Abstract
Many VISA (vancomycin intermediately resistant Staphylococcus aureus) strains are characterized by increased cell wall biosynthesis and decreased cross-linking of the peptide side chains, leading to accumulation of free d-alanyl-d-alanine termini in the peptidoglycan, which act as false target sites for vancomycin. A spontaneous mutant of methicillin-resistant VISA strain SA137/93A (vancomycin MIC [E-test], 8 μg/ml), called SA137/93G, showed increased resistance to vancomycin (MIC [E-test], 12 μg/ml). Analysis of the resistance profile of the mutant revealed a loss of β-lactam resistance with a concomitant increase in resistance to glycopeptides. In both strains, cell wall thickness was 1.4-fold greater than that of control isolates. However, cross-linking of the cell wall was drastically lower in SA137/93A than in SA137/93G. The sensitivity of strain SA137/93G to β-lactams was due to loss of the β-lactamase plasmid and a deletion that comprises 32.5 kb of the methicillin resistance cassette SCCmec, as well as 65.4 kb of chromosomal DNA. A spontaneous mutant of SA137/93G with higher sensitivity to vancomycin displayed a cell wall profile similar, in some respects, to that of an fmhB mutant. Results described here and elsewhere show that the only feature common to all VISA strains is a thickened cell wall, which may play a central role in the vancomycin resistance mechanism.
Staphylococcus aureus is one of the most common pathogens and is a trigger of community-acquired and nosocomial disease. The drugs of choice against methicillin-resistant S. aureus (MRSA) strains are the glycopeptide antibiotics vancomycin and teicoplanin. Since 1996, emergence of MRSA strains resistant to vancomycin has been reported in many countries (4-6, 13, 16, 21, 30, 36, 41, 45, 53). According to the NCCLS, strains for which the MIC of vancomycin is less than or equal to 4 μg/ml are considered susceptible whereas those for which the MIC is greater than or equal to 32 μg/ml are considered resistant. Strains for which the MICs are 8 to 16 μg/ml are intermediately resistant. Detailed characterization of clinical isolates and in vitro-selected vancomycin-resistant mutants showed that the resistance phenotype of at least some vancomycin intermediately resistant S. aureus (VISA) strains is caused by activation of cell wall biosynthesis and a significant increase in free d-alanyl-d-alanine termini in the cell wall, which represent false target sites for vancomycin. Resistance was shown to go along with thickening of the cell wall, changes in the composition of the peptidoglycan, and the expression of penicillin-binding proteins (PBPs) (7, 8, 18, 31, 37, 44, 46-48). Some authors have suggested that the genes involved in glycopeptide resistance are probably under the control of a comprehensive regulatory system (8, 33). However, the reported changes in cell wall biochemistry are not restricted to glycopeptide-resistant strains and the magnitude of the changes does not always correlate with the resistance level (7, 39).
In this study, we characterize two VISA isolates and one revertant strain. SA137/93A was identified by the screening of 457 clinical isolates of MRSA from the strain collection of the Reference Center for Staphylococci of the University of Bonn (4). A spontaneous β-lactam-susceptible mutant of SA137/93A with an increased vancomycin MIC was named SA137/93G. SA137/93G1 is a vancomycin-susceptible revertant of strain SA137/93G. We examined the three strains with regard to the previously reported characteristics of glycopeptide-resistant S. aureus strains and with emphasis on the differences between SA137/93A and SA137/93G.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and passaging procedure.
SA137/93A was isolated in 1993 from a tracheal secretion, and its intermediate resistance to vancomycin was recognized during a retrospective screening of the early MRSA from the strain collection of the National Reference Center for Staphylococci in Bonn, Germany (4). Northern German epidemic MRSA strain SA1450/94 and S. aureus NCTC 8325 were provided by the National Reference Center for Staphylococci in Wernigerode, Germany. Japanese VISA isolate Mu50 was isolated by Hiramatsu et al. (21). S. aureus strains were stored as frozen stocks in 50% glycerol at −70°C and cultured in brain heart infusion (BHI) medium (Oxoid, Wesel, Germany) at 37°C with aeration unless indicated otherwise. For each experiment, an overnight culture was diluted 100-fold in fresh BHI broth and further incubated to ensure exponential growth conditions. Cell growth was monitored by measuring the culture optical density at 600 nm (OD600). To evaluate the hemolysis phenotype, strains were streaked onto sheep blood agar, incubated at 37°C overnight, and kept in a refrigerator for 24 h. Serial passaging was done daily on BHI agar by choosing several colonies from each plate and passaging them on BHI agar again. SA137/93G1 was detected by picking colonies on BHI agar with or without vancomycin.
Antimicrobial susceptibility testing.
MIC determinations were performed in BHI broth and cation-supplemented Mueller-Hinton broth by the broth microdilution method with an inoculum of 5 × 105 CFU/ml (42) and by E-test on BHI agar (Oxoid) by applying an inoculum of 2 McFarland. Growth was read after 24 and 48 h of incubation at 37°C. The exact MICs were determined by a modified broth microdilution method using arithmetic instead of twofold dilution. The population analysis was carried out as described previously (21).
PFGE and phage typing.
Chromosomal DNA for the SmaI and ApaI (Roche, Mannheim, Germany) restriction digests was purified as described previously (15). Pulsed-field gel electrophoresis (PFGE) was performed on the DRIII contour-clamped homogeneous electric field system (Bio-Rad, Munich, Germany) by using Pulsed-Field Certified Agarose (1%; Bio-Rad), 6 V/cm, a field angle of 120°, a switch time of 5 to 15 s for 7 h, and a switch time of 15 to 60 s for a further 19 h. S. aureus NCTC 8325 served as the mass standard.
Phage typing was performed with the international set for phage typing employed at the routine test dilution and 100 times the routine test dilution in accordance with the standard rules agreed on by the International Union of Microbiological Societies Subcommittee on Phage Typing of Staphylococci.
Characterization of the SCCmec type and chromosomal deletion of strain SA137/93G by PCR.
Chromosomal DNA of the strains was purified after lysis of the cells in the presence of 5 μg of lysostaphin per ml for 16 h at 37°C by employing genomic tip20 columns (Qiagen, Hilden, Germany).
The presence of mecA was probed by PCR with primers mecAI (5′-AAA ATC GAT GGT AAA GGT TGG C-3′) and mecAII (5′-AGT TCT GCA GTA CCG GAT TTG C-3′). The ccr (cassette chromosome recombinase) complex was identified with primer β2, which is common to all three ccrB genes, and three primers specific for each ccrA gene, α2 (ccrA1) α3 (ccrA2), and α4 (ccrA3) (25). The chromosomal deletion of strain SA137/93G was analyzed by starting with primers cR4 (25), which corresponds to the 5′ end of orfX, and mr8 (28). It was finally characterized by using primers E040 3′ (5′-ACT TTA GCC ATT GCT ACC TTC-3′), which anneals to the IS431 transposase gene of the type I SCCmec of S. aureus NCTC 10442 (EMBL accession no. AB033763), and drm 3′ (5′-CTG GCA AGC GAT CAT CGA AA-3′), which anneals to the phosphopentomutase gene in the nucleotide sequence of S. aureus N315 (EMBL accession no. AP003129). The 2.7-kb PCR product that was obtained with these primers and Pwo polymerase (AGS, Heidelberg, Germany) was purified (Qiagen PCR purification kit) and sequenced with primer DEL SEQ 2 (5′-CGT TTT AAA TTG TTT ATA TCT G-3′) (Sequiserve, Vaterstetten, Germany). CE010 of the type I SCCmec was identified with primers 5′-TTT ATT ATT TGG TTT ATC TGT TTT G-3′ (forward) and 5′-GAT AGC GAC TCT GAT GCG G-3′ (reverse).
Determination of the agr type.
Classification of the two VISA strains into the four agr specificity groups of S. aureus was done by PCR as described by Jarraud et al. (26). Primers 5′-ATG CAC ATG GTG CAC ATG CA (forward) and 5′-CAT AAT CAT GAC GGA ACT TGC GC (reverse) were used to amplify an ∼1.2-kb DNA fragment that comprises a hypervariable region including the 3′ terminus of agrB, all of agrD, and the 5′ terminus of agrC. The PCR was performed with Pwo polymerase (AGS) by using a standard protocol. The PCR products were purified (Qiagen PCR purification kit) and sequenced (Sequiserve).
Transmission electron microscopy.
The VISA strains were cultured in the absence and in the presence of vancomycin (0.5 × MIC). The cells were fixed in 3% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 6.8, containing 0.1 M sucrose and 1% tannin for 4 h at 4°C. For contrast amplification, the cells were treated with 2% uranyl acetate in 0.9% NaCl for 3 h at 20°C in the dark. Cells were dehydrated with 70% ethanol for 18 h and embedded in acrylic resin (LR White, medium grade; London Resin Company, Berkshire, United Kingdom). Ultrathin sections were stained with lead citrate and then examined with a transmission electron microscope (Zeiss EM10A).
Calculation of cell wall thickness was performed by means of photographs taken at a final magnification of ×63,000. To measure cell wall thickness, 15 cells that had been cut equatorially were chosen from each strain and the ratio of cell wall diameter to cell size was calculated.
Analysis of peptidoglycan by high-performance liquid chromatography (HPLC).
Mutanolysin-digested muropeptides were prepared from S. aureus isolates, and the muropeptide mixture was separated by HPLC as described previously (51), except that the cells were broken by homogenizing in the presence of glass beads (diameter, 0.45 to 0.5 mm; Braun).
Membrane purification and analysis of PBPs.
Membranes of S. aureus were prepared from 6 liters of cell culture with an OD600 of 0.6 to 0.8. The cells were harvested by centrifugation, washed with cold 10 mM Tris maleate buffer (pH 7.8), and resuspended in 6 ml of buffer for disruption in the presence of glass beads (diameter, 0.45 to 0.5 mm; Braun). The cell lysate was centrifuged at 12,000 × g for 10 min to remove cell debris and glass beads. The supernatant was collected, and the pellet was washed twice with buffer. The total volume of the supernatant was ultracentrifuged at 150,000 × g for 60 min. The membrane pellets were washed twice with 10 mM potassium phosphate buffer, pH 6.8, and finally resuspended in a total volume of 500 μl of 50 mM potassium phosphate buffer, pH 7, containing 10 mM magnesium chloride. The protein concentration of the membranes was determined by the bicinchoninic acid method (Pierce, Perbio Science, Bonn, Germany), and the preparations were stored at −20°C.
Detection of PBPs was based on the method developed by Spratt (50). The membrane preparations were incubated for 30 min at 37°C with biotinylated ampicillin (11), which binds to the PBPs (250 μg of protein and 2.5 μg of biotinylated ampicillin in a total volume of 35 μl of phosphate buffer, pH 7). The reaction was stopped by adding penicillin G (120 μg in 2 μl). Aliquots of the samples were subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. The separated proteins were blotted onto a nitrocellulose membrane, and the PBPs were visualized on an X-ray film by enhanced chemiluminescence detection with a streptavidin-horseradish peroxidase antibody conjugate (11).
Sequencing of pbp4.
The pbp4 gene of the VISA strains and its promoter region (2,076 bp) were PCR amplified as previously described (44), with Pwo polymerase (AGS). The PCR products were purified (Qiagen PCR purification kit) and sequenced (Sequiserve).
Time-to-regrowth assay.
For each strain, 250 ml of BHI in a 1,000-ml culture flask was inoculated with 2.5 ml of an overnight culture. When the cultures had reached an OD600 of 0.4, vancomycin was added to a final concentration of 30 μg/ml. At the end of each experiment, the cultures were checked for contamination by plating on blood agar and determining the MIC. CFU were determined by serial dilution and plating on Columbia agar. For the vancomycin bioassay, a culture of Micrococcus luteus ATCC 4698 was grown to an OD600 of 0.3 and used to inoculate 1,000 ml of 0.6% Luria-Bertani agar at 45°C. Thin agar plates with seven wells per plate were prepared. Culture samples were centrifuged, and the supernatant was pasteurized at 80°C for 10 min. From each sample, six 50-μl aliquots were pipetted into the wells. As standards, 22 vancomycin concentrations ranging from 1 to 30 μg/ml in BHI were used. Additionally, one well per plate was filled with BHI containing 10 μg of vancomycin per ml in order to adjust for plate thickness. After 48 h of incubation at 37°C, the diameters of the inhibition zones were read.
RESULTS
Resistance determination and stability of the resistance phenotype.
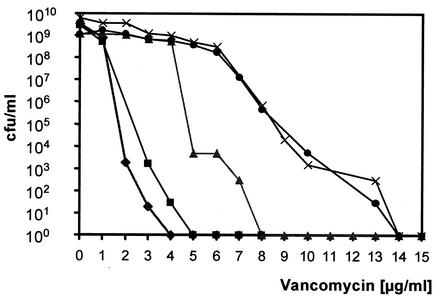
Analysis of the vancomycin resistance level by E-test, broth microdilution, and population analysis revealed that both isolates are VISA strains (Table 1; Fig. 1). Strain SA137/93G, which had coincidentally been isolated from a glycerol stock culture of SA137/93A, showed greater resistance than SA137/93A. The population analyses demonstrated similar resistance profiles for SA137/93G and Mu50, which is a homogeneously resistant strain (Fig. 1). However, the MIC of vancomycin for SA137/93G in cation-supplemented Mueller-Hinton broth was lower than that of Mu50 and reached only 4 μg/ml, which is typical for heterogeneously resistant strains. By comparison, all VISA strains reached the highest MICs with the E-test procedure. The vancomycin resistance phenotype of both VISA isolates was stable for at least 30 passages without selective pressure. However, a single vancomycin-susceptible revertant of strain SA137/93G (MIC, 2 μg/ml in BHI) called SA137/93G1 was obtained by screening 50 colonies after the first passage. SA137/93A is an MRSA strain (Table 1), while SA137/93G has lost its resistance to penicillin and methicillin (Table 1) and a PCR experiment with mecA primers did not yield a product.
TABLE 1.
Determination of MICs by different susceptibility test methods
| Strain | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Vancomycin twofold microdilution
|
Vancomycin arithmetic microdilution, BHI | Vancomycin E-test, BHI | Teicoplanin twofold microdilution
|
Oxacillin twofold microdilution, BHI | Penicillin twofold microdilution, BHI | |||
| BHI | CAMHa | BHI | CAMH | |||||
| NCTC 8325 | 2 | 1 | 2 | NDb | 2 | 1 | 0.5 | 0.06 |
| SA1450/94 | 2 | 1 | 2 | ND | 2 | 4 | 16 | ND |
| SA137/93A | 8 | 2 | 5 | 8 | 16 | 8 | 256 | 32 |
| SA137/93G | 8/16 | 4 | 6 | 12 | 32 | 32 | 0.5 | 0.06 |
| SA137/93G1 | 2 | 1/2 | ND | ND | 2 | ND | 0.5 | ND |
| Mu50 | 8 | 4/8 | 6 | 12-16 | 16 | 16 | 512 | 4 |
CAMH, cation-supplemented Mueller-Hinton broth.
ND, not determined.
FIG. 1.
Population analysis of VISA isolates SA137/93A (▴) and SA137/93G (×) and control strains NCTC 8325 (♦), SA1450/94 (▪), and Mu50 (•). The points on the abscissa refer to zero CFU values.
MRSA strain SA1450/94, which is clonally related to the VISA isolates, showed complete beta-hemolysis on sheep blood agar. In contrast, only single colonies of SA137/93A and SA137/93G showed beta-hemolysis.
PFGE and phage typing.
Analysis of VISA isolates by PFGE had previously shown a close relationship to northern German epidemic strain SA1450/94 (4). SA137/93G displayed a three-band difference from SA137/93A (Fig. 2). Band B was slightly larger, while band F was missing and there seemed to be a doublet at the position of band H. Vancomycin-susceptible revertant SA137/93G1 displayed the same banding pattern as its parent, strain SA137/93G (data not shown). Phage typing confirmed the close relationship of strains SA137/93A and SA137/93G, while strains SA137/93G1 and SA137/93G displayed identical phage patterns (data not shown).
FIG. 2.
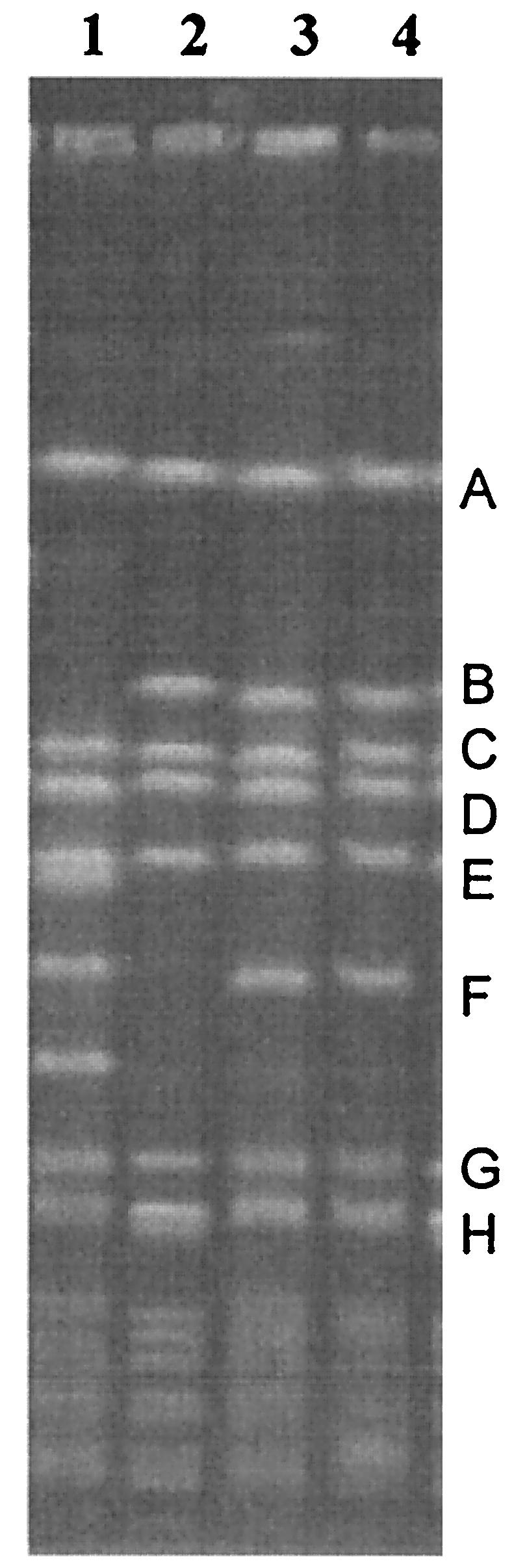
PFGE of the strains employed in this study. Lanes: 1, NCTC 8325; 2, SA137/93G; 3, SA1450/94; 4, SA137/93A.
Characterization of the SCCmec and agr types and mapping of the chromosomal deletion of strain SA137/93G by PCR.
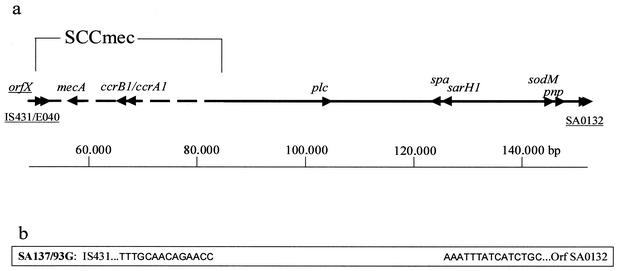
Since the mecA PCR had not yielded a product with chromosomal DNA of SA137/93G, the strain was screened for the presence of the surrounding open reading frames (ORFs). In S. aureus, four types of SCCmec with substantial differences in nucleotide sequence and size have been described and can be discerned by PCR (25, 35). SA1450/94 and SA137/93A both possessed a ccr1 (cassette chromosome recombinase) complex and ORF CE010 (fibrinogen-binding protein), which is present only in type I SCCmec. Neither of the strains gave a product with primers mI1 and mI2 (29), which anneal within mecI, and thereby indicated the presence of a class B mec complex. The combination of ccr1 and a class B mec complex is also typical of type I MRSA strains (25, 35). In contrast, primers cR4 and mR8, which amplify the downstream junction of SCCmec and chromosomal DNA, gave a product with all three strains, indicating that SCCmec had not been fully excised by the ccr complex in SA137/93G. However, the upstream junction of SCCmec was not present in SA137/93G. The presence of various upstream chromosomal ORFs was probed by PCR assay, and the first ORF that yielded a PCR fragment in SA137/93G was drm, which encodes phosphopentomutase and is located about 67 kb upstream of SCCmec in S. aureus N315. The gap that is formed by the deletion in SA137/93G was closed with a primer pair annealing in E040, the IS431 transposase gene in the type I SCCmec, and drm, which yielded a 2.7-kb product. The sequence present in SA137/93G is shown in Fig. 3b. The deletion in SA137/93G starts exactly downstream of the IS431 indirect repeat in SCCmec, ends within SA0132, and encompasses about 98 kb of DNA (Fig. 3a). Additionally, a large plasmid, which probably carries the gene for β-lactamase, is not present in SA137/93G (data not shown) and the loss of β-lactamase production was obvious in an agar diffusion assay with a penicillin disk (10 μg) and culture supernatant (50 μl) of both strains with S. aureus NCTC 8325 as the indicator strain.
FIG. 3.
(a) The ca. 100-kb chromosomal fragment that is present in MRSA strains SA1450/94 and SA137/93A and that was deleted in methicillin-susceptible S. aureus SA137/93G. (b) Nucleotide sequence present in SA137/93G. The 5′ end comprises the indirect repeat of IS431, which is directly attached to the 3′ end of ORF SA0132.
The agr locus regulates the expression of most staphylococcal exoproteins and other virulence factors and represents a density-dependent autoinducible signal-transducing system. The staphylococcal agr locus is extensively conserved, but there is some sequence variation, which can serve to define distinct interfering specificity groups (27). Amino acid sequence comparison of agr specificity groups I to IV and VISA strains SA137/93A and SA137/93G revealed identity with the conserved AgrD motif of the group I agr locus (U85097) (26; sequence not shown). The fact that the entire nucleotide sequences of SA137/93A and SA137/93G were identical serves as additional confirmation that SA137/93G is a mutant of strain SA137/93A. Mu50 and most other VISA strains belong to agr specificity group II, while most clinical S. aureus isolates belong to agr group I (34, 43).
Transmission electron microscopy.
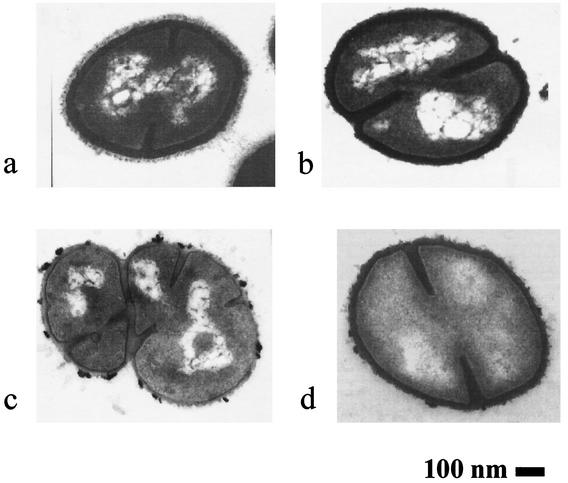
Figure 4 shows photographs of ultrathin sections of both VISA isolates, vancomycin-susceptible mutant SA137/93G1 and control strain NCTC 8325, grown in the absence of vancomycin. The cell wall thickness of the VISA strains was approximately 1.4-fold that of the control strain (Table 2). Additionally, cells of strain SA137/93G (Fig. 4b) were significantly smaller. However, the cell wall took up 8.4% of the total cell diameter (control, 6.1%). After growth in the presence of vancomycin (0.5 × MIC), only strain SA137/93A showed changes in cell size. The cell diameter decreased from 0.82 to 0.72 μm, but the cell wall remained thickened (8.2%) (photo not shown). An increase in the cell wall diameter of the VISA strains only in the presence of vancomycin as described by Pfeltz et al. was not observed (39).
FIG. 4.
Electron micrographs of ultrathin sections of VISA strains SA137/93A (a) and SA137/93G (b), susceptible revertant SA137/93G1 (c), and control strain NCTC 8325 (d). Magnification, ×63,000.
TABLE 2.
Cell wall diameter of VISA strains and vancomycin-susceptible revertant SA137/93G1 compared to that of control strain NCTC 8325
| Strain | Cell diam (μm) | Cell wall diam (nm) | % Cell wall | Fold increase in cell wall thickness |
|---|---|---|---|---|
| NCTC 8325 | 0.87 | 26.4 | 6.1 | 1 |
| SA137/93A | 0.82 | 35.6 | 8.7 | 1.4 |
| SA137/93G | 0.72 | 30.0 | 8.4 | 1.4 |
| SA137/93G1 | 0.88 | 24.5 | 5.6 | 0.9 |
Cell size and cell wall diameter of vancomycin-susceptible mutant SA137/93G1 were similar to those of the control strain (Fig. 4c; Table 2). However, the cell division process of this strain was incomplete and the cells built up large aggregates. Despite this, the growth of the strain did not seem impaired.
Cross-linking of the peptidoglycan.
Analysis of the cell wall muropeptides revealed that the sensitive control strain S. aureus NCTC 8325 was characterized by a normal cross-linking value of 75.4% (Table 3). Only a slightly decreased cross-linking value was found for SA137/93G. However, in the other isolate, strain SA137/93A, the cross-linking was decreased to a low value of 65.9%. In this strain, the amount of highly cross-linked peptidoglycan (oligomers) was significantly reduced and the amount of monomers and dimers was increased compared to those of the other strains. Growth of the cells in the presence of vancomycin did not have any significant influence on the results.
TABLE 3.
Peptidoglycan analysisa
| Strain | % Cross-linkingb | % Monomers | % Dimers | % Trimers | % Oligomers | % Free d-Ala-d-Ala residues |
|---|---|---|---|---|---|---|
| NCTC 8325 | 75.4 | 9.0 | 11.9 | 7.7 | 71.4 | 24.6 |
| SA137/93A | 65.9 | 17.1 | 17.2 | 7.8 | 57.9 | 34.1 |
| SA137/93G | 73.4 | 10.5 | 13.3 | 8.0 | 68.2 | 26.6 |
| SA137/93G1 | 75.0 | 10.2 | 10.0 | 7.7 | 72.1 | 24.9 |
After digestion with mutanolysin, the muropeptides were separated by reversed-phase HPLC. The data represent the percent are a under the elution peaks and thus the amounts of the respective muropeptides.
Calculated by 0.5 × dimer (%) + 0.67 × trimer (%) + 0.9 × oligomer (%).
Muropeptide analysis of phenotypic revertant strain SA137/93G1 revealed drastic changes in the composition of the peptidoglycan compared to that of its parent, strain SA137/93G. There was an increased proportion of monomers with no pentaglycine chain and dimers that seemed to be directly cross-linked (data not shown). The presence of these unusual peaks in the HPLC profile indicated that phenotypic revertant SA137/93G1 probably harbors a defect in the biosynthesis of the glycine interpeptide bridges. This may be the reason for its decreased resistance to vancomycin and the morphological defects shown in Fig. 4c. The gene fmhB, which codes for the enzyme that adds the first glycine residue to the stem pentapeptide, was sequenced. However, the nucleotide sequence of SA137/93G1 was identical to the sequence of S. aureus NCTC 8325 (EMBL accession no. 105976) and to the sequence of SA137/93G, and thus, the reason for the observed alteration in cell wall biochemistry remains unclear.
Analysis of PBPs and sequencing of pbp4.
In S. aureus, five PBPs have been identified. PBP1 to -4 are present in all S. aureus strains, while PBP2a, which confers methicillin resistance (24), is only found in MRSA.
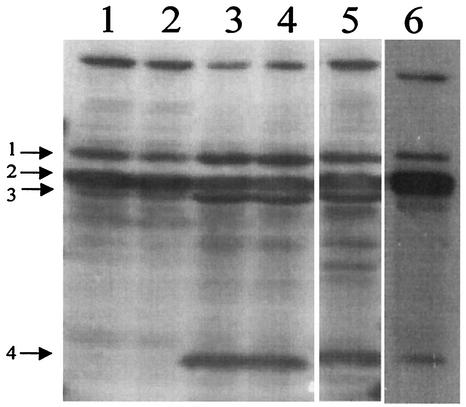
PBPs catalyze the transpeptidation and transglycosylation reactions of bacterial cell wall biosynthesis (38). Figure 5 shows the PBP profiles of the VISA isolates and control strains NCTC 8325 and SA1450/94. In SA137/93G, the PBPs were expressed in amounts that were comparable to those in the control strains. In contrast, in SA137/93A, the enzymatic activities of PBP3 and -4 appeared to be lower or not present, respectively. PBP4 has transpeptidase and dd-alanine carboxypeptidase activity, thereby removing the terminal d-alanine residue of the pentapeptide side chain (32). Thus, loss of activity of PBP4 has been shown to lead to lower cross-linking and a concomitant increase in false target sites (12, 48, 55) and explains the lower cross-linking detected in strain SA137/93A than in strain SA137/93G. Sequencing of pbp4 and its promoter region demonstrated that the nucleotide sequences of the two VISA isolates were identical, indicating that the weak PBP4 band in the PBP profile of SA137/93A was not caused by any mutation in the structural gene or the promoter region of pbp4 (12, 48).
FIG. 5.
PBP patterns of control strains NCTC 8325 (lane 5) and SA1450/94 (lane 6) and VISA isolates SA137/93A (lanes 1 and 2) and SA137/93G (lanes 3 and 4) cultured in the absence (lanes 1 and 3) and presence (lanes 2 and 4) of vancomycin. About 125 μg of separated membrane proteins was loaded onto the gel. The PBPs were labeled with biotinylated ampicillin and detected by Western blotting. The numbers on the left correspond to PBP1 to -4.
Binding of vancomycin to living cells—time to regrowth.
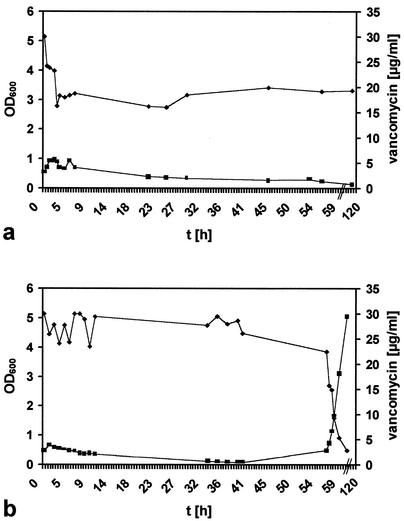
After addition of 30 μg of vancomycin per ml, culture growth was monitored by determination of OD600 and CFU. Vancomycin binding to the cells was determined by measuring the vancomycin concentration in the supernatant. SA137/93A (2 × 109 cells/ml) was able to remove 12 μg of the antibiotic per ml from the medium. The OD600 oscillated between 0.5 and 1 in the first 20 h of the experiment and dropped to 0.1 in the following 5 days (Fig. 6a). After this time, viable cells could not be recovered from the culture medium. SA137/93G bound less vancomycin from the medium (7 μg/2 × 109 cells; Fig. 6b) than did SA137/93A. While the OD600 of the culture decreased from 0.6 to 0.1 over 40 h, the vancomycin concentration in the supernatant averaged 27 μg/ml. After 2 days, the culture started to regrow and the vancomycin concentration in the supernatant decreased proportionally to the increase in cell mass. Thus, although able to remove about 12 μg of vancomycin per ml from the medium, SA137/93A was not able to restart growth in the presence of 18 μg of vancomycin per ml, whereas SA137/93G started to regrow without binding great amounts of the antibiotic from the supernatant. The different capacities of the strains to bind vancomycin were consistent with the results of the muropeptide analysis of the two VISA strains (Table 3) and the presence of a considerably greater amount of free d-Ala-d-Ala groups in SA137/93A that act as binding sites for vancomycin.
FIG. 6.
Uptake of vancomycin from medium by VISA cells. Vancomycin at 30 μg/ml was added to a growing culture (OD600, 0.4), and binding of vancomycin to the cells was monitored by determining the concentration of vancomycin in the supernatant. a, SA137/93A; b, SA137/93G.
DISCUSSION
On the basis of their glycopeptide MICs, Boyle-Vavra et al. divided clinical glycopeptide intermediately resistant S. aureus (GISA) isolates into three groups and postulated that the vancomycin resistance phenotype is unstable in these clinical strains (6). Analysis of the resistance phenotype of VISA isolates SA137/93A and SA137/93G revealed that they are members of group A, which is intermediately resistant to vancomycin and teicoplanin. In contrast to other reports (1, 6), the resistance phenotype of our isolates was stable for at least 30 passages on drug-free medium, as was also reported for some laboratory mutants (39). However, a single strain with sensitivity to vancomycin (SA137/93G1; MIC, 2 μg/ml) was isolated from strain SA137/93G (MIC, 8 μg/ml). Peptidoglycan analysis of SA137/93G1 revealed that it was not a genotypic revertant but gained at least one additional mutation. In this strain, the percentage of pentaglycine interpeptide bridges was relatively low and direct cross-linking of the peptidoglycan precursors with no interpeptide bridge was observed. The latter phenomenon points to profound changes in PBP activity. The reasons for this are still unclear; however, these results indicate that correct biosynthesis of the peptidoglycan may be as essential for vancomycin resistance as for methicillin resistance (3). A loss of teicoplanin resistance was also observed by Sieradzki et al. after inactivation of femB and femC (47). Obviously, loss of vancomycin resistance can be caused by additional mutations and phenotypic revertants may be genetically quite distinct from the sensitive parent strain of the vancomycin-resistant isolate.
The vancomycin resistance mechanism in Mu50 is based on increased cell wall turnover and a thickened cell wall with decreased cross-linking, which leads to an increase in false target sites for vancomycin (13, 17). Actually, a thickened cell wall was observed in all of the VISA strains characterized so far and was also seen in the isolates described here (5, 8, 9, 30, 36). Analysis of the peptidoglycan revealed that the cell wall cross-linking of SA137/93A was drastically decreased to 65.9%, which is even lower than the values obtained for Japanese isolate Mu50, with 72% cross-linking, and American VISA isolate NJ, which is characterized by 73.9% cross-linking (7). SA137/93A was also characterized by increased uptake of vancomycin from the culture medium compared to the other strain. The uptake of vancomycin is presumably mediated by the free d-Ala-d-Ala groups, which act as false targets for vancomycin and could prevent access of further antibiotic to the lipid membrane by clogging the cell wall (8). However, SA137/93A, which displayed the lowest cross-linking, was not the strain that was characterized by the highest resistance to vancomycin. Similar phenomena have been described before, for example, resistant American isolate IL, which did not show low cross-linking (81%). On the other hand, a femC mutant was characterized that contains a high percentage of free d-Ala-d-Ala groups without being automatically vancomycin resistant (7). In conclusion, the above-described results indicate that low cross-linking and a high content of d-Ala-d-Ala groups may increase the resistance of the strain but are not the only mechanisms that can provide insensitivity to vancomycin (6).
PBP analysis revealed that PBP4 activity was absent or greatly reduced in SA137/93A. PBP4 has been shown to have transpeptidase and dd-carboxypeptidase activities (32), and knockout of PBP4 resulted in a 7 to 14% reduction in cross-linking of the peptidoglycan (12), whereas overproduction of PBP4 led to an increase in peptidoglycan cross-linking and to increased resistance to methicillin (10, 19, 20). In SA137/93A, the loss of PBP4 activity was consistent with very low cross-linking of the peptidoglycan, which is alleviated in its descendant SA137/93G, which displays normal PBP4 activity. In a recent study by Finan et al., loss of PBP4 activity, as demonstrated by PBP profiles, was a consistent finding in clinical VISA isolates (12). This loss of activity was shown to be directly related to vancomycin resistance by demonstrating a two- to threefold decrease in vancomycin resistance when PBP4 activity was provided in trans on a high-copy-number plasmid. However, Western blotting demonstrated an immunoreactive PBP4 band in all VISA isolates and sequencing did not yield any obvious changes that might be deleterious to the enzymatic activity (12). The nucleotide sequences of pbp4 and a 400-bp stretch covering the proximal noncoding region of both of the VISA isolates in this study were identical and therefore do not explain the observed differences in activity. In conclusion, the activity of PBP4 seems to play a role in vancomycin resistance; however, the mechanism of regulation of PBP4 activity, which may be one of the key features of vancomycin resistance in clinical strains, remains unclear.
The data presented above demonstrate that (i) SA137/93G emerged from SA137/93A and is characterized by the loss of a large chromosomal fragment including mecA, (ii) the cell wall cross-linking of SA137/93G is greater than that of SA137/93A and PBP4 is active and (iii), despite this, its resistance to vancomycin is increased. The resistance mechanism of SA137/93G is still unclear and—since it is a laboratory mutant—may be incompatible with survival in a clinical setting. However, the most intriguing question is whether the chromosomal deletion could be the cause for the changes in cell wall biochemistry and/or resistance.
Loss of methicillin resistance has been described for highly vancomycin-resistant mutant VM50, in which the mecA gene is disrupted by sequence duplication and the insertion of a stop codon (49). In contrast, SA137/93G lacks nearly the complete SCCmec region and an additional fragment comprising chromosomal genes. Similar deletions of the mecA region in connection with the loss of a β-lactamase plasmid have already been reported (40, 54), and it has been proposed that penicillinase plasmids may play an important role in the stability and phenotypic expression of the mecA gene (22). It remains unclear whether the presence of PBP2a inhibits full expression of the vancomycin resistance phenotype (2, 49).
On the other hand, loss of other genes located on the deleted fragment detected in SA137/93G or independent mutations elsewhere on the chromosome may have caused the increase in vancomycin resistance and/or changes in cell wall biochemistry in this strain. In addition to several conserved hypothetical proteins, the chromosomal deletion of SA137/93G comprises four ORFs coding for proteins with similarity to transcriptional regulators, the genes spa (protein A) and sodM (superoxide dismutase) (23), and four genes involved in exopolysaccharide and capsular polysaccharide synthesis. One of the transcriptional regulators is staphylococcal accessory regulator A homologue 1, SarH1, which is an agr-dependent global regulator of virulence gene expression in S. aureus (52). It has a strong repressive effect on hla (α-toxin) transcription and an activating effect on spa (protein A) transcription and affects the transcription of several other exoproteins (52). Since it belongs to a global regulatory system, an influence on the vancomycin resistance phenotype or cell wall cross-linking and activity of PBP4 might be possible, especially since the expression of spa is downregulated in Mu50 (33). Most research on the resistance mechanism of GISA has been performed on highly resistant laboratory mutants and clinical VISA isolate Mu50 (8, 14, 17, 18, 39, 48). Data explaining the mechanism of glycopeptide resistance of clinical strains from other sources are still rare (7, 12). However, results described here and elsewhere show that there seems to be no resistance mechanism common to all VISA isolates and, with the exception of cell wall thickness, reports on resistance stability, changes in cell wall biochemistry, enzyme activities, and gene expression seem to be true only of single isolates (7). Therefore, every opportunity to study new and especially genetically related VISA isolates should be taken advantage of in order to elucidate similarities and differences between the strains. These studies are the basis for the generation of insight into the evolution of VISA strains and the development of novel strategies by which to combat this adaptive form of antibiotic resistance.
Acknowledgments
This work was supported by grants Bi 504/4-1 and Bi 504/4-2 from the Deutsche Forschungsgemeinschaft to G.B.
We are grateful to Keiichi Hiramatsu for kindly providing strain Mu50 and to the National Reference Center for Staphylococci in Wernigerode, Germany, which provided strain SA1450/94 and S. aureus NCTC 8325. We thank Christiane Szekat and Simone Masberg for expert technical assistance with electron microscopy and Brigitte Buchen of the Botanical Department of the University of Bonn for providing the technical equipment for electron microscopy.
REFERENCES
- 1.Aeschlimann, J. R., E. Hershberger, and M. J. Rybak. 1999. Analysis of vancomycin population susceptibility profiles, killing activity, and postantibiotic effect against vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avison, M. B., P. M. Bennett, R. A. Howe, and T. R. Walsh. 2002. Preliminary analysis of the genetic basis for vancomycin resistance in Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 49:255-260. [DOI] [PubMed] [Google Scholar]
- 3.Berger-Bächi, B. 1999. Genetic basis of methicillin resistance in Staphylococcus aureus. CMLS Cell Mol. Life Sci. 56:764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierbaum, G., K. Fuchs, W. Lenz, C. Szekat, and H.-G. Sahl. 1999. Presence of Staphylococcus aureus with reduced susceptibility to vancomycin in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 18:691-696. [DOI] [PubMed] [Google Scholar]
- 5.Bobin-Dubreux, S., M.-E. Reverdy, C. Nervi, M. Rougier, A. Bolmström, F. Vandenesch, and J. Etienne. 2001. Clinical isolate of vancomycin-heterointermediate Staphylococcus aureus susceptible to methicillin and in vitro selection of a vancomycin-resistant derivative. Antimicrob. Agents Chemother. 45:349-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., S. K. Berke, J. C. Lee, and R. S. Daum. 2000. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob. Agents Chemother. 44:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle-Vavra, S., H. Labischinski, C. C. Ebert, K. Ehlert, and R. S. Daum. 2001. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daum, R. S., S. Gupta, and R. Sabbagh. 1992. Characterization of Staphylococcus aureus isolates with decreased susceptibility to vancomycin and teicoplanin: isolation and purification of a constitutively produced protein associated with decreased susceptibility. J. Infect. Dis. 166:1066-1072. [DOI] [PubMed] [Google Scholar]
- 10.Domanski, T. L., B. L. M. De Jonge, and K. W. Bayles. 1997. Transcriptional analysis of the Staphylococcus aureus gene encoding penicillin-binding protein 4. J. Bacteriol. 179:2651-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehlert, K. 1995. Kopplung von Phospholipid- und Mureinsynthese in Escherichia coli. Ph.D. thesis. Universität Tübingen, Tübingen, Germany.
- 12.Finan, J. E., G. L. Archer, M. J. Pucci, and M. W. Climo. 2001. Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:3070-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisel, R., F.-J. Schmitz, L. Thomas, G. Berns, O. Zetsche, B. Ulrich, A. C. Fluit, H. Labischinski, and W. Witte. 1999. Emergence of heterogeneous intermediate vancomycin resistance in Staphylococcus aureus isolates in the Düsseldorf area. J. Antimicrob. Chemother. 43:846-848. [DOI] [PubMed] [Google Scholar]
- 14.Geisel, R., F. J. Schmitz, A. C. Fluit, and H. Labischinski. 2001. Emergence, mechanism, and clinical implications of reduced glycopeptide susceptibility in Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 20:658-697. [DOI] [PubMed] [Google Scholar]
- 15.Goering, R. V., and R. D. Duensing. 1990. Rapid field inversion gel electrophoresis in combination with an rRNA gene probe in the epidemiological evaluation of staphylococci. J. Clin. Microbiol. 28:426-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerin, F., A. Buu-Hoï, J.-L. Mainardi, G. Kac, N. Colardelle, S. Vaupré, L. Gutmann, and I. Podglajen. 2000. Outbreak of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in a Parisian hospital. J. Clin. Microbiol. 38:2985-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 18.Hanaki, H., H. Labischinski, Y. Inaba, N. Kondo, H. Murakami, and K. Hiramatsu. 1998. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 42:315-320. [DOI] [PubMed] [Google Scholar]
- 19.Henze, U. U., and B. Berger-Bächi. 1996. Penicillin-binding protein 4 overproduction increases β-lactam resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:2121-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henze, U. U., M. Roos, and B. Berger-Bächi. 1996. Effects of penicillin-binding protein 4 overproduction in Staphylococcus aureus. Microb. Drug Resist. 2:193-199. [DOI] [PubMed] [Google Scholar]
- 21.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-146. [DOI] [PubMed] [Google Scholar]
- 22.Hiramatsu, K., E. Suzuki, H. Takayama, Y. Katayama, and T. Yokota. 1990. Role of penicillinase plasmids in the stability of the mecA gene in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 34:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horst, J.-P., T. Wu, and M. G. Marinus. 1999. Escherichia coli mutator genes. Trends Microbiol. 7:29-36. [DOI] [PubMed] [Google Scholar]
- 24.Inglis, B., P. R. Matthews, and P. R. Stewart. 1988. The expression in Staphylococcus aureus of cloned DNA encoding methicillin resistance. J. Gen. Microbiol. 134:1465-1469. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarraud, S., G. J. Lyon, A. M. S. Figueiredo, L. Gérard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 28.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, M.-N., C. H. Pai, J. H. Woo, J. S. Ryu, and K. Hiramatsu. 2000. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 38:3879-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komatsuzawa, H., K. Ohta, S. Yamada, K. Ehlert, H. Labischinski, J. Kajimura, T. Fujiwara, and M. Sugai. 2002. Increased glycan chain length distribution and decreased susceptibility to moenomycin in a vancomycin-resistant Staphylococcus aureus mutant. Antimicrob. Agents Chemother. 46:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozarich, J. W., and J. L. Strominger. 1978. A membrane enzyme from Staphylococcus aureus which catalyzes transpeptidase, carboxypeptidase, and penicillinase activities. J. Biol. Chem. 253:1272-1278. [PubMed] [Google Scholar]
- 33.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda, M., T. Oht, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Q. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 35.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchese, A., G. Balistreri, E. Tonole, E. A. Debbia, and G. C. Schito. 2000. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J. Clin. Microbiol. 38:866-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreira, B., S. Boyle-Vavra, B. L. M. De Jonge, and R. S. Daum. 1997. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, W., and M. Matsuhashi. 1984. Staphylococcus aureus and Micrococcus luteus peptidoglycan transglycosylases that are not penicillin-binding proteins. J. Bacteriol. 157:538-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeltz, R. F., V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal, and B. J. Wilkinson. 2000. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 44:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poston, S. M., G. R. Glancey, J. E. Wyatt, T. Hogan, and T. J. Foster. 1993. Co-elimination of mec and spa genes in Staphylococcus aureus and the effect of agr and protein A production on bacterial adherence to cell monolayers. J. Med. Microbiol. 39:422-428. [DOI] [PubMed] [Google Scholar]
- 41.Rotun, S. S., V. McMath, D. J. Schoonmaker, P. S. Maupin, F. C. Tenover, B. C. Hill, and D. M. Ackman. 1999. Staphylococcus aureus with reduced susceptibility to vancomycin isolated from a patient with fatal bacteremia. Emerg. Infect. Dis. 5:147-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahm, D. F., and J. A. Washington II. 1991. Antibacterial susceptibility tests: dilution methods, p. 1105-1116. In A. Balows, W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 43.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 45.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 46.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sieradzki, K., and A. Tomasz. 1998. Suppression of glycopeptide resistance in a highly teicoplanin-resistant mutant of Staphylococcus aureus by transposon inactivation of genes involved in cell wall synthesis. Microb. Drug Resist. 4:159-168. [DOI] [PubMed] [Google Scholar]
- 48.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sieradzki, K., S. W. Wu, and A. Tomasz. 1999. Inactivation of the methicillin resistance gene mecA in vancomycin-resistant Staphylococcus aureus. Microb. Drug Resist. 5:253-257. [DOI] [PubMed] [Google Scholar]
- 50.Spratt, B. G. 1977. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur. J. Biochem. 72:341-352. [DOI] [PubMed] [Google Scholar]
- 51.Stranden, A. M., K. Ehlert, H. Labischinski, and B. Berger-Bächi. 1997. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 53.Trakulsomboon, S., S. Danchaivijitr, Y. Rongrungruang, C. Dhiraputra, W. Susaemgrat, T. Ito, and K. Hiramatsu. 2001. First report of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Thailand. J. Clin. Microbiol. 39:591-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wada, A., Y. Katayama, K. Hiramatsu, and T. Yokota. 1991. Southern hybridization analysis of the mecA deletion from methicillin-resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 176:1319-1325. [DOI] [PubMed] [Google Scholar]
- 55.Wyke, A. W., B. J. Ward, M. V. Hayes, and N. A. C. Curtis. 1981. A role in vivo for penicillin-binding protein-4 of Staphylococcus aureus. Eur. J. Biochem. 119:389-393. [DOI] [PubMed] [Google Scholar]