Abstract
A conjugative plasmid, pOLA52, conferring resistance to the antibiotic growth promoter olaquindox has been isolated from Escherichia coli from swine manure. It also confers resistance to ampicillin and chloramphenicol and has a high frequency of transfer between strains of E. coli. Plasmid-borne olaquindox resistance has not been demonstrated before.
The synthetic chemotherapeutic agent olaquindox has found wide use as a growth promoter in pig farming. It is active against coliform bacteria (3), where it inhibits DNA synthesis (10). Resistance has been defined with a breakpoint of 64 μg/ml (3) or 50 μg/ml (7). Until 2000, the compound has been allowed in concentrations of up to 100 mg/kg in feed for pigs younger than 4 months.
Since the introduction of the compound in the 1980s, there has been some concern about whether resistance would arise and, if so, whether this resistance would be transferable and whether it would be linked to other resistance determinants (2, 5, 7). A survey study in Denmark has demonstrated the presence of a small fraction of olaquindox-resistant coliform bacteria in farm animals (3). Other studies have demonstrated increased resistance to olaquindox on pig farms using olaquindox. There was a slight correlation between resistance to olaquindox and resistance to chloramphenicol or ampicillin (5, 7). The aim of the present study was to find out if plasmid-bound olaquindox resistance exists.
Isolation of a resistant bacterium.
Swine manure from a farm using olaquindox as a feed additive was tested for the presence of bacteria able to grow on Gould S1 medium (4) with 100 μg of olaquindox per ml. Diluted manure corresponding to 10 μl of undiluted manure gave rise to three uniform colonies that did not show the fluorescence characteristic of fluorescent pseudomonads. One of the colonies was restreaked several times on Levine EMB plates (GIBCO Products for Microbiology: Technical Manual and Catalog; GIBCO Laboratories, Madison, Wis.) and Luria-Bertani (LB) agar plates (9), both containing 100 μg of olaquindox per ml. Olaquindox (98% pure; ICN, Costa Mesa, Calif.) was added as a 10-mg/ml stock solution in 2.5 M NaOH; HCl was added to the final medium to counteract the high pH. All incubations were done at 37°C. The isolate was identified as Escherichia coli by use of the API 20E system (BioMérieux, Marcy l'Etoile, France).
Olaquindox resistance was tested with an agar dilution test, while resistance to an array of other antibiotics was tested with Sensititre plates (1). The isolate was found to be resistant to ampicillin (MIC, >32 μg/ml), kanamycin (MIC, >64 μg/ml), chloramphenicol (MIC, >64 μg/ml), nitrofurantoin (MIC, 128 μg/ml), streptomycin (MIC, 128 μg/ml), olaquindox (MIC, 128 μg/ml), sulfamethoxazole (MIC, >512 μg/ml), trimethoprim (MIC, >32 μg/ml), and carbadox (MIC, >128 μg/ml). The strain was sensitive to apramycin (MIC, 4 μg/ml), ciprofloxacin (MIC, 0.125 μg/ml), colistin (MIC, 1 μg/ml), gentamicin (MIC, 0.5 μg/ml), nalidixic acid (MIC, 16 μg/ml), and tetracycline (MIC, 1 μg/ml). In addition, the isolate did not grow on LB agar plates containing 100 μg of rifampin per ml.
Conjugal transfer of the plasmid.
Tentative resistance plasmids were transferred to E. coli CSH26 (8) Rifr (resistant to rifampin at 100 μg/ml) by filter conjugation. Overnight cultures of the donor and recipient were harvested by centrifugation (6,000 × g, 5 min, 4°C), washed, and resuspended in phosphate-buffered saline. Equal amounts of the two cultures were mixed and spread on sterile membrane filters (0.2-μm pore size) placed on LB agar plates. The filters were incubated at 37°C for 4 h, after which they were washed in phosphate-buffered saline. The experiment gave rise to approximately 10−4 transconjugants per recipient (T/R). The transconjugants had the rifampin resistance of the recipient combined with resistance to ampicillin (MIC, >32 μg/ml), chloramphenicol (MIC, 64 μg/ml), and olaquindox (MIC, 128 μg/ml). It had achieved intermediate resistance to nitrofurantoin (MIC, 64 μg/ml) relative to the recipient and was sensitive to kanamycin, streptomycin, sulfamethoxazole, trimethoprim, and carbadox. The recipient strain did not grow on plates containing 100 μg of olaquindox per ml. Some of the resistance determinants of the isolate, including olaquindox resistance, had thus been transferred to E. coli CSH26, while others had not. This is consistent with earlier observations (5) that olaquindox resistance can be linked to chloramphenicol resistance. In this case, it is also linked to ampicillin resistance and—to some extent—nitrofurantoin resistance.
Plasmid DNA preparations (6) showed that the transconjugants had received one of the plasmid bands of the donor (data not shown). This plasmid was named pOLA52.
Restriction map.
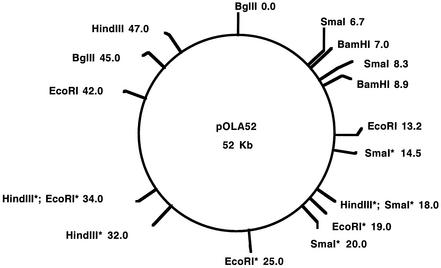
A restriction map of the plasmid has been constructed (Fig. 1) by use of the enzymes EcoRI, BglII, HindIII, BamHI, and SmaI. A QIAprep Spin Miniprep Kit and a QIAGEN Plasmid Midi Kit were used for purification of plasmid DNA (QIAGEN GmbH, Hilden, Germany). In order to improve the separation of large DNA fragments, these were subjected to pulsed-field gel electrophoresis with the Gene Navigator System (with a version 18-1019-19 electrophoresis unit and a HEX electrode) from Pharmacia LKB Biotechnology AB, Uppsala, Sweden.
FIG. 1.
Restriction map of pOLA52. Sites marked with asterisks have not been located exactly. The numbers following the names of the restriction enzymes are distances (in kilobases) from the BglII site defined to be the numbering starting point.
Frequency of conjugal transfer.
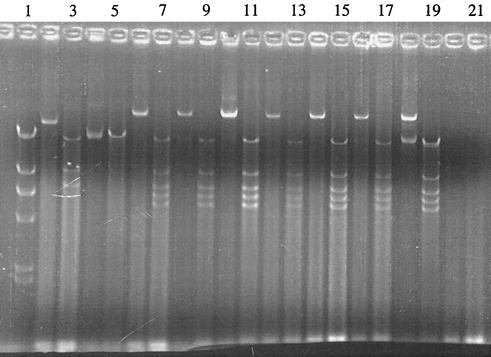
Conjugational transfer of pOLA52 between E. coli CSH26 Rifr and E. coli CSH26 Nalr (resistant to nalidixic acid at 100 μg/ml) was tested further. The frequency of transfer was 0.0069 to 0.055 T/R (mean, 0.026 T/R; standard deviation, 0.015; n = 9). Fifteen transconjugants were tested for resistance to olaquindox (100 μg/ml), rifampin (100 μg/ml), nalidixic acid (100 μg/ml), ampicillin (100 μg/ml), and chloramphenicol (20 μg/ml). Fourteen of the transconjugants tested had the antibiotic resistance profile expected (nalidixic acid, ampicillin, chloramphenicol, and olaquindox), but in one case, the ampicillin resistance was lost. Plasmid DNA was purified from eight transconjugants by use of QIAprep spin columns (QIAGEN GmbH). The DNA was eluted from the spin columns with water preheated to 70°C. Aliquots of the plasmid DNA preparations were digested with EcoRI, followed by gel electrophoresis. The gels were photographed in a Bio-Rad Gel Doc 1000 setup controlled with the Quantity One software, version 4.0.3. Plasmid DNAs from the transconjugants gave identical band patterns on agarose gels, except for the strain that had lost its ampicillin resistance. A similar loss of ampicillin resistance and an altered band pattern were found in transconjugants from several preliminary conjugation experiments. Figure 2 shows examples of band patterns from the donor and from transconjugants with and without loss of ampicillin resistance.
FIG. 2.
Plasmid preparations from strains with and without pOLA52. All preparations were run with and without prior digestion with EcoRI. Lanes: 1, λ DNA digested with HindIII; 2 to 5, plasmid preparations from two transconjugants that had lost the ampicillin resistance of the plasmid (each is shown undigested and digested); 6 to 17, plasmid preparations from six transconjugants (each is shown undigested and digested); 18 and 19, plasmid preparation from the donor, undigested and digested; 20 and 21, “plasmid preparation” from the recipient (no plasmids), undigested and digested.
Concluding remarks.
In this work, we have isolated a highly conjugative plasmid conferring resistance to olaquindox, ampicillin, and—to some extent—chloramphenicol and nitrofurantoin. Plasmid-borne olaquindox resistance has not been demonstrated before. The fact that the plasmid was readily found in pig manure shows that there is a potential for spread of olaquindox resistance among coliform bacteria and that this can be accompanied by the spread of other antibiotic resistance determinants.
Acknowledgments
This work was supported by the Danish Ministry of Food, Agriculture and Fisheries, project MIL96-2.
REFERENCES
- 1.Aarestrup, F. M., Y. Agersø, P. Ahrens, J. C. Ø. Jørgensen, M. Madsen, and L. B. Jensen. 2000. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet. Microbiol. 74:353-364. [DOI] [PubMed] [Google Scholar]
- 2.Corpet, D. E. 1984. The effect of bambermycin, carbadox, chlortetracycline and olaquindox on antibiotic resistance in intestinal coliforms: a new model. Ann. Microbiol. (Paris) 135A:329-339. [DOI] [PubMed] [Google Scholar]
- 3.DANMAP. 1999. DANMAP 98—consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Statens Serum Institut, Danish Veterinary and Food Administration, Danish Medicines Agency, Danish Veterinary Laboratory, Copenhagen, Denmark.
- 4.Gould, W. D., C. Hagedorn, T. R. Bardinelli, and R. M. Zablotowicz. 1985. New selective media for enumeration and recovery of fluorescent pseudomonads from various habitats. Appl. Environ. Microbiol. 49:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedges, A. J., and A. H. Linton. 1988. Olaquindox resistance in the coliform flora of pigs and their environment: an ecological study. J. Appl. Bacteriol. 64:429-443. [DOI] [PubMed] [Google Scholar]
- 6.Ish-Horowicz, D., and J. F. Burke. 1981. Rapid and efficient cosmid cloning. Nucleic Acids Res. 9:2989-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linton, A. H., A. J. Hedges, and P. M. Bennett. 1988. Monitoring for the development of antimicrobial resistance during the use of olaquindox as a feed additive on commercial pig farms. J. Appl. Bacteriol. 64:311-327. [DOI] [PubMed] [Google Scholar]
- 8.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 9.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Suter, W., A. Rosselet, and F. Knüsel. 1978. Mode of action of quindoxin and substituted quinoxaline-di-N-oxides on Escherichia coli. Antimicrob. Agents Chemother. 13:770-783. [DOI] [PMC free article] [PubMed] [Google Scholar]