Abstract
SPM-1 is a new metallo-β-lactamase recently identified in Pseudomonas aeruginosa strain 48-1997A, isolated in Sao Paulo, Brazil. Kinetic analysis demonstrated that SPM-1 has a broad hydrolytic profile across a wide range of β-lactam antibiotics. Considerable variation was observed within the penicillin, cephalosporin, and carbapenem subfamilies; however, on the whole, SPM-1 appears to preferentially hydrolyze cephalosporins. The highest kcat/Km ratios (in micromolar per second) overall were observed for this subgroup. The hydrolytic profile of SPM-1 bears the most similarity to that of the metallo-β-lactamase IMP-1, yet for the most part, SPM-1 has kcat/Km values higher than those of IMP-1. Zinc chelator studies established that progressive inhibition of SPM-1 by EDTA, dipicolinic acid, and 1-10-o-phenanthroline demonstrated a biexponential pattern in which none of the chelators completely inhibited SPM-1. A homology model of SPM-1 was developed on the basis of the IMP-1 crystal structure, which showed the protein folding and active-site structure characteristic of metallo-β-lactamases and which provides an explanation for the kinetic profiles observed.
The metallo-β-lactamases are a group of clinically important hydrolytic enzymes, not least because of their hydrolysis of the carbapenems and the absence of a clinically useful inhibitor (25). Metallo-β-lactamases have been identified from several environmental and pathogenic bacteria, including Bacillus cereus (41), Bacteroides fragilis (31), Stenotrophomonas maltophilia (35), Chryseobacterium meningosepticum (34, 47), Chryseobacterium indologenes (3, 4), Legionella gormanii (6, 28), Janthinobacterium lividium (33), Aeromonas spp. (26, 45), and Caulobacter crescentus (39).
To date four metallo-β-lactamase crystal structures are available: S. maltophilia L1, B. cereus BCII, B. fragilis CcrA, and the mobile IMP-1 from Pseudomonas aeruginosa (7, 9, 10, 44). All possess the same overall αββα structural fold. In addition, with the exception of the metallo-β-lactamases from Aeromonas spp. (18, 45), all require two zinc ions in the active site for catalytic activity. Furthermore, key residues are conserved, namely, HXHXD (residues 116 to 120) (14). This motif and other residues coordinate zinc ions, which are required for the bridging of water molecules involved in the hydrolytic pathway.
Metallo-β-lactamases encoded by genes carried on mobile genetic elements pose a greater clinical threat than chromosomally encoded enzymes, as these genes have the ability to transfer from one bacterium to another intergenerically (5). Two metallo-β-lactamase genes, imp and vim, have been shown to be mobilized by integrons (19, 24, 29, 38, 48). As many as nine IMP variants have now been identified and isolated from organisms throughout Southeast Asia (8, 17, 20, 49, 50) and more recently in Europe (11, 43) and Canada (15). Three variants of VIM have been documented. VIM-1 and VIM-2 have been identified in strains of P. aeruginosa across Europe (12, 27, 29, 30); VIM-3 was isolated from a P. aeruginosa strain in Taiwan (48). Kinetically, enzymes of the VIM and IMP families have broad substrate profiles and are capable of hydrolyzing penicillins, cephalosporins, and carbapenems (12, 22, 29). The metal chelators EDTA, dipicolinic acid, and 1-10-o-phenanthroline inhibit IMP-1 and VIM-1, and the inhibition exhibits pseudo-first-order inactivation.
Recently, we reported the sequence structure of a new type of metallo-β-lactamase, SPM-1 (42), which, although it has a high level of amino acid identity (35.5%) compared to the sequence of IMP-1, has a unique 23-amino-acid insertion that may influence the enzyme's kinetic profile. SPM-1 has been shown to represent a unique enzyme subclass, and therefore, this report describes the purification and characterization of the recently reported SPM-1 from P. aeruginosa together with the steady-state kinetic profiles for a range of β-lactam substrates and inhibitors. A homology model of the active site of SPM-1 based on the IMP-1 crystal structure is also presented and is used as a hypothesis to explain the kinetics observed.
MATERIALS AND METHODS
Bacterial strain.
The P. aeruginosa strain used in this study, strain 48-1997A, was a clinical isolate obtained from a patient with lymphoblastic leukemia in Sao Paulo, Brazil, as part of the SENTRY program. The strain demonstrated high-level resistance to a range of broad-spectrum β-lactam antibiotics (42).
Antibiotics.
The following antibiotics were used in this study and were acquired from the indicated sources: benzylpenicillin, ampicillin, azlocillin, piperacillin, cephloridine, cefalothin, cefotaxime, cefuroxime, cefoxitin, and moxalactam, Sigma-Aldrich (Poole, United Kingdom); ticarcillin, ceftazidime, and clavulanic acid, GlaxoSmithKline (Greenford, United Kingdom); imipenem, Merck Sharp & Dohme (Huddesdon, United Kingdom); meropenem, Zeneca (Cheshire, United Kingdom); cefepime and aztreonam, Bristol-Myers Squibb (Wallingford, Conn.); nitrocefin, Becton Dickinson (Cockeysville, Md.); and tazobactam, Wyeth (Maidenhead, United Kingdom).
Analytical IEF.
Purified protein was subjected to isoelectric focusing (IEF) as described by Bellais et al. (3). Isoelectric points were estimated by comparison to those for reference proteins by using a pI 3 to 10 calibration kit (Bio-Rad, Watford, United Kingdom).
Purification of SPM-1.
SPM-1 was purified from P. aeruginosa strain 48-1997A (42). An overnight culture (100 ml) of the cells was diluted in 4 liters of nutrient broth and grown aerobically for 18 h at 37°C with orbital shaking. The cells were harvested by centrifugation (3,500 × g for 20 min at 4°C) and then lysed by resuspension of the pellets in 100 ml of Bugbuster (Novagen, Nottingham, United Kingdom) at pH 6.0. The cell debris was removed by centrifugation (9,000 rpm for 20 min at 4°C). Metallo-β-lactamase activity was purified from the resulting supernatant by the method of Avison et al. (2). During purification carbapenemase activity was monitored by using 150 μM meropenem as a substrate in 50 mM cacodylate buffer containing 100 μM ZnCl2 (pH 7.0) at 20°C.
Protein separation techniques.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by the procedure described by Laemmli (21) with a Mini-Protean II apparatus (Bio-Rad).
Mass spectrometric determination of molecular weight.
A solution of approximately 20 μM enzyme was prepared for mass spectrometry as described by Winston and Fitzgerald (46). Matrix-assisted laser desorption ionization-time of flight mass spectrometry was performed with a Voyager DE-PRO/STR mass spectrometer (Perkin-Elmer Instruments, Warrington, United Kingdom).
N-terminal sequencing.
Protein was prepared for N-terminal sequencing by the same procedure used for mass spectrometry. The N-terminal sequence of the protein was determined with a 477A amino acid analyzer (Applied Biosystems, Foster City, Calif.).
Zinc assay.
Purified SPM-1 was dialyzed against 500 volumes of zinc-free 50 mM cacodylate buffer (pH 6.0) for 2 days with two changes of buffer. The resulting enzyme was concentrated to approximately 20 μM with a Centricon column (molecular weight cutoff, >10,000; Millipore Corporation, Watford, United Kingdom). The concentrated protein was acid hydrolyzed with an equal volume of HNO3 (Aristar grade; BDH) at 20°C overnight and analyzed with a Unicam 919 atomic absorption spectrometer at 213.9 nm.
Determination of kinetic parameters.
The concentration of each substrate used was determined by incubating 0.34 μM (50 μl) purified enzyme with the substrate and allowing hydrolysis to continue until completion. Hydrolysis was measured by observing the changes in absorption as a result of the opening of the β-lactam ring of the antibiotic and was measured with a Lambda 35 spectrophotometer (Perkin-Elmer, Cambridge, United Kingdom). The extinction coefficients and wavelengths used in the assays were those described previously (2, 12, 13, 22). Substrate hydrolysis by the purified enzyme was ascertained by observing the changes in absorbance for a variety of substrates at a range of concentrations, and the steady-state kinetic parameters Km (in micromolar) and kcat (in seconds−1) were deduced from the initial rates of hydrolysis by using the Hanes-Woolf plot (37).
For each assay the total reaction volume was 2,050 μl (2 ml of substrate and 50 μl of purified enzyme). All assays were carried out at 20°C in 50 mM cacodylate buffer containing 100 μM ZnCl2 (pH 7.0).
The inhibition of SPM-1 by aztreonam and the serine β-lactamase inhibitor clavulanic acid was investigated by separately combining 15 μM aztreonam and 20 μM clavulanic acid with a range of nitrocefin concentrations (3 to 100 μM). A total of 50 μl (0.34 μM) of purified SPM-1 was added to each mixture, and initial rates of nitrocefin hydrolysis were measured. The final reaction volume for each assay was 1,050 ml, and the conditions for the study were the same as those described above for the kinetic studies. The Ki of each substrate was deduced by determining Km by using the Eadie-Hofstee plot and applying equation 1 (37):
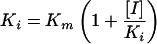 |
(1) |
where [I] is the concentration of the inhibitor.
Inhibition by chelating agents.
The chelating agents EDTA, dipicolinic acid, and 1-10-o-phenanthroline were purchased from Sigma-Aldrich. The progressive inhibition of SPM-1 by EDTA (200 μM), dipicolinic acid (200 μM), and 1-10-o-phenanthroline (200 μM) was investigated by analysis of the time-dependent inhibition of hydrolysis of 100 μM nitrocefin, as performed by Laraki et al. (22) and Franceschini et al. (12) for the IMP-1 and VIM-1 metallo-β-lactamases, respectively. The reaction conditions used for this study were the same as those used for the kinetic studies described above, except that zinc-free buffer was used.
Homology modeling of the SPM-1 metallo-β-lactamase.
The level of sequence identity between SPM-1 and IMP-1 (35.5%) allowed a homology model of SPM-1 to be created (40). The primary sequence of SPM-1 was threaded into the IMP-1 single-chain crystal structure (chain A; protein database [pdb] identification number, 1JJE) (9) by using the program Swiss Protein model (16) and the alignment in Fig. 1. The loop region from R113 to S131 was not modeled, as there was no comparable sequence in the IMP-1 crystal structure. Side-chain torsion angles were altered manually to avoid steric clashes and unfavorable φ-ϕ orientations. The resultant molecule was energy minimized by using the program Gromos96 with parameter set 43B1 (36) until the average absolute derivative of coordinates with respect to energy fell below 0.01 kcal Å−1.
FIG. 1.
Alignment used to generate the homology model of SPM-1. Asterisks denote identical residues in IMP-1 and SPM-1, and dots denote residues of similar function in IMP-1 and SPM-1. Residues for the IMP-1 crystal structure of the sequence with pdb identification number 1JJE are numbered in accordance with the Swiss Protein pdb file. The sequence with pdb identification number 2BBK that was used to model the inserted loop region (SPM-1 residues 113 to 131) was LLVDQRDGWRHKTASRFV (residues 283 to 301) in the pdb file.
RESULTS
Purification and structural properties of the SPM-1 enzyme.
IEF carried out with a crude protein extract showed the pI of SPM-1 to be 7.5 (data not shown). SDS-PAGE analysis showed purified SPM-1 to have a purity of 99.5% and an approximate molecular mass of just under 30 kDa (Fig. 2). Mass spectrometry data for SPM-1 gave a molecular mass of 27.517 kDa (predicted molecular mass, 27.514 kDa). The N-terminal amino sequence of SPM-1 was SDHVDLPYNL, which is in precise agreement with the predicted amino acid sequence of the mature protein. The Zn content was 1.48 μM per μM protein.
FIG. 2.
SDS-PAGE of the purified SPM-1 metallo-β-lactamase. Lane 1, molecular weight markers (in kilodaltons; Broad Range Molecular Weight Standards; Bio-Rad) are indicated by arrows; lane 2, purified SPM-1 metallo-β-lactamase as eluted from the gel filtration column.
Kinetic parameters of SPM-1.
The kinetic parameters of SPM-1 encompassing Km, kcat, and kcat/Km ratios were derived for a series of representative β-lactam antibiotics and serine β-lactamase inhibitors, as shown in Table 1. Among the cephalosporins, the lowest Km values for SPM-1 were seen with cefoxitin, cephalothin, and cefuroxime, with Km values of 2, 4, and 4 μM, respectively. The weakest binding was seen with ceftazidime, with a Km of 46 μM, similar to that reported for IMP-1 (22). SPM-1 gave kcat values greater than 8 s−1 for all cephalosporins, the best activity being seen with cephalothin, cefuroxime, and ceftazidime, with kcat values of 43, 37, and 28 s−1, respectively. The highest kcat/Km value for the cephalosporins was seen with cephalothin (11.7 μM · s−1).
TABLE 1.
Kinetic parameters of SPM-1 with a range of β-lactam antibiotics in comparison with those of other metallo-β-lactamases
| Antibiotic | SPM-1
|
IMP-1
|
VIM-1
|
VIM-2
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (S−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (S−1) | Km (μM) | kcat/Km (μM−1 s−1) | |
| Benzylpenicillin | 108 ± 4 | 38 ± 1 | 2.8 | 320 | 520 | 0.62 | 29 | 841 | 0.034 | 56 | 49 | 1.14 |
| Ampicillin | 117 ± 2 | 72 ± 3 | 1.6 | 950 | 200 | 4.8 | 37 | 917 | 0.04 | b | b | b |
| Carbenicillin | 74 ± 4 | 814 ± 29 | 0.09 | a | a | 0.02 | 167 | 75 | 2.2 | b | b | b |
| Azlocillin | 53 ± 2 | 147 ± 17 | 0.35 | b | b | b | 1,525 | 123 | 12 | b | b | b |
| Piperacillin | 117 ± 6 | 59 ± 5 | 2 | a | a | 0.72 | 1,860 | 3,500 | 0.53 | 33 | 72 | 0.45 |
| Ticarcillin | a | <0.35 | a | 1.1 | 740 | 0.0015 | 452 | 1,117 | 0.41 | 32 | 46 | 0.69 |
| Nitrocefin | 0.53 ± 0.01 | 4 ± 1 | 0.12 | 63 | 27 | 2.3 | 95 | 17 | 5.6 | b | b | b |
| Cephaloridine | 14 ± 1 | 18 ± 3 | 0.8 | 53 | 22 | 2.4 | 313 | 30 | 10 | b | b | b |
| Cephalothin | 43 ± 2 | 4 ± 1 | 11.7 | 48 | 21 | 2.4 | 281 | 53 | 5.3 | 57 | 44 | 1.28 |
| Cefuroxime | 37 ± 3 | 4 ± 1 | 8.8 | 8 | 37 | 0.22 | 324 | 42 | 7.7 | 12 | 22 | 0.55 |
| Ceftazidime | 28 ± 2 | 46 ± 2 | 0.6 | 8 | 44 | 0.18 | 60 | 794 | 0.076 | 89 | 98 | 0.90 |
| Cefotaxime | 16 ± 0.5 | 9 ± 1 | 1.9 | 1.3 | 4c | 0.35 | 169 | 247 | 0.68 | 28 | 32 | 0.86 |
| Cefoxitin | 8 ± 0.5 | 2 ± 1 | 4 | 16 | 8c | 2 | 26 | 131 | 0.2 | 3 | 24 | 0.12 |
| Cefepime | 18 ± 1 | 18 ± 3 | 1 | 7 | 11c | 0.66 | 549 | 145 | 3.8 | 5 | 184 | 0.03 |
| Imipenem | 33 ± 2 | 37 ± 4 | 1 | 46 | 39 | 1.2 | 2.0 | 1.5 | 1.3 | 10 | 10 | 0.99 |
| Meropenem | 63 ± 3 | 281 ± 27 | 0.22 | 50 | 10 | 0.12 | 13 | 48 | 0.27 | 1.0 | 5 | 0.28 |
| Moxalactam | 13 ± 1 | 97 ± 10 | 0.13 | 88 | 10c | 8.8 | b | b | b | 15 | 80 | 0.18 |
| Aztreonam | a | <0.3 | a | >0.01 | >1,000 | <0.0001 | <0.01 | >1,000 | <0.0001 | <0.5 | a | a |
| Clavulanic acid | a | <0.1 | a | b | b | b | b | b | b | b | b | b |
| Tazobactam | 0.6 ± 0.5 | 3 ± 1 | 0.2 | >1,000 | >3.98 | 0.0039 | 5.3 | 337 | 0.016 | b | b | b |
Data could not be determined.
No data were available.
Km, was obtained as the Ki value.
With respect to the penicillins, SPM-1 had the lowest Km values for penicillin, piperacillin, and ampicillin, with Km values of 38, 59, and 72 μM, respectively. The highest Km was seen for carbenicillin (814 μM). These values differed significantly from those for both IMP-1 (22) and VIM-1 (12), which generally have higher Km values for the penicillins. SPM-1 showed good hydrolytic activity with all penicillins, the highest values being for piperacillin, ampicillin, and penicillin, with kcat values of 117, 117, and 108 s−1, respectively. The highest kcat/Km ratio was 2.8 μM · s−1 for benzylpenicillin, which is significantly higher than those for IMP-1 (22) and VIM-1 (12). SPM-1 was unable to hydrolyze ticarcillin at all, whereas IMP-1 (22), VIM-1 (12), and VIM-2 (29) showed moderate to high levels of activity, with kcat values of 1.1, 452, and 0.32 s−1, respectively.
SPM-1 was found to have high Km values for the carbapenems, with meropenem having a Km of 281 μM, whereas the Km values for IMP-1 (22), VIM-1 (12), and VIM-2 (29) were 10, 48, and 5 μM, respectively. The Km value for imipenem was 37 μM, similar to that reported for IMP-1 (39 μM) (22), while VIM-1 and VIM-2 have been reported to have significantly lower Km values for imipenem: 1.5 and 10 μM, respectively (12, 29). SPM-1 was found to have kcat values of 63 s−1 for meropenem and 33 s−1 for imipenem, which are significantly higher than the kcat values of VIM-1 (for imipenem, 2 s−1; for meropenem, 13 s−1) (12) and VIM-2 (for imipenem, 10 s−1; for meropenem, 1 s−1) (29). The kcat/Km values of SPM-1 for both imipenem and meropenem (1 and 0.22 μM · s−1, respectively) are comparable to those of IMP-1, VIM-1, and VIM-2.
SPM-1 showed no hydrolysis of the monobactam aztreonam or the serine β-lactamase inhibitor clavulanic acid. A 3-h incubation at 37°C with these compounds at a concentration of 250 μM and with 2 μM enzyme did not reveal any significant hydrolysis. The serine β-lactamase inhibitor tazobactam is recognized and hydrolyzed by SPM-1, albeit with a low catalytic efficiency (kcat, 0.6 s−1). This value contrasts with that of IMP-1, which has a kcat >1,000 s−1 (22). Although VIM-1 has a similar kcat for tazobactam (5.3 s−1), it weakly binds to tazobactam (Km, 337 μM) (12) compared to the level of binding of SPM-1, which has a Km for tazobactam of 3 μM.
Inhibition studies with nitrocefin as the reporter substrate showed that aztreonam and clavulanic acid each act as a competitive inhibitor of SPM-1. The Kis of aztreonam and clavulanic acid were calculated according to equation 1 and were found to be 36 and 67 μM, respectively.
Inhibition by chelating agents.
Attempts to fit the time profiles of EDTA, dipicolinic acid, and 1-10-o-phenanthroline inhibition to kinetic models based on identical zinc sites of the sort used by Laraki et al. (22) and Franceschini et al. (12) for IMP-1 and VIM-1, respectively, were unsuccessful because the progressive inactivation of SPM-1 did not follow pseudo-first-order kinetics. However, the time profiles did fit a biexponential model of the form
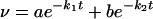 |
(2) |
where v is the rate of the reaction at time t. Interestingly, even after a 1-h incubation with any of the three chelators, a measurable rate of β-lactam hydrolysis was observed. The values for the fitted constants for both inhibitors are shown in Table 2.
TABLE 2.
Inhibition kinetics for SPM-1 and zinc chelatorsa
| Chelator | a | k1 | b | k2 |
|---|---|---|---|---|
| EDTA | 0.7665 | −0.03692 | 0.7051 | 0.01164 |
| Dipicolinic acid | 0.9554 | −0.0871 | 0.4551 | 0.00352 |
| 1-10-o-Phenanthroline | 1.9 | −0.003758 | 0.0001929 | 0.08838 |
The constants are those required to fit the data in Fig. 2 to equation 1. In all cases the sum of squares due to error was <0.004.
Homology modeling with the IMP-1 structure.
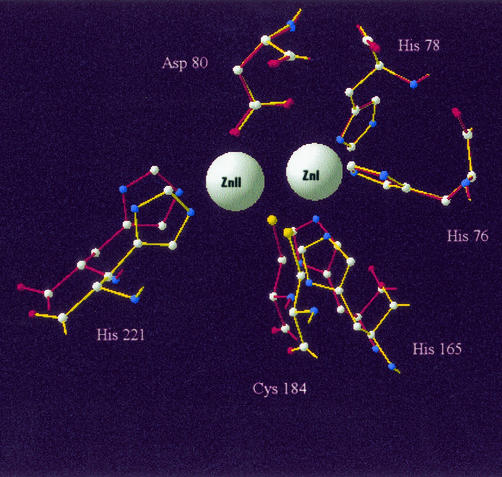
As expected, given the high degree of sequence identity (35.5%) and overall chemical similarity between the IMP-1 and SPM-1 primary structures, the SPM-1 amino acid sequence threaded well through the IMP-1 structure with the exception of the loop region, for which no homologous sequence was present. After energy minimization, the root mean square deviation of the α-carbon backbone for SPM-1 compared with that for IMP-1 was 0.66 Å for the N-terminal region up to the start of the inserted loop (residues 3 to 110 in Fig. 1) and 1.17 Å for the carboxy half of the molecule after the loop (residues 146 to 246 in Fig. 1). The overall structure clearly showed the characteristic αββα fold of the metallo-β-lactamase family; the position and orientation of active-site zinc ligands showed remarkable similarities to those for IMP-1. A comparison of the two active-site regions is shown in Fig. 3. The root mean square deviation for all the atoms of the zinc ligand side chains shown in Fig. 3 was <0.01 Å. However, there is a potential displacement of three of the atoms from the zinc coordinating residues in the SPM-1 model. The key zinc coordinating atoms, H165N(epsilon) (zinc 1), H221N(epsilon) (zinc 2), and C158S(gamma) (zinc 2) are all significantly farther from the relevant zinc atoms in the model than in the IMP-1 structure. These displacements are summarized in Table 3.
FIG. 3.
Zinc-coordinating ligands of the IMP-1 crystal structure of the sequence with pdb identification number 1JJE (red) compared with those from the SPM-1 homology model (yellow). SPM-1 residues are numbered as described in the legend to Fig. 1.
TABLE 3.
Relative displacement of key zinc ligand atoms in the SPM-1 homology model compared with the IMP-1 crystal structure
| Coordinating atom for SPM-1 (IMP-1) | Distance (Å) for SPM-1 (IMP-1)a | Zinc atom |
|---|---|---|
| C158SG (C158SG) | 3.72 (2.28) | 2 |
| H165NE (H139NE) | 2.56 (1.99) | 1 |
| H221NE (H197NE) | 2.97 (2.15) | 2 |
Distance from the atom center in column 1 to the center of the relevant zinc atom indicated in the last column. Appropriate atoms and equivalent distances for IMP-1 are shown in parentheses.
DISCUSSION
The gene encoding SPM-1, like both imp and vim, has been shown to be associated with a mobile genetic element and, thus, may have originated in another bacterium (1, 23, 24, 29, 32).
SPM-1, like IMP-1, VIM-1, and VIM-2, is also capable of hydrolyzing all three classes of β-lactams. However, while SPM-1 shares similar Km and kcat ratios for some substrates, the ratios differ significantly for particular compounds, most notably, tazobactam and nitrocefin. The common factor detected when the kinetic parameters of SPM-1, IMP-1, VIM-1, and VIM-2 are compared is that they are all unable to hydrolyze aztreonam. Inhibition assays demonstrated that aztreonam and clavulanic acid are both competitive inhibitors of SPM-1, with Kis of 67 and 36 μM, respectively; no such data are available for IMP-1, VIM-1, and VIM-2.
SPM-1 showed unusual inhibition kinetics with respect to the three metal chelators. IMP-1 and VIM-1 were found to exhibit pseudo-first-order kinetics with regard to metal chelator inhibition (12, 22). This implies that the two zinc sites have equal affinities and that a chelator can thus remove zinc equally from either site. The chelator inhibition kinetics for SPM-1 are clearly inconsistent with a pseudo-first-order kinetics profile; however, all chelators were found to fit a biexponential model with very unequal rate constants. This suggests that the inhibition process is composed of one relatively fast step in which a low-affinity zinc is removed reasonably quickly and one slower step in which a more tightly bound zinc is removed. The moderately low zinc content of 1.48 μmol of Zn per μM protein would tend to support this interpretation, suggesting that the enzyme is not fully binuclear.
The displacement of zinc coordinating atoms in the SPM-1 model provides a potential explanation for this, as displaced zinc ligands could result in altered binding constants for the zinc atoms. It is not clear at this stage whether the unusual inhibition kinetics are due to differential accessibilities of the zinc sites to specific chelators or reflect more fundamental differences in the binding of zinc. This clearly merits further, more detailed study.
Acknowledgments
We thank the Medical Research Council for supporting T. A. Murphy and the Biotechnology and Biological Sciences Research Council for supporting A. M. Simm. This work was partly supported by a grant from the British Society of Antimicrobial Chemotherapy to A. M. Simm (grant GA418).
We thank W. Mawby (Department of Biochemistry, University of Bristol) for assistance with mass spectrometry. We also thank Douglas Biedenbach (JONES Group/JMI Laboratories) for technical assistance.
REFERENCES
- 1.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avison, M. B., C. S. Higgins, C. J. von Heldreich, P. M. Bennett, and T. R. Walsh. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellais, S., S. Leotard, L. Poirel, T. Naas, and P. Nordmann. 1999. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol. Lett. 171:127-132. [DOI] [PubMed] [Google Scholar]
- 4.Bellais, S., L. Poirel, S. Leotard, T. Naas, and P. Nordmann. 2000. Genetic diversity of carbapenem-hydrolysing metallo-β-lactamases from Chryseobacterium (Flavobacterium) indologenes. Antimicrob. Agents Chemother. 44:3028-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, P. M. 1999. Integrons and gene cassettes: a genetic construction kit for bacteria. J. Antimicrob. Chemother. 43:1-4. [PubMed] [Google Scholar]
- 6.Boschi, L., P. S. Mercuri, M. L. Riccio, G. Amicosante, M. Galleni, J. M. Frere, and G. M. Rossolini. 2000. The Legionella (Fluoribacter) gormanii metallo-β-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 β-lactamases. Antimicrob. Agents Chemother. 44:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carfi, A., S. Pares, E. Duee, M. Galleni, C. Duez, J. M. Frere, and O. Dideberg. 1995. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14:4914-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu, Y. W., M. Afzal-Shah, E. T. Houang, M. I. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Concha, N. O., C. A. Janson, P. Rowling, S. Pearson, C. A. Cheever, B. P. Clarke, C. Lewis, M. Galleni, J. M. Frere, D. J. Payne, J. H. Bateson, and S. S. Abdel-Meguid. 2000. Crystal structure of the IMP-1 metallo-β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39:4288-4298. [DOI] [PubMed] [Google Scholar]
- 10.Concha, N. O., B. A. Rasmussen, K. Bush, and O. Herzberg. 1996. Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Structure 4:823-836. [DOI] [PubMed] [Google Scholar]
- 11.Cornaglia, G., M. L. Riccio, A. Mazzariol, L. Lauretti, R. Fontana, and G. M. Rossolini. 1999. Appearance of IMP-1 metallo-β-lactamase in Europe. Lancet 353:899-900. [DOI] [PubMed] [Google Scholar]
- 12.Franceschini, N., B. Caravelli, J. D. Docquier, M. Galleni, J. M. Frere, G. Amicosante, and G. M. Rossolini. 2000. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galleni, M., N. Franceschini, B. Quinting, L. Fattorini, G. Orefici, A. Oratore, J. M. Frere, and G. Amicosante. 1994. Use of the chromosomal class A β-lactamase of Mycobacterium fortuitum D316 to study potentially poor substrates and inhibitory β-lactam compounds. Antimicrob. Agents Chemother. 38:1608-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, and J. M. Frere. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele. blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 17.Hawkey, P. M., J. Xiong, H. Ye, H. Li, and F. H. M'Zali. 2001. Occurrence of a new metallo-β-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People's Republic of China. FEMS Microbiol. Lett. 194:53-57. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez Valladares, M., A. Felici, G. Weber, H. W. Adolph, M. Zeppezauer, G. M. Rossolini, G. Amicosante, J. M. Frere, and M. Galleni. 1997. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry 36:11534-11541. [DOI] [PubMed] [Google Scholar]
- 19.Ito, H., Y. Arakawa, S. Ohsuka, R. Wacharotayankun, N. Kato, and M. Ohta. 1995. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob. Agents Chemother. 39:824-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh, T. H., G. S. Babini, N. Woodford, L. H. Sng, L. M. Hall, and D. M. Livermore. 1999. Carbapenem hydrolysing IMP-1 β-lactamase in Klebsiella pneumoniae from Singapore. Lancet 353:2162.. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Laraki, N., N. Franceschini, G. M. Rossolini, P. Santucci, C. Meunier, E. de Pauw, G. Amicosante, J. M. Frere, and M. Galleni. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43:902-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J. M. Frere, and G. M. Rossolini. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 26.Massidda, O., G. M. Rossolini, and G. Satta. 1991. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J. Bacteriol. 173:4611-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavroidi, A., A. Tsakris, E. Tzelepi, S. Pournaras, V. Loukova, and L. S. Tzouvelekis. 2000. Carbapenem-hydrolysing VIM-2 metallo-β-lactamase in Pseudomonas aeruginosa from Greece. J. Antimicrob. Chemother. 46:1041-1042. [DOI] [PubMed] [Google Scholar]
- 28.Mercuri, P. S., F. Bouillenne, L. Boschi, J. Lamotte-Brasseur, G. Amicosante, B. Devreese, J. van Beeumen, J. M. Frere, G. M. Rossolini, and M. Galleni. 2001. Biochemical characterization of the FEZ-1 metallo-β-lactamase of Legionella gormanii ATCC 33297T produced in Escherichia coli. Antimicrob. Agents Chemother. 45:1254-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its Plasmid- and Integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prats, G., E. Miro, B. Mirelis, L. Poirel, S. Bellais, and P. Nordmann. 2002. First isolation of a carbapenem-hydrolysing β-lactamase in Pseudomonas aeruginosa in Spain. Antimicrob. Agents Chemother. 46:932-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen, B. A., Y. Gluzman, and F. P. Tally. 1990. Cloning and sequencing of the class B β-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob. Agents Chemother. 34:1590-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossolini, G. M., M. A. Condemi, F. Pantanella, J. D. Docquier, G. Amicosante, and M. C. Thaller. 2001. Metallo-β-lactamase producers in environmental microbiota: new molecular class B enzyme in Janthinobacterium lividum. Antimicrob. Agents Chemother. 45:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossolini, G. M., N. Franceschini, M. L. Riccio, P. S. Mercuri, M. Perilli, M. Galleni, J. M. Frere, and G. Amicosante. 1998. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing a broad substrate profile. Biochem. J. 332:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanschagrin, F., J. Dufresne, and R. C. Levesque. 1998. Molecular heterogeneity of the L1 metallo-β-lactamase family from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 42:1245-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott, W. R. P., P. H. Hunenberger, I. G. Tironi, A. E. Mark, S. R. Bileter, J. Fennen, A. E. Torda, T. Huber, and P. Kruger. 1999. The GROMOS biomolecular simulation package. J. Phys. Chem. 103:3596-3607. [Google Scholar]
- 37.Seigel, I. H. 1975. Enzyme kinetics behaviour and analysis of rapid equilibrium and steady-state enzyme systems. John Wiley & Sons, Inc., New York, N.Y.
- 38.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simm, A. M., C. S. Higgins, S. T. Pullan, M. B. Avison, P. Niumsup, O. Erdozain, P. M. Bennett, and T. R. Walsh. 2001. A novel metallo-β-lactamase, Mbl1b, produced by the environmental bacterium Caulobacter crescentus. FEBS Lett. 509:350-354. [DOI] [PubMed] [Google Scholar]
- 40.Srinivasan, N., K. Guruprasad, and T. L. Blundell. 1996. Comparative modelling of proteins, vol. 170. Oxford University Press Inc., New York, N.Y.
- 41.Thomson, J. S., and M. H. Malamy 1990. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus β-lactamase II. J. Bacteriol. 172:2584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toleman, M., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed]
- 43.Tysall, L., M. W. Stockdale, P. R. Chadwick, M. F. Palepou, K. J. Towner, D. M. Livermore, and N. Woodford. 2002. IMP-1 carbapenemase detected in an Acinetobacter clinical isolate from the UK. J. Antimicrob. Chemother. 49:217-218. [DOI] [PubMed] [Google Scholar]
- 44.Ullah, J. H., T. R. Walsh, I. A. Taylor, D. C. Emery, C. S. Verma, S. J. Gamblin, and J. Spencer. 1998. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J. Mol. Biol. 284:125-136. [DOI] [PubMed] [Google Scholar]
- 45.Walsh, T. R., W. A. Neville, M. H. Haran, D. Tolson, D. J. Payne, J. H. Bateson, A. P. MacGowan, and P. M. Bennett. 1998. Nucleotide and amino acid sequences of the metallo-β-lactamase, ImiS, from Aeromonas veronii bv. sobria. Antimicrob. Agents Chemother. 42:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winston, R. L., and M. C. Fitzgerald. 1998. Concentration and desalting of protein samples for mass spectrometry analysis. Anal. Biochem. 262:83-85. [DOI] [PubMed] [Google Scholar]
- 47.Woodford, N., M. F. Palepou, G. S. Babini, B. Holmes, and D. M. Livermore. 2000. Carbapenemases of Chryseobacterium (Flavobacterium) meningosepticum: distribution of blaB and characterization of a novel metallo-β-lactamase gene, blaB3, in the type strain, NCTC 10016. Antimicrob. Agents Chemother. 44:1448-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan, J. J., P. R. Hsueh, W. C. Ko, K. T. Luh, S. H. Tsai, H. M. Wu, and J. J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, J. J., W. C. Ko, and J. J. Wu. 2001. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-β-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:2368-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yano, H., A. Kuga, R. Okamoto, H. Kitasato, T. Kobayashi, and M. Inoue. 2001. Plasmid-encoded metallo-β-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob. Agents Chemother. 45:1343-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]