cluster.
Heterologous expression of glmA and glmB in E. coli and S. lividans.
To verify the assumed glutamate mutase activity, we intended to express glmA and glmB in E. coli heterologously. DNA fragments containing the glmA or the glmB gene were generated by PCR with NdeI-BglII or BamHI-HindIII restriction sites at their 3′- and 5′-terminal ends, respectively. Since the cofactor B12-binding site is located at the N-terminal end of glmA (Fig. 3), it is more likely that a His tag at the C-terminal end would have no negative effect on the enzyme activity. Therefore, glmA was cloned as a NdeI-BglII fragment in expression plasmid pJOE2775cat (15, 58) under the control of the rhamnose-inducible promoter rhaP (and by using of the ribosome binding site of vector pRSETB). The resulting plasmid was called pEHGA2 (Table 1).
In contrast, the glmB gene was cloned as a BamHI-HindIII fragment in BglII-HindIII-digested pRSETB, resulting in plasmid pEHGB2 (Table 1). Expression of the gene was under the control of the T7 promoter and resulted in the production of a GlmB protein with an N-terminal His tag (HisglmB).
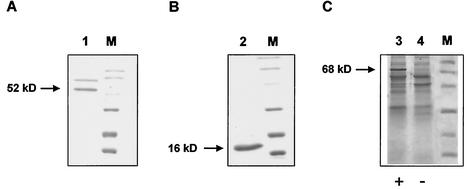
Whereas it was not possible to express hisglmB in E. coli, HisglmA was produced in only an insoluble and inactive form (Fig. 5B). Modifications of the induction conditions and an approach that used renaturation by use of an urea gradient did not result in a soluble GlmA protein (data not shown). Similar problems were also reported for the expression of other Streptomyces genes, e.g., the chloroperoxidase gene from S. lividans (3), the peptide synthetase gene snbC from Streptomyces pristinaspiralis (13), and the aconitase-like gene pmi from Streptomyces viridochromogenes (15), which failed or resulted in inactive proteins in E. coli. Therefore, we proceeded to express both His-tagged genes in S. lividans T7 (Table 1). The hisglmA gene was cloned as an NdeI-HindIII fragment of pEHGA2 9 bp downstream of a conserved ribosome binding site in pRSETB, resulting in plasmid pEHGA3. Then, plasmids pEHGA3 and pEHGB2, carrying hisglmA and hisglmB under the control of the T7 promoter, were cloned as HindIII fragments into vector pGM9, resulting in the Streptomyces and E. coli shuttle plasmids pEHGA4 and pEHGB3, respectively (Table 1). S. lividans T7 was transformed with these plasmids, and after induction with thiostrepton, the production of HisglmA and HisglmB was investigated by an SDS-polyacrylamide gel electrophoresis (PAGE). While it was possible to purify HisglmB by metal chelate affinity chromatography with Ni-NTA resin (Fig. 5A), the production of HisglmA was not detectable under either native or denaturing conditions.
FIG. 5.
Heterologous production of His-tagged GlmA, GlmB, and GlmBA proteins in E. coli and S. lividans T7. Production of glutamate mutase proteins were examined by SDS-PAGE after purification with Ni-NTA agarose (A and B) or in crude cell extracts (C). (A) Production of insoluble HisglmB in E. coli (lane 1); (B) production of HisglmA in S. lividans T7 (lane 2); (C) production of HisglmBA in S. lividans T7 (pEHG*3) (lane 3) and S. lividans T7(pEHK) as a control (lane 4). Lanes M, low molecular weight marker (Bio-Rad).
Fusion of glmA and glmB by recombinant PCR.
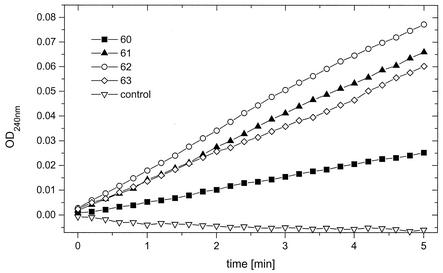
To overcome the problems with the glmA and glmB expression in S. lividans and E. coli, we intended to fuse both genes in order to increase the solubility and stability of the proteins produced. Fusion of the glutamate mutase subunits of C. tetanomorphum was successfully carried out in experiments previously described by Chen and Marsh (11), and this fusion resulted in an active form of the protein. The linkage of glmA and glmB was carried out via recombinant PCR, as described in Materials and Methods. The resulting expression plasmid was named pEHG*3. After transformation of S. lividans T7 with this plasmid and after induction with thiostrepton, overproduction of HisglmBA with a molecular mass of approximately 68 kDa was detected in crude cell extracts (Fig. 5C). During the purification of the corresponding protein by affinity chromatography under native conditions, four fractions (fractions 60, 61, 62, and 63) containing the HisglmBA protein were found. Enzyme activity was detectable by the spectroscopic glutamate mutase assay (Fig. 6), and specific enzyme activities between 0.3 U/mg (fraction 60) and 1.2 U/mg (fraction 62) were calculated within the protein-containing fractions. By a control approach [S. lividans T7(pEHK)], no glutamate mutase activity was detectable in the corresponding fractions (Fig. 6).
FIG. 6.
Determination of the glutamate mutase activity of GlmBA. The GlmBA-containing fractions (fractions 60, 61, 62, and 63) from the protein purification were examined for glutamate mutase activity, as described in Material and Methods. The formation of mesaconate is linked to an increase in the extinction coefficient at 240 nm. In a corresponding fraction of a control approach (S. lividans T7 carrying only the expression plasmid pEHK), no activity was found (control). OD240, optical density at 240 nm.
Together with the results of the genetic experiments described above, the biochemical characterization of the heterologously produced protein verifies the assumed function of GlmA-GlmB as a glutamate mutase involved in providing the unusual amino acid methylaspartate in friulimicin biosynthesis.
DISCUSSION
Several interesting metabolites are synthesized by members of the biotechnologically important genera Actinoplanes: Actinoplanes teichomyceticus and Actinoplanes sp. strain ATCC 33076 produce the antibiotics teicoplanin (49) and ramoplanin (35), respectively; and Actinoplanes sp. strain SE50/110 (U.S. patent 4,062,950; Bayer AG, February 1976) secretes the α-glucosidase inhibitor acarbose. Despite several efforts, up to now neither a host-vector system nor methods for DNA uptake have been developed for any Actinoplanes spp.
The A. friuliensis protocol described in this paper used the principle of intergeneric conjugation between E. coli and gram-positive bacteria, which has successfully been used for the genetic manipulation of streptomycetes and related bacteria (32). The conjugation efficiency of 10−6 to 10−7 for A. friuliensis is comparable to the values reported for the use of integrative plasmids in conjugation experiments with streptomycetes (14). Interestingly, it seems to be possible to adapt the A. friuliensis protocol to other Actinoplanes strains. After slight modifications, it was possible to introduce a targeted mutation in an acarbose biosynthetic gene of Actinoplanes sp. strain SE50/110 by a gene disruption experiment (E. Heinzelmann, unpublished data).
The usefulness of the protocol was proved by the generation of friulimicin biosynthetic mutants. Whereas the peptide synthetase mutant is a null mutant, glutamate mutase mutant GM18 still formed a small inhibition zone in the bioassay. Since the natural precursor methylaspartic acid is not available in the glutamate mutase mutant, it can be speculated that the respective peptide synthetase module may incorporate the structurally similar amino acid aspartic acid to a certain degree. From the chemical analysis of naturally occurring friulimicin derivatives, it is known that a small portion of the antimicrobial lipopeptides contains aspartic acid instead of methylaspartic acid (L. Vértesy, personal communication).
Such reduced specificity leading to side products was described for several peptide synthetases, e.g., the cylosporin synthetase from Tolypocladium niveum (Weber et al., 1994). The small inhibition zone can then easily be explained by a combination of diminished lipopeptide production and reduced antibiotic activity, which has been determined for friulimicin derivatives harboring aspartic acid instead of methylaspartic acid (Vértesy, personal communication).
The occurrence of nonproteinogenic amino acids in nonribosomally synthesized peptides has been documented in many cases (23). Whereas the proteinogenic amino acids are derived from primary metabolism, the biosynthesis of the nonproteinogenic amino acids is part of secondary metabolism. Therefore, the genes encoding the enzymes required for the synthesis of those amino acids are often located in the antibiotic biosynthetic clusters, as reported for dihydroxyphenylglycine (balhimycin biosynthesis) (37), l-pipecolate (rapamycin biosynthesis) (20), and phosphinothricin (phosphinothricin tripeptide biosynthesis) (15, 48). In contrast to the examples mentioned above, which are typically found in secondary metabolites, methylaspartic acid represents a well-known intermediate within the glutamate fermentation of the strictly or facultatively anaerobically growing Clostridium spp. or members of the family Enterobacteriaceae, respectively. The enzymes and genes involved in this process have been well characterized and resemble those of the friulimicin gene cluster. The presence of secondary metabolite-specific glutamate mutase genes which are similar to related primary metabolic genes raises the question about the origins of genes involved in the secondary metabolic pathway. The enzyme recruitment model for evolution of metabolic pathways (18) suggests that enzymes which are somewhat loose in their substrate specificity could initially function in multiple pathways and later evolve into two separate, more specific enzymes. In Streptomyces, the occurrence of secondary metabolism genes that have a similar counterpart in primary metabolism has been described for various proteins such as the acyl carrier protein in actinorhodin and fatty acid biosynthesis in Streptomyces coelicolor (42) or the aconitase-like protein Pmi in Streptomyces viridochromogens (15). Unlike these examples, it seems unlikely that a glutamate mutase involved in primary metabolism is found in actinomycetes. So far, no biochemical evidence for glutamate fermentation has been described for these aerobically growing microorganisms. In addition, no glutamate mutase-like genes were found in the DNA sequence of the S. coelicolor genome (5). The sequence of the A. friuliensis genome is not known, but hybridization with nikU-nikV or glmA-glmB as the probe and chromosomal DNA as the template clearly indicated the presence of only the glutamate mutase genes glmA and glmB in this organism. This excludes the possibility that the friulimicin-specific glutamate mutase genes were derived from a gene duplication in the producing organism. Other cofactor B12-dependent mutases such as isobutyryl coenzyme A mutase (41) and methylmalonyl coenzyme A mutase (63) have been described in Streptomyces strains. These primary metabolic enzymes show structural and functional similarities to glutamate mutases. Whereas isobutyryl coenzyme A mutase consists of two subunits, the methylmalonyl coenzyme A mutase represents one protein that comprises two fused subunits. The amino acid sequences of the subunits or domains, respectively, show only a low level of similarity to the primary structure of the GlmA and GlmB proteins. For example, the cofactor B12-binding protein IcmB from Streptomyces cinnamonensis shows an identity of 16% to the deduced GlmA protein from A. friuliensis. Despite the limited homology to glutamate mutases (and assuming that the isobutyryl coenzyme A mutase and methylmalonyl coenzyme A mutase genes are present in A. friuliensis), it cannot be excluded that the secondary metabolism-specific GlmA and GlmB proteins have been developed from these enzymes during evolution. As an alternative to a gene duplication event, the glmA and glmB genes may have originated from a horizontal gene transfer between actinomycetes and Clostridium spp. or members of the family Enterobacteriaceae. However, DNA characteristics such as the G+C content or codon usage provide no evidence for such an event.
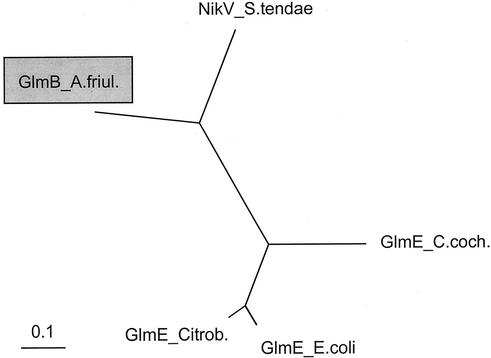
Comparison of the large subunits of glutamate mutases and related enzymes (Fig. 7) yielded the highest degree of similarity for GlmB to the NikV protein from S. tendae. Similar results were also detected by comparison of the small subunits (Fig. 3). These in silico findings were strengthened by a genetic complementation experiment with an S. tendae NikV mutant (NV1) (24) as the recipient. Introduction of glmA and glmB under the control of the constitutive PermE promoter into mutant NV1 restored production of the antibiotic nikkomycin, although at a reduced level compared to that in the wild type (Heinzelmann, unpublished). This indicates that the GlmA and GlmB proteins of A. friuliensis seem to have a relaxed substrate specificity and can catalyze the isomerization of both glutamate and α-ketoglutarate, whereas the highly specific glutamate mutases from Clostridium spp. are unable to carry out the latter reaction (43).
FIG. 7.
Phylogenetic tree of the large subunits of glutamate mutases and related enzymes including GlmB from A. friuliensis. GlmB_A.friul., GlmB from A. friuliensis; GlmE_E.coli, GlmE from E. coli; GlmE_C.coch., GlmE from C. cochlearium, NikV_S.tendae, NikV protein from S. tendae; GlmE_Citrob., GlmE from Citrobacter amalonaticus. The GlmB protein from A. friuliensis is marked by a grey box.
Acknowledgments
This research was supported by DFG (Graduiertenkolleg Mikrobiologie and Graduiertenkolleg Infektionsbiologie) and by BMBF (ZSP Bioverfahrenstechnik, D 3.2 E and GenoMik network). E. Heinzelmann was supported by a grant from the Landesgraduiertenkolleg Baden-Württemberg.
We are very grateful to J. Altenbuchner for providing the S. lividans T7 strain, to W. Buckel for providing the enzyme methylaspartase, and to H. Decker (Aventis) for helpful discussions. We are also grateful to C. Bormann for the suggestion to screen for glutamate mutase genes by using the S. tendae nikU and nikV genes as a probe.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aretz, W., G. Meiwes, G. Seibert, G. Vobis, and J. Wink. 2000. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. I. Taxonomic studies of the producing microorganism and fermentation. J. Antibiot. (Tokyo) 53:807-815. [DOI] [PubMed] [Google Scholar]
- 3.Bantleon, R., J. Altenbuchner, and K. H. van Pée. 1994. Chloroperoxidase from Streptomyces lividans: isolation and characterization of the enzyme and the corresponding gene. J. Bacteriol. 176:2339-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, H. A., V. Rooze, F. Suzuki, and A. A. Iodice. 1964. The glutamate mutase system. J. Biol. Chem. 239:3260-3266. [PubMed] [Google Scholar]
- 5.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bibb, M. J., P. R. Findlay, and M. W. Johnson. 1984. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30:157-166. [DOI] [PubMed] [Google Scholar]
- 7.Buckel, W. 2001. Unusual enzymes involved in five pathways of glutamate fermentation. Appl. Microbiol. Biotechnol. 57:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta galactosidase selection. Focus 5:376-378. [Google Scholar]
- 9.Campelo, A. B., and J. A. Gil. 2002. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 148:51-59. [DOI] [PubMed] [Google Scholar]
- 10.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H.-P., and E. N. G. Marsh. 1997. Adenosylcobalamin-dependent glutamate mutase: examination of substrate and coenzyme binding in a engineered fusion protein possessing simplified subunit structure and kinetic properties. Biochemistry 36:14939-14945. [DOI] [PubMed] [Google Scholar]
- 12.Davanloo, P., A. H. Rosenberg, J. J. Dunn, and F. W. Studier. 1984. Cloning and expression of the gene for bacteriophage T7 polymerase. Proc. Natl. Acad. Sci. USA 81:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Crécy-Lagrad, V., W. Saurin, D. Thibaut, P. Gil, L. Naudin, J. Crouzet, and V. Blanc. 1997. Streptogramin B biosynthesis in Streptomyces pristinaspiralis and Streptomyces virginiae: molecular characterization of the last structural peptide synthetase gene. Antimicrob. Agents Chemother. 41:1904-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 15.Heinzelmann, E., G. Kienzlen, S. Kaspar, J. Recktenwald, W. Wohlleben, and D. Schwartz. 2001. The phosphinomethylmalate isomerase gene pmi, encoding an aconitase-like enzyme, is involved in the synthesis of phosphinothricin tripeptide in Streptomyces viridochromogenes. Appl. Environ. Microbiol. 67:3603-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann, B., R. Konrat, H. Bothe, W. Buckel, and B. Krautler. 1999. Structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium cochlearium. Eur. J. Biochem. 263:178-188. [DOI] [PubMed] [Google Scholar]
- 17.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. John Innes Foundation, Norwich, United Kingdom.
- 18.Irvin, S. D., and J. K. Bhattacharjee. 1998. A unique fungal lysine biosynthesis enzyme shares a common ancestor with tricarboxylic acid cycle and leucine biosynthetic enzymes found in diverse organisms. J. Mol. Evol. 46:4401-4408. [DOI] [PubMed] [Google Scholar]
- 19.Kato, Y., and Y. Asano. 1997. 3-Methylaspartate ammonia-lyase as a marker enzyme of the mesaconate pathway for (S)-glutamate fermentation in Enterobacteriaceae. Arch. Microbiol. 168:457-463. [DOI] [PubMed] [Google Scholar]
- 20.Khaw, L. E., G. A. Bohm, S. Metcalfe, J. Staunton, and P. F. Leadlay. 1998. Mutational biosynthesis of novel rapamycins by a strain of Streptomyces hygroscopicus NRRL 5491 disrupted in rapL, encoding a putative lysine cyclodeaminase. J. Bacteriol. 180:809-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieser, T., D. A. Hopwood, H. M. Wright, and C. J. Thompson. 1982. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol. Gen. Genet. 185:223-228. [DOI] [PubMed] [Google Scholar]
- 22.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 23.Konz, D., and M. A. Marahiel. 1999. How do peptide synthetases generate structural diversity? Cem. Biol. 6:R39-R48. [DOI] [PubMed] [Google Scholar]
- 24.Lauer, B., R. Russwurm, W. Schwarz, A. Kálmánczhelyi, C. Bruntner, A. Rosemeier, and C. Bormann. 2001. Molecular characterization of co-transcribed genes from Streptomyces tendae Tü901 involved in the biosynthesis of the peptidyl moiety and assembly of the peptidyl nucleoside antibiotic nikkomycin. Mol. Gen. Genet. 264:662-673. [DOI] [PubMed] [Google Scholar]
- 25.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 26.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2674. [DOI] [PubMed] [Google Scholar]
- 27.Matsushima, P., M. C. Broughton, J. R. Turner, and R. H. Baltz. 1994. Conjugal transfer of cosmid DNA from Escherichia coli to Saccharopolyspora spinosa: effects of chromosomal insertions on macrolide A83543 production. Gene 146:39-45. [DOI] [PubMed] [Google Scholar]
- 28.Mazodier, P., R. Petter, and C. Thompson. 1989. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 171:3583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moellering, R. C., Jr. 1998. The specter of glycopeptide resistance: current trends and future considerations. Am. J. Med. 104:3S-6S. [DOI] [PubMed]
- 30.Muth, G., W. Wohlleben, and A. Pühler. 1988. The minimal replicon of the Streptomyces ghanaensis plasmid pSG5 identified by subcloning and Tn5 mutagenesis. Mol. Gen. Genet. 211:424-429. [DOI] [PubMed] [Google Scholar]
- 31.Muth, G., B. Nuβbaumer, W. Wohlleben, and A. Pühler. 1989. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol. Gen. Genet. 219:341-348. [Google Scholar]
- 32.Muth, G., D. F. Brolle, and W. Wohlleben. 1998. Genetics of Streptomyces, p. 353-367. In J. E. Davies and A. L. Demain (ed.), Manual of industrial microbiology and biotechnology. ASM Press, Washington, D.C.
- 33.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 34.Normark, S., S. Bergström, T. Edlund, T. Grundström, B. Jaurin, F. P. Lindberg and O. Olsson. 1983. Overlapping genes. Annu. Rev. Genet. 17:499-525. [DOI] [PubMed] [Google Scholar]
- 35.O'Hare, M. D., G. Ghosh, D. Felmingham, and R. N. Gruneberg. 1990. In-vitro studies with ramoplanin (MDL 62, 198): a novel lipoglycopeptide antimicrobial. J. Antimicrob. Chemother. 25:217-220. [DOI] [PubMed] [Google Scholar]
- 36.Page, R. D. H. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 37.Pfeifer, V., G. J. Nicholson, J. Ries, J. Recktenwald, A. B. Schefer, R. M. Shawky, J. Schröder, W. Wohlleben, and S. Pelzer. 2001. A polyketide synthase in glycopeptide biosynthesis: the biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J. Biol. Chem. 276:38370-38377. [DOI] [PubMed] [Google Scholar]
- 38.Pigac, J., and H. Schrempf. 1995. A simple and rapid method of transformation of Streptomyces rimosus R6 and other streptomycetes by electroporation. Appl. Environ. Microbiol. 61:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quiros, L. M., I. Aguirrezabalaga, C. Olano, C. Mendez, and J. A. Salas. 1998. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol. Microbiol. 28:1177-1185. [DOI] [PubMed] [Google Scholar]
- 40.Rao, R. N., M. A. Richardson, and S. Kuhstoss. 1987. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 153:166-198. [DOI] [PubMed] [Google Scholar]
- 41.Ratnatilleke, A., J. W. Vrijbloed, and J. A. Robinson. 1999. Cloning and sequencing of the coenzyme B12-binding domain of isobutyryl-CoA mutase from Streptomyces cinnamonensis, reconstitution of mutase activity, and characterization of the recombinant enzyme produced in Escherichia coli. J. Biol. Chem. 274:31679-31685. [DOI] [PubMed] [Google Scholar]
- 42.Revill, W. P., M. J. Bibb, and D. A. Hopwood. 1996. Relationship between fatty acid and polyketide synthases from Streptomyces coelicolor A3(2): characterization of the fatty acid synthase acyl carrier protein. J. Bacteriol. 178:5660-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roymoulik, I., H.-P. Chen, and E. N. G. Marsh. 1999. The reaction of the substrate analog 2-ketoglutarate with adenosylcobalamin-dependent glutamate mutase. J. Biol. Chem. 274:11619-11622. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., T. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schäfer, A., J. Kalinowski, R. Simon, A.-H. Seep-Feldhaus, and A. Pühler. 1990. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J. Bacteriol. 172:1663-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrempf, H., H. Bujard, D. A. Hopwood, and W. Göbel. 1975. Isolation of covalently closed circular deoxyribonucleic acid from Streptomyces coelicolor A3(2). J. Bacteriol. 121:416-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz, D., J. Recktenwald, S. Pelzer, and W. Wohlleben. 1998. Isolation and characterization of the PEP-phosphomutase and the phosphonopyruvate decarboxylase genes from the phosphinothricin tripeptide producer Streptomyces viridochromogenes Tü494. FEMS Microbial. Lett. 163:149-157. [DOI] [PubMed] [Google Scholar]
- 49.Somma, S., L. Gastelado, and A. Corti. 1984. Teicoplanin, a new antibiotic from Actinoplanes teichomyceticus nov. sp. Antimicrob. Agents Chemother. 26:917-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stachelhaus, T., H. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 51.Staden, R., and A. D. McLachlan. 1982. Codon preference and its use in identifying protein coding regions in large DNA sequences. Nucleic Acids Res. 10:141-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Studier, F. W., and B. A. Moftt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Evol 189:113.. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tollinger, M., C. Eichmüller, R. Konrat, M. S. Huhta, E. N. G. Marsh, and B. Kräutler. 2001. The B12-binding subunit of glutamate mutase from Clostridium tetanomorphum traps the nucleotide moiety of coenzyme B12. J. Mol. Biol. 309:777-791. [DOI] [PubMed] [Google Scholar]
- 55.Vara, J., M. Lewandowska-Skarbek, Y.-G. Wang, S. Donadio, and C. R. Hutchinson. 1989. Cloning of the genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythreae (Streptomyces erythreus). J. Bacteriol. 171:5872-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vértesy, L., E. Ehlers, H., Kogler, M., Kurz, J. Meiwes, G. Seibert, M. Vogel, and P. Hammann. 2000. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. II. Isolation and structural characterization. J. Antibiot. (Tokyo) 53:816-827. [DOI] [PubMed] [Google Scholar]
- 57.Voeykova, T., L. Emelyanova, V. Tabakov, and N. Mkrtumyan. 1998. Transfer of plasmid pTO1 from Escherichia coli to various representatives of the order Actinomycetales by intergeneric conjugation. FEMS Microbiol. Lett. 162:47-52. [DOI] [PubMed] [Google Scholar]
- 58.Volff, J. N., C. Eichenseer, P. Viell, W. Piendl, and J. Altenbuchner. 1996. Nucleotide sequence and role in DNA amplification of the direct repeats composing the amplifiable element AUD1 of Streptomyces lividans 66. Mol. Microbiol. 5:1037-1047. [DOI] [PubMed] [Google Scholar]
- 59.Vrijbloed, J. W., J. Madon, and L. Dijkhuizen. 1994. A plasmid from the methylotrophic actinomycete Amycolatopsis methanolica capable of site-specific integration. J. Bacteriol. 176:7087-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vrijbloed, J. W., J. Madon, and L. Dijkhuizen. 1995. Transformation of the methylotrophic actinomycete Amycolatopsis methanolica with plasmid DNA: stimulatory effect of a pMEA300-encoded gene. Plasmid 34:96-104. [DOI] [PubMed] [Google Scholar]
- 61.Wohlleben, W., and A. Pielsticker. 1989. Investigation of plasmid transfer between Escherichia coli and Streptomyces lividans, p. 301-305. In D. Behrens and A. J. Driesel (ed.), Dechema Bio/Technology Conferences, Vol. 3. VCH Verlagsgesellschaft, Weinheim, Germany.
- 62.Zhang, J. H., N. B. Quigley, and D. C. Gross. 1997. Analysis of the syrP gene, which regulates syringomycin synthesis by Pseudomonas pv. syringae. Appl. Environ. Microbiol. 63:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, W., and K. Reynolds. 2001. MeaA, a putative coenzyme B12-dependent mutase, provides methylmalonyl coenzyme A for monensin biosynthesis in Streptomyces cinnamonensis. J. Bacteriol. 183:2071-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]