Abstract
Most Yersinia enterocolitica strains are resistant to β-lactam antibiotics due to the production of one or two chromosomally encoded β-lactamases. Strain Y56 is a Y. enterocolitica O:3 serotype natural isolate that is resistant to moderate amounts of penicillins and that produces a single class A β-lactamase. To select mutants with increased levels of resistance to β-lactam antibiotics, strain Y56 was grown on plates containing increasing amounts of ampicillin, and variants resistant to up to 500 μg of ampicillin per ml were obtained. Chromosomal DNA from hyperresistant isolates was analyzed by Southern hybridization with a blaA-specific probe to detect gene rearrangements. The use of pulsed-field gel electrophoresis revealed that the increase in the resistance level correlated with the amplification in tandem of a DNA fragment of about 28 kb containing the blaA gene. The phenotype of these isolates was not stable, and they recovered the basal low resistance level when the ampicillin used for selection was withdrawn from the growth medium. This loss of resistance was followed by the recovery of the original chromosomal structure. To understand this amplification process, the 28-kb amplification unit was cloned, and the ends were sequenced. The analysis of these sequences did not reveal the presence of either repeats or transposable elements to explain this process. However, we found short sequences similar to some DNA gyrase target sequences that have been described. In addition, we observed that the frequency of appearance of ampicillin-hyperresistant isolates by amplification of the blaA locus was lowered in the presence of the gyrase inhibitor novobiocin. These findings suggest that the DNA gyrase could be involved in this amplification event.
The evolution of antibiotic resistance in bacteria is a complex and very effective process responsible for the many clinical problems in the treatment of bacterial infections found today. Among the multiple genetic mechanisms that contribute to antibiotic resistance, gene amplification is not very frequent, but several examples have been already reported, such as those affecting the ampC gene for the chromosomal cephalosporinase gene (6, 19) or the sulfonamide resistance gene (18) in Escherichia coli. In general terms, genetic amplification produces an increase in the number of copies of an antibiotic resistance gene, and this may result in an increase in the level of resistance to the drug or related antibiotics to which the bacteria was formerly susceptible. This is observed when single-copy bacterial genes are cloned in multicopy plasmids, as occurred with the appearance of resistance to gentamicin by the cloning of the aac2′ gene from Providencia (26) and Mycobacterium fortuitum (1) and the resistance to bacitracin after the cloning of the E. coli bacA gene (3).
Gene amplification is very frequently the origin of the resistance to chemotherapeutic agents in tumor cells (9).
From an energetic point of view, gene amplification should be very similar to the acquisition of a plasmid. It may be seen as an inefficient way to obtain resistance, since the organism must increase the size of its genome and needs to replicate additional genetic material, with a considerable burden of energy. Increased gene expression can be more easily gained by point mutations in the gene promoter or other transcription regulatory signals. However, gene amplification has an advantage over plasmid acquisition or promoter mutations because it is often a reversible process and the chromosome recovers the basal structure upon removal of the antibiotic from the culture medium, while the other mechanisms persist in the absence of the antibiotic. Reversible amplification of a gene is a very efficient mechanism for adaptation to ambient conditions. Furthermore, genotypic reversal can be beneficial when the resistance protein that is overproduced has some negative effect on cell growth, as could be the case with β-lactamases.
Class A β-lactamases constitute a broad and phylogenetically compact family of enzymes. In enterobacteria they are predominantly plasmid or transposon bound. Yersinia enterocolitica is a member of the family Enterobacteriaceae, but most of the strains are resistant to β-lactam antibiotics due to the production of one or two chromosomally encoded β-lactamases (4, 30). One of these β-lactamases belongs to molecular class A (31). Comparison of the sequence of the enzyme from Y. enterocolitica with the sequences of other members of the same class allowed the separation of two subgroups with a few signature residues. One subgroup consists of chromosomally encoded enzymes, while the second group contains enzymes from plasmids and transposons (31).
Most Y. enterocolitica isolates produce a second β-lactamase that belongs to molecular class C; however, this β-lactamase is not active in strain Y56 (5, 32), which was used in the present study.
The capacity of β-lactamase-producing strains to easily improve their resistance profile is well known (21, 22). Class A enzymes acquire the capability to hydrolyze new β-lactam antibiotics as a result of a few amino acid substitutions (14, 34). Strains containing class C enzymes are able to increase the level of enzyme expression by stable derepression (23, 29) by amplification of the gene (6, 19) and by mutations in their promoter regions (20).
In this study, we have isolated and characterized mutants of Y. enterocolitica strain Y56 with increased levels of resistance to β-lactams (ampicillin MIC, 16 μg/ml). The ampicillin MIC was up to 20 times greater for the mutants than for the parental strain. The hyperresistant phenotype was unstable, and the basal susceptibility level was recovered after withdrawal of the ampicillin from the growth medium. These variants showed the amplification in tandem of a chromosomal DNA fragment of about 28 kb containing the blaA gene. This amplification unit has been characterized and its ends have been sequenced. The role of the DNA gyrase inhibitor novobiocin in the amplification event was also investigated.
MATERIALS AND METHODS
Selection of Y. enterocolitica Y56 variants hyperresistant to ampicillin.
Y. enterocolitica Y56 was successively grown at 37°C for 24 h in Luria-Berani (LB) broth (Difco) containing 25, 50, 100, 200, 300, and 500 μg of ampicillin (Sigma) per ml and then plated on LB agar containing ampicillin at 500 μg/ml. Several colonies for which the ampicillin MIC was greater than 256 μg/ml were selected from these plates. This process was also performed in the presence of the DNA gyrase inhibitor novobiocin at 25, 50, or 100 μg/ml. In this case, the number of mutants with increased resistance levels in plates containing the same concentration of ampicillin as the broth culture was determined daily.
Preparation of chromosomal DNA in agarose blocks and pulsed-field gel electrophoresis (contour-clamped homogeneous electric field [CHEF] gel electrophoresis).
Intact chromosomal DNA was prepared from bacteria included in low-melting-point agarose blocks by an adaptation of a method described previously (12). Lysis was accomplished by treatment with 0.2% sodium deoxycholate-0.5% sodium dodecyl sulfate-1 mg of lysozyme per ml in 10 mM Tris-50 mM NaCl-100 mM EDTA (pH 7.2) for 6 h at 37°C, followed by removal of the buffer and one rinse with wash buffer (20 mM Tris-HCl, 50 mM EDTA [pH 8.0]). Next, the agarose plugs were incubated with proteinase K (1 mg/ml) in 100 mM EDTA (pH 8.0)-1% sodium dodecyl sulfate-0.2% sodium deoxycholate for 18 h at 50°C with gentle shaking.
For restriction endonuclease digestion of chromosomal DNA in agarose blocks, proteinase K was inactivated by treating the agarose plugs with 0.5 mM phenylmethylsulfonyl fluoride in 20 mM Tris-50 mM EDTA (pH 8.0) for 1 h. The agarose blocks were washed at least twice in wash buffer. Prior to restriction endonuclease digestion, the EDTA concentration in the agarose plugs was lowered by incubation for 0.5 h in wash buffer diluted 10 times. Then, the blocks were incubated with the restriction endonucleases under the conditions recommended by the supplier.
Pulsed-field gel electrophoresis was performed with the CHEF DR-III system (Bio-Rad). Gels were made with multipurpose (MP) agarose (Roche) in 0.5× Tris-borate buffer (0.5× Tris-borate buffer is 45 mM Tris-borate plus 1 mM EDTA); Tris-borate EDTA buffer was also used as the running buffer for electrophoresis. The total run time and the pulse times were as stated in the appropriate figure legends. Bacteriophage lambda DNA concatemers (Sigma) and a pulse marker (Sigma) were used as molecular weight markers.
Southern blot hybridization.
Chromosomal DNA, isolated as described previously (2) and digested with the appropriate restriction endonucleases, was separated in 0.8% agarose gels and blotted onto positively charged nylon membranes. DNA separated in pulsed-field gels was similarly blotted by capillarity. To ensure transfer of the high-molecular-weight fragments, the gels were UV (312 nm) irradiated, and the transfer was made in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for at least 24 h. The 500-bp NruI-HincII blaA fragment from pSU611 (30) was isolated, labeled with digoxigenin (Roche), and used to probe the blots. After hybridization, the filters were washed under stringent conditions, further incubated with a phosphatase-coupled antidigoxigenin antibody, incubated with the luminescent substrate CDP-star (Roche), and exposed to film.
DNA cloning, sequencing, and sequence analysis.
Plasmid preparations, DNA ligation, and transformation were carried out basically as described elsewhere (28). DNA sequencing was performed with a DNA automatic sequencer (Vistra 725; Amersham). M13 forward and M13 reverse sequencing primers were used for sequencing. The sequences were analyzed with the program BLASTN (version 2.2.3) in the Blastserver at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST).
Nucleotide sequence accession numbers.
The nucleotide sequences a/b, c/b, and c/d described herein have been deposited in the GenBank nucleotide sequence database under accession numbers AY143385, AY143386, and AY143387, respectively.
RESULTS
Identification and characterization of DNA amplification events.
Individual colonies of strain Y56 growing on plates containing 500 μg of ampicillin per ml were isolated and the ampicillin MIC's were determined. Those colonies for which the ampicillin MIC was greater than 256 μg/ml were selected for further study.
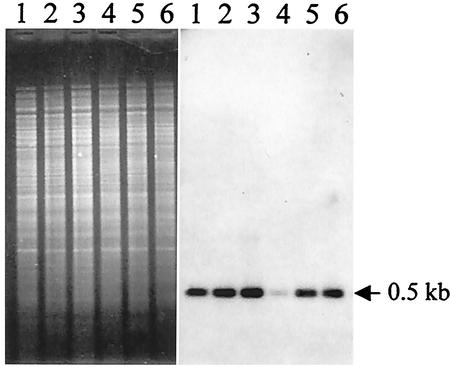
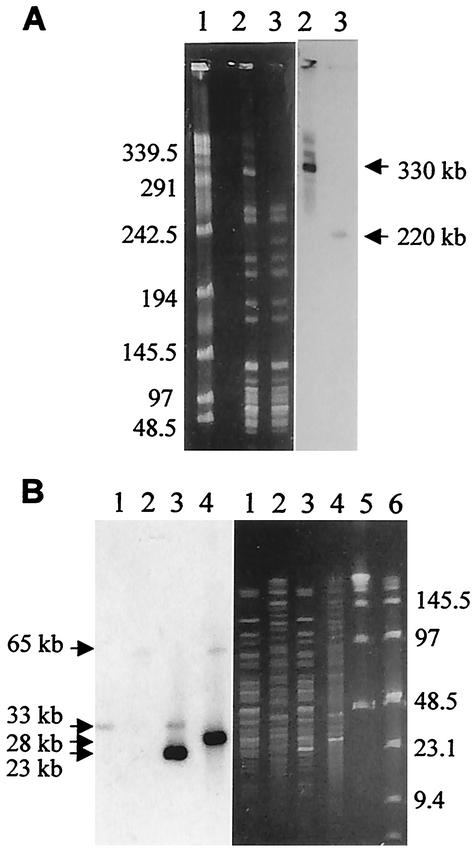
Chromosomal DNA from these strains was digested with HincII and NruI and then blotted and hybridized with an intragenic blaA-specific probe. The bands containing this gene in the hyperresistant strains were of the same size as those from the original strain Y56. However, the intensities of the bands were notably and consistently stronger in the hyperresistant variants than in Y56 (Fig. 1). The increase in band intensity observed in the blots of DNA in the hyperresistant strains suggested that the increased resistance was due to amplification of the blaA gene and that the DNA amplified was a large segment whose limits were beyond the restriction sites used in this analysis. To detect a large genomic rearrangement, we analyzed the genome of one of these hyperresistant strains (strain Y100) by restriction with rarely cutting endonucleases and CHEF gel electrophoresis. When the chromosomes were digested with NotI, a 220-kb NotI band that was observed in the chromosome of Y56 was absent from mutant Y100. In this mutant, we could see instead a strong band of about 330 kb accompanied by some fainter surrounding bands. This gel was blotted and hybridized with the blaA-specific probe. The 220-kb NotI fragment from Y56 hybridized with the probe, indicating that it contained the blaA gene. Hybridization in the mutant revealed that the band of 330 kb as well as the fainter accompanying bands observed in the stained gel also contained the blaA gene (Fig. 2A). The multiplicity of the bands containing the blaA gene suggested that cultures of hyperresistant mutants could be heterogeneous and contain bacteria with different gene arrangements around the blaA gene.
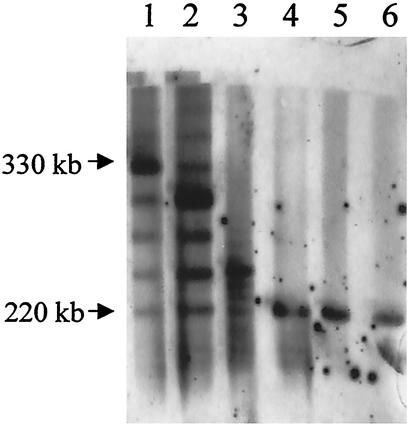
FIG. 1.
Southern blot analysis of several derivatives from strain Y56 for which ampicillin MICs are greater than 250 μg/ml. Chromosomal DNA (2 μg) was digested with HincII and NruI, and the blot was hybridized with the HincII-NruI blaA-specific probe. Lanes 1, 2, 3, 5 and 6, hyperresistant isolates; lane 4, strain Y56.
FIG. 2.
Separation by CHEF gel electrophoresis of chromosomal restriction fragments and hybridization with the blaA-specific probe. (A) Chromosomal DNA digested with NotI was separated in 1% agarose gels run for 15 h at 6 V/cm and 14°C. The initial switch time was 10 s, and the final switch time was 50 s. Lane 1, bacteriophage lambda ladder concatemers; lanes 2, hyperresistant isolate Y100; lanes 3, strain Y56. (B) Chromosomal XbaI and SfiI fragments were separated in 1% agarose gels at 6 V/cm by using two consecutive sets of pulse conditions. In block 1, the initial switch time was 5 s and the final switch time was 10 s for 6 h. In block two, the initial switch time was 1 s and the final switch time was 6 s for 12 h. Lanes 1, unamplified strain Y56 digested with SfiI; lanes 2, strain Y56 digested with XbaI; lanes 3, hyperresistant isolate Y100 digested with SfiI; lanes 4; Y100 digested with XbaI; lanes 5 and 6, bacteriophage lambda ladder concatemers and pulse marker, respectively.
Analysis of the chromosomes digested with XbaI and separated in gels revealed a very intense band of about 28 kb in the mutant but not in strainY56 (Fig. 2B). When this gel was blotted and hybridized with the blaA-specific probe, a single 65-kb XbaI hybridization band was seen in Y56. This band was also present in the mutant but was accompanied by a second, more intense band of about 28 kb. When the chromosomes were digested with the restriction enzyme SfiI, a single 33-kb SfiI hybridization band was present in Y56. This band was accompanied in Y100 by a more intense band of 23 kb (Fig. 2B).
Stability of the hyperresistant phenotype.
To analyze the phenotypic stability of the hyperresistant isolates, a mutant for which the ampicillin MIC was 250 μg/ml was repeatedly grown in ampicillin-free LB broth. After 1 week of daily subculture, colonies for which the ampicillin MIC had gone down to the original value of 16 μg/ml were isolated. Variants for which the ampicillin MICs were low and intermediate were isolated, and their chromosomes were analyzed by hybridization after NotI digestion and CHEF gel electrophoresis. Multiple bands were seen around a more abundant 330-kb band for isolates for which the ampicillin MIC was greater than 125 μg/ml, as happened with mutant Y100. The sizes of the bands decreased with the ampicillin MIC, and only the 220-kb band was seen in the cultures for which the ampicillin MIC was at the basal level (Fig. 3). The sizes of the hybridization bands in this gel followed a regular pattern consistent with the gain or loss of discrete DNA units of 25 to 30 kb as the origin of the polymorphism observed in the NotI bands containing blaA.
FIG. 3.
Hybridization with a blaA-specific probe of NotI-digested CHEF gel electrophoresis-separated chromosomal fragments from Y56 isolates for which ampicillin MICs were different, showing the structural reversibility of the amplification process. Electrophoresis conditions were adjusted to separate DNA fragments in the 200- to 500-kb range. A 0.8% agarose gel was run for 20 h at 6 V/cm and 14°C. The initial switch time was 20 s, and the final switch time was 80 s. Lane 1, strain for which the MIC is 250 μg/ml; lane 2, strain for which the MIC is 125 μg/ml; lane 3, strain for which the MIC is 64 μg/ml; lane 4, strain for which the MIC is 32 μg/ml; lane 5, strain for which the MIC is 16 μg/ml; lane 6, unamplified Y56.
Cloning of amplification unit and sequencing of terminal regions.
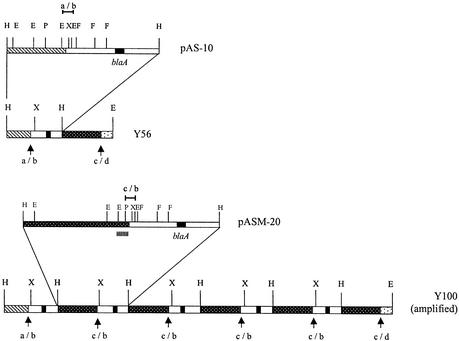
Chromosomal DNA from Y56 and hyperresistant strain Y100 was digested with HindIII and cloned into plasmid vector pACYC184, and recombinants were selected on plates containing ampicillin (100 μg/ml). In this way we obtained two plasmids, called pAS-10 and pASM-20, containing DNA fragments of 22 and 28 kb, respectively, from Y56 and amplified strain Y100, respectively. A map of these chromosomal fragments with several restriction endonucleases is presented in Fig. 4. The two plasmids were different from their left ends to the XbaI central restriction site, but they were identical from that site to their right ends. Since the 28-kb HindIII fragment cloned in pASM-20 was of the same size as the chromosomal XbaI fragment from Y100 and this size coincided with the estimation done from Fig. 3, we concluded that the size of the amplification unit was 28 kb and that both HindIII and XbaI cut only once in the amplification unit. Using the data from the two plasmids, we could construct the map of the amplification unit, as shown in Fig. 4. Next, we determined three different DNA sequences, namely, a/b, b/c, and c/d. The a/b sequence contains the junction between the nonamplified chromosome and the left end of the amplification unit. It was present in both strains Y56 and Y100. It was cloned in a 1.5-kb EcoRI fragment from plasmid pAS-10, whose sequence was completely determined. The c/b sequence contains the tail-to-head junction of two consecutive amplification unit copies. The c/b junction was cloned and sequenced in a 0.9-kb PstI-EcoRI fragment from plasmid pASM-20. The c/d sequence contains the junction between the right end of the amplification unit and the adjacent nonamplified chromosomal DNA. To obtain the c/d sequence, we walked a probe consisting of the EcoRI-PstI fragment of 1.2 kb from pASM-20 from the left end of the amplification unit on the Y56 chromosome. In this way, a 3-kb EcoRI fragment whose right end (a 1.8-kb PstI-EcoRI fragment) was cloned and sequenced was obtained. A representation of the structures of the chromosomes of wild-type strain Y56 and amplified strain Y100 is also shown in Fig. 4. When the nucleotide sequences were analyzed, we could precisely determine the start and the end of the amplification unit. However, we could not find in the proximity either directed or inverted repeats or evidence of insertion sequences which could be implicated in the amplification events. The sequences in the amplification unit borders showed weak similarity to the sites used by DNA gyrase in illegitimate recombination events (16). For instance, the sequence found in c/d (CAT/TTGTAC) is similar to E. coli DNA gyrase cleavage site 3 in pBR322 DNA (TAT/TTGTTT) (13).
FIG. 4.
Restriction maps of the chromosomal region containing the blaA genes from Y56 and amplified strain Y100. The DNA inserts in plasmids pAS-10 and pASM-20 are enlarged for further detail. Plasmid pAS-10 carries a 22-kb HindIII fragment from the chromosome of wild-type strain Y56, and pASM-20 carries a 28-kb HindIII chromosomal DNA fragment from amplified strain Y100. The fragments cloned to obtain sequences a/b and c/b are indicated by horizontal lines. The EcoRI-PstI fragment of 1.2 kb used as a probe to obtain the c/d sequence is also indicated. The amplification unit in Y56 is found between the junctions of sequences a/b and c/d and is marked with vertical arrows. In Y100 the junction between adjacent copies of the amplification unit is also marked with vertical arrows. E, EcoRI; H, HindIII; P, PstI; F; SfiI and X, XbaI.
Effect of novobiocin on the amplification process.
To examine the possible involvement of DNA gyrase-mediated recombination in the amplification process, we grew Y. enterocolitica Y56 in LB broth containing amounts of ampicillin that increased from 25 to 500 μg/ml in the presence of subinhibitory concentrations of the antibiotic novobiocin (100, 50, or 25 μg/ml). This antibiotic is a DNA gyrase inhibitor and specifically targets the ATP-binding B domain of the enzyme, depriving it of energy and resulting in a loss of negative supercoiling activity. We observed that novobiocin exerted a dose-dependent effect on the number of hyperresistant mutants obtained in its presence. No colonies were obtained when Y56 was growing in the presence of ampicillin at 300 μg/ml and novobiocin at 100 μg/ml. The number of hyperresistant mutants obtained was also significantly decreased with novobiocin at 50 and 25 μg/ml (Fig. 5). Next, we isolated chromosomal DNA from five colonies obtained in 300 μg of ampicillin per ml with each different novobiocin concentration (0, 25, and 50 μg/ml). The DNA was digested with NruI-HincII, blotted, and hybridized with the blaA-specific probe. The five hyperresistant strains isolated in the absence of novobiocin showed a fully amplified blaA locus. However, the locus was fully amplified from only two of the hyperresistant isolates obtained in the presence of 25 μg of novobiocin per ml, and a low level of amplification was seen for the three remaining colonies. The blaA locus was not amplified from two of the hyperresistant isolates obtained in the presence of 50 μg of novobiocin per ml, and three colonies had a low level of amplification (Fig. 5). This result suggests that novobiocin has an effect on the amplification process. In addition, it shows that amplification is the predominant mechanism giving rise to ampicillin-hyperresistant isolates under normal conditions but that other mechanisms come into play in the presence of novobiocin to increase the ampicillin resistance level.
FIG. 5.
Effect of novobiocin on the amplification process. The effect of sublethal concentrations of the DNA gyrase inhibitor novobiocin (Novo) on the frequency of appearance of hyperresistant mutants was assayed by determination of the number of CFU per milliliter on plates containing different concentrations of ampicillin. A dose-dependent decrease in the frequency of appearance of hyperresistant mutants was seen in the presence of novobiocin. Data are represented as the logarithm of the mean plus the standard deviation from three independent experiments. The amplification status of the blaA locus was monitored by hybridization with a blaA-specific probe in the hyperresistant mutants obtained in 300 μg of ampicillin per ml and different amounts of novobiocin. Both the frequency and the degree of amplification were affected by increasing amounts of novobiocin. C, control (strain Y56).
DISCUSSION
Gene amplification is not a rare process in bacteria. Most cases of amplification in bacteria may be explained by the action of the general recombination pathway or the interplay of transposable elements (27). The ampC gene from E. coli was apparently amplified by recA-dependent recombination between short direct repeats (7). Similarly, amplification of 7- to 37-kb regions including the lac region from E. coli has also been described (35). These amplifications have been shown to occur at short sequence repeats, but the enzymes involved in this process remain unknown (36). Some Streptomyces species are characterized by a high degree of genomic instability usually manifested as amplifications and deletions. In this case amplification has been correlated with the presence of specific genetic elements characterized by the presence of long repeats and specific gene products (24). Genetic instability has been reported in members of the genus Yersinia, especially in Y. pestis, in which pathogenicity genes are often lost as result of a big chromosomal deletion affecting the 102-kb fragment flanked by a repetitive element (8) that could be homologous to the insertion sequence IS100 (25).
Gene amplification is involved in adaptation to environmental stresses such as the presence of antibiotics (6, 18) and heavy metals (15). Overexpression of gene products through gene amplification may confer the phenotypic advantages needed for survival. The amplified state remains as long as the selective condition exists, and the copy number is adjustable according to changes in the environment. For instance, the level of resistance to sulfathiazole was markedly decreased upon the loss of amplified DNA after growth under nonselective conditions (18), and the loss of copies during growth under nonselective conditions suggested that the amplified DNA was organized as tandem repeats.
In this study, we have shown amplification of a DNA region containing the blaA gene in tandem in Y. enterocolitica strain Y56 when the bacteria were exposed to elevated concentrations of ampicillin. The hyperresistant strains that resulted from the amplification event could grow on plates containing 500 μg of ampicillin per ml. On the basis of the analysis of the chromosomes of Y56 and hyperresistant strain Y100 digested with XbaI, SfiI, and NotI, we estimated that the size of the amplification unit would be 28 kb and that the amplification produced copies of the amplification unit in tandem according to the model represented in Fig. 4. The results in Fig. 2A showed that the 220-kb NotI band present in the chromosome of Y56 was replaced in the mutant by a larger band of about 330 kb, indicating a size increase of about 110 kb. This suggested the presence of at least five copies of the amplification unit in the Y100 chromosome. This value is in agreement with those from the densitometric quantification of films like the one shown in Fig. 1 and the in vitro measurement of β-lactamase activity (data not shown).
This amplification process seems likely to occur in a stepwise manner, depending on the strength of the selective pressure, and was found to be reversible. Accordingly, a culture of a hyperresistant variant was not homogeneous and contained cells with intermediate degrees of amplification. This can explain the heterogeneity of the large NotI bands seen in the mutants. These bands would correspond to the presence of bacteria with different degrees of amplification in the same culture. On the other hand, the flexibility of the process allows a good adaptation to ambient conditions without the need to permanently maintain and replicate an unnecessary number of copies of the amplification unit, although it could be more finely tuned by a reduction in the size of the amplification unit.
The reversibility of the process was manifested by the reversal to the basal resistance level when the selective pressure was removed. The phenotypic reversal was the consequence of the loss of amplified units revealed by the progressive decrease in the sizes of the hybridization bands to regain the original configuration of the chromosome.
Assays with plasmid pAS-10 and especially plasmid pASM-20, obtained from the chromosome of an amplified strain, confirmed the size of the amplification unit. The cloning of the amplification unit allowed us to sequence their ends as well as the left and right chromosomal flanking sequences. Upon analysis, we did not see the involvement of either directed or invert repeats or insertion sequences in the amplification events. In addition to general recombination and transposition, amplification may also be the result of different illegitimate recombination processes that generate free DNA ends. It is known that illegitimate recombination at small regions of homology (1 to 5 bp) occurs via DNA gyrase-catalyzed reactions (16). In E. coli, these events are mediated by subunit exchange between different DNA gyrase molecules (11, 33). Furthermore, it has been shown that adaptive evolution in Salmonella is constrained by signals transmitted from the external environment via changes in the activity of DNA gyrase (17). To gain some insight into the mechanism of amplification of the bla locus observed in Y. enterocolitica Y56, we studied the amplification process in the presence of novobiocin, an inhibitor of DNA gyrase. We found that in the presence of novobiocin the frequency of occurrence of hyperresistant variants decreased in a dose-dependent manner and that amplification was almost abolished in the presence of 100 μg of novobiocin per ml. These data suggest a possible role for the DNA gyrase in the amplification process; however, the effect of novobiocin could also be attained as a result of indirect effects of the drug on DNA supercoiling or transcription. The reversible amplification in tandem of a 28-kb region from the Y. enterocolitica chromosome containing the blaA gene described herein constitutes an example of an adaptive genetic change in Y. enterocolitica. Adaptive amplification and adaptive mutation are parallel routes of inducible genetic instability that allow rapid evolution under stress and escape from growth inhibition (10). This amplification mechanism provides Y. enterocolitica, an opportunistic pathogen, with an adaptive pathway that could be relevant in relation to its resistance to antibiotics, a phenotype of great importance for animal or human colonization. Identification of the precise mechanism for this amplification process can also be exploited by biotechnological methods, since genes of interest placed in the appropriate context could expand when needed and adapt closely to changing ambient circumstances.
Acknowledgments
This work was supported by grant 99/1224 from the Fondo de Investigaciones Sanitarias of the Spanish Ministry of Health.
REFERENCES
- 1.Ainsa, J. A., C. Martin, B. Gicquel, and R. Gomez-Lus. 1996. Characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase gene from Mycobacterium fortuitum. Antimicrob. Agents Chemother. 40:2350-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F., M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Cain, B. D., P. J. Norton, W. Eubanks, H. S. Nick, and C. M. Allen. 1993. Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J. Bacteriol. 175:3784-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis, G. 1981. Antibiotic resistance in Yersinia enterocolitica, p. 55-71. In E. J. Bottone (ed.), Yersinia enterocolitica. CRC Press, Inc., Boca Raton, Fla.
- 5.De la Prieta, M. C., A. Seoane, J. Diaz, J. Navas, and J. M. Garcia Lobo. 1995. Beta-lactamase genes and beta-lactamic susceptibility in Yersinia enterocolitica. Contrib. Microbiol. Immunol. 13:184-187. [PubMed] [Google Scholar]
- 6.Edlund, T., T. Grundström, and S. Normark. 1979. Isolation and characterization of DNA repetitions carrying the chromosomal β-lactamase gene of Escherichia coli K-12. Mol. Gen. Genet. 173:115-125. [DOI] [PubMed] [Google Scholar]
- 7.Edlund, T., and S. Normark. 1981. Recombination between short DNA homologies causes tandem duplication. Nature 292:269-271. [DOI] [PubMed] [Google Scholar]
- 8.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 9.Gorre, M. E., M. Mohammed, K. Ellwood, N. Hsu, R. Paquette, P. N. Rao, and C. L. Sawyers. 2001. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293:876-880. [DOI] [PubMed] [Google Scholar]
- 10.Hastings, P. J., H. J. Bull, J. R. Klump, and S. M. Rosenberg. 2000. Adaptive amplification: an inducible chromosomal instability mechanism. Cell 103:723-731. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda, H., K. Aoki, and A. Naito. 1982. Illegitimate recombination mediated in vitro by DNA gyrase of Escherichia coli: structure of recombinant DNA molecules. Proc. Natl. Acad. Sci. USA 79:3724-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iteman, I., C. Baril, I. Saint Girons, and E. Carniel. 1991. Pulsed field electrophoresis of the chromosome of the pathogenic Yersiniae. Contrib. Microbiol. Immunol. 12:198-202. [PubMed] [Google Scholar]
- 13.Kirkegaard, K., and J. C. Wang. 1981. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell 23:721-729. [DOI] [PubMed] [Google Scholar]
- 14.Knox, J. R. 1995. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob. Agents Chemother. 39:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondratyera, T. F., L. N. Muntyan, and G. I. Karavaiko. 1995. Zinc- and arsenic-resistant strains of Thiobacillus ferrooxidans have increased copy numbers of chromosomal resistance genes. Microbiology 141:1157-1162. [DOI] [PubMed] [Google Scholar]
- 16.Marvo, S. L., S. R. King, and S. R. Jaskunas. 1983. Role of short regions of homology in intermolecular illegitimate recombination events. Proc. Natl. Acad. Sci. USA 80:2452-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massey, R. C., P. B. Rainey, B. J. Sheehan, O. M. Keane, and C. J. Dorman. 1999. Environmentally constrained mutation and adaptive evolution in Salmonella. Curr. Biol. 9:1477-1480. [DOI] [PubMed] [Google Scholar]
- 18.Nichols, B. P., and G. G. Guay. 1989. Gene amplification contributes to sulfonamide resistance in Escherichia coli. Antimicrob. Agents Chemother. 12:2042-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Normark, S., T. Edlund, T. Grundström, S. Bergström, and H. Wolf-Watz. 1977. Escherichia coli K-12 mutants hyperproducing chromosomal beta-lactamase by gene repetitions. J. Bacteriol. 132:912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsson, O., S. Bergström, and S. Normark. 1982. Identification of a novel ampC β-lactamase promoter in a clinical isolate of Escherichia coli. EMBO J. 1:1411-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrosino, J., C. Cantu III, and T. Palzkill. 1998. β-Lactamases: protein evolution in real time. Trends Microbiol. 6:323-327. [DOI] [PubMed] [Google Scholar]
- 22.Philippon, A., R. Labia, and G. Jacoby. 1989. Expanded spectrum β-lactamases. Antimicrob. Agents Chemother. 33:1131-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philippon, A., G. Arlet, and G. Jacoby. 2002. Plasmid-determined ampC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piendl, W., C. Eichenseer, P. Viel, J. Altenbuchner, and J. Cullum. 1994. Analysis of putative DNA amplification genes in the element AUD1 of Streptomyces lividans 66. Mol. Gen. Genet. 244:439-443. [DOI] [PubMed] [Google Scholar]
- 25.Portnoy, D. A., and S. Falkow. 1981. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J. Bacteriol. 148:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rather, P. N., E. Orosz, K. J. Shaw, R. Hare, and G. Miller. 1993. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J. Bacteriol. 175:6492-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero, D., and R. Palacios. 1997. Gene amplification and genomic plasticity in prokaryotes. Annu. Rev. Genet. 31:91-111. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sanders, C. C. 1987. Chromosomal cephalosporinases responsible for multiple resistance to newer β-lactam antibiotics. Annu. Rev. Microbiol. 41:573-593. [DOI] [PubMed] [Google Scholar]
- 30.Seoane, A., and J. M. García-Lobo. 1991. Cloning of chromosomal β-lactamase genes from Yersinia enterocolitica. J. Gen. Microbiol. 137:141-146. [DOI] [PubMed] [Google Scholar]
- 31.Seoane, A., and J. M. García-Lobo. 1991. Nucleotide sequence of a new classA β-lactamase gene from the chromosome of Yersinia enterocolitica. Implications for the evolution of class A β-lactamases. Mol. Gen. Genet. 228:215-220. [DOI] [PubMed] [Google Scholar]
- 32.Seoane, A., M. V. Francia, and J. M. García-Lobo. 1992. Nucleotide sequence of the ampC-ampR region from the chromosome of Yersinia enterocolitica. Antimicrob. Agents Chemother. 36:1049-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimuzu, S., H. Yamaguchi, Y. Ashizawa, Y. Kohno, M. Asami, J. Kato, and H. Ikeda. 1997. Short-homology-independent illegitimate recombination in Escherichia coli: distinct mechanism from short-homology-dependent illegitimate recombination. J. Mol. Biol. 266:297-305. [DOI] [PubMed] [Google Scholar]
- 34.Sougakoff, W., S. Goussard, G. Gerbaud, and P. Courvalin. 1988. Plasmid mediated resistance to third generation cephalosporins caused by point mutations in TEM type penicillinase genes. Rev. Infect. Dis. 10:879-884. [DOI] [PubMed] [Google Scholar]
- 35.Tlsty, T. D., A. M. Albertini, and J. H. Miller. 1984. Gene amplification in the lac region of E. coli. Cell 37:217-224. [DOI] [PubMed] [Google Scholar]
- 36.Whoriskey, S. K., V. Nghiem, P. Leong, J. M. Mason, and J. H. Miller. 1987. Genetic rearrangements and gene amplification in Escherichia coli: DNA sequences at the junctures of amplified gene fusions. Genes Dev. 1:227-237. [DOI] [PubMed] [Google Scholar]