Abstract
The genetic determinants of resistance to mefloquine in malaria parasites are unclear. Some studies have implied that amplification of, or mutations in, the multidrug resistance gene pfmdr1 in Plasmodium falciparum may be involved. Using the rodent malaria model Plasmodium chabaudi, we investigated the role of the orthologue of this gene, pcmdr1, in a stable mefloquine-resistant mutant, AS(15MF/3), selected from a sensitive clone. pcmdr1 exists as a single copy gene on chromosome 12 of the sensitive clone. In AS(15MF/3), the gene was found to have undergone duplication, with one copy translocating to chromosome 4. mRNA levels of pcmdr1 were higher in the mutant than in the parent sensitive clone. A partial genetic map of the translocation showed that other genes in addition to pcmdr1 had been cotranslocated. The sequences of both copies of pcmdr1 of AS(15MF/3) were identical to that of the parent sensitive clone. A cross was made between AS(15MF/3) and an unrelated mefloquine-sensitive clone, AJ. Phenotypic and molecular analysis of progeny clones showed that duplication and overexpression of the pcmdr1 gene was an important determinant of resistance. However, not all mefloquine-resistant progeny contained the duplicated gene, showing that at least one other gene was involved in resistance.
Quinoline-ring containing drugs such as chloroquine and mefloquine are among the most widely used antimalarial chemotherapeutic agents. Mutants of Plasmodium falciparum resistant to chloroquine which emerged in the 1960s have now spread to most areas of the world where this drug has been used. Mefloquine was developed with the primary purpose of treating chloroquine-resistant malaria, but parasites resistant to this drug were detected soon after its introduction, even being recorded in areas where the drug had not been used previously (49).
The genes determining resistance to mefloquine have not yet been identified. In contrast, the genetic basis of chloroquine resistance in P. falciparum is now becoming well understood. A recently identified gene denoted pfcrt that encodes a membrane transporter protein has been shown to be uniquely associated with this type of resistance. An allele of this gene (pfcrt-76T) encoding threonine (T) at amino acid position 76 of the gene product has been shown to be linked to chloroquine-resistance in a genetic cross between resistant and sensitive clones of P. falciparum (19). Field studies have confirmed this association (16). In addition, much attention has been paid to the possible role in chloroquine resistance of the P. falciparum pfmdr1 gene, which encodes a membrane P glycoprotein denoted Pgh-1. Several field studies have shown that an allele of this gene encoding tyrosine (Y) at position 86 in Pgh-1 (pfmdr1-86Y) appears to be associated with high chloroquine resistance in P. falciparum isolates containing pfcrt-76T (1, 16), although mutations in pfmdr1 alone are not predictive of chloroquine susceptibility. Transfection experiments have also shown that the chloroquine response of a resistant P. falciparum clone can be modulated when its pfmdr1 is replaced by a different allele of this gene (38).
The idea that pfmdr1 could be involved in mefloquine resistance has been investigated by several research groups but with inconclusive results. Early studies examined whether amplification and/or overexpression of the gene could cause resistance. Clones of P. falciparum selected in vitro for high mefloquine resistance showed evidence of amplification and overexpression of pfmdr1 (14, 31), whereas a decrease in resistance was associated with deamplification of the gene (2). However, in other studies no amplification was found in parasites selected for increased resistance (27, 40, 42). An association between amplification of pfmdr1 and mefloquine resistance has been claimed in some field investigations (34, 35, 51) but not in others (3, 9).
Other studies have examined whether specific alleles of the pfmdr1 gene could be associated with the response to mefloquine. In a genetic cross between two P. falciparum clones differing in response to the drug, low resistance to mefloquine cosegregated with an allele encoding Y at position 184 and asparagine (N) at position 1042 (18). Phenylalanine (F) was reported to have replaced Y at position 86 after selection of a highly mefloquine-resistant line of P. falciparum from a sensitive clone (31). In a field study (35), it was claimed that 86N in pfmdr1 appeared to be associated with increased mefloquine tolerance in P. falciparum in the Western border area of Thailand. Most recently, transfection work (38) showed that allelic replacement of one pfmdr1 allele with another affected the response of the transfected parasites to mefloquine, as well as to chloroquine. Furthermore, mefloquine has been shown functionally to inhibit the activity of the human mdr1 P-glycoprotein by decreasing the rates of extrusion of fluorescent dyes from drug-resistant cells and decreasing the resistance of these cells to other drugs (39).
Classical linkage analysis is a critical procedure in tracking down genes involved in phenotypes such as drug resistance. In malaria, this is exemplified by the P. falciparum cross used to identify the pfcrt gene determining chloroquine resistance (19). Here we make use of the rodent malaria model Plasmodium chabaudi to investigate the genetics of mefloquine resistance. Genetic crossing work is much easier with this species than with P. falciparum, and it is a good model for P. falciparum because there are many biological similarities between them, as well as conservation of synteny between linkage groups (5-7). We describe the production of a stable mefloquine-resistant clone of P. chabaudi by using a continuous low pressure and multistep drug selection procedure. The sequence of pcmdr1, the P. chabaudi orthologue of pfmdr1, is the same in the mefloquine-resistant mutant as in its sensitive parent. However, we have found that a cluster of genes, including pcmdr1, has undergone duplication in the resistant clone and that translocation of the copied genes to another chromosome has occurred. We also show that a higher expression of pcmdr1 occurs in the resistant compared to the sensitive parent clone. We show in a genetic cross between the mefloquine-resistant parasite and an unrelated sensitive clone that all progeny with duplicated pcmdr1 have resistant phenotypes, whereas all sensitive progeny possess only a single copy. However, some resistant progeny lack the duplication, showing that duplicated pcmdr1 is not the only mechanism conferring resistance and that, consequently, at least one other gene is involved in this character.
MATERIALS AND METHODS
P. chabaudi.
The P. chabaudi lines used in this work were clones of two genetically distinct drug-sensitive isolates denoted AS and AJ, originally taken from naturally infected thicket-rats (Thamnomys rutilans) from the Central African Republic (4). They were maintained in laboratory mice and transmitted through the mosquito Anopheles stephensi as described previously (48).
Selection for mefloquine resistance.
Mefloquine hydrochloride (MF) was obtained as a gift from the Walter Reed Army Institute Research, Washington, D.C. Dilutions of the drug were made in dimethyl sulfoxide and administered orally to mice.
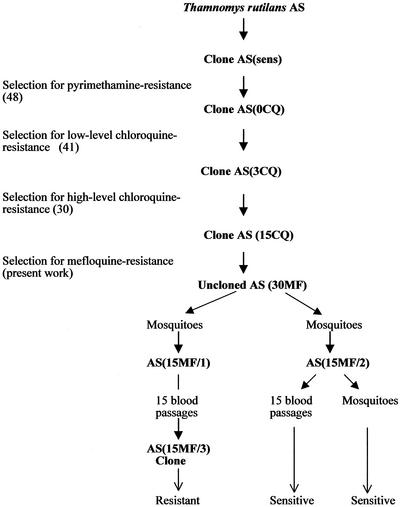
Mefloquine resistance was selected in a clone of AS previously selected for resistance to pyrimethamine and chloroquine (Fig. 1). Briefly, the original AS isolate was cloned by limiting dilution (8), selected for resistance to pyrimethamine, and then recloned (48). This clone, designated AS(0CQ), was then selected for resistance to chloroquine, and a line resistant to six daily doses of this drug at 3 mg kg−1 was obtained (41). This line was cloned and denoted AS(3CQ). AS(3CQ) was then subjected to further stepwise increases of chloroquine, producing a line resistant to six daily doses of chloroquine at 15 mg kg−1 (30). This parasite was cloned [AS(15CQ)] before being used in the present selection experiments.
FIG. 1.
History and drug sensitivities of clones of P. chabaudi used in this work. AS is an uncloned isolate (4).
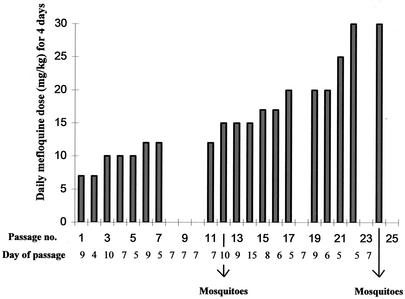
To select for mefloquine resistance, the AS(15CQ) clone was exposed to gradual increases in mefloquine treatment over 23 passages in mice (Fig. 2). A total of 106 parasites of AS(15CQ) were first inoculated into groups of 4- to 6-week-old C57BL mice. Mefloquine at 7 mg kg (mouse body weight)−1 was administered orally 3 h later, and at the same time on the following 3 days. Parasites that survived drug treatment were passaged into other mice, and the treatment was repeated. In subsequent passages, the drug pressure was gradually increased (Fig. 2) up to a maximum of 30 mg kg−1. The parasite line finally obtained [AS(30MF)] was transmitted through mosquitoes on two occasions to produce the parasite lines AS(15MF/1) and AS(15MF/2) (Fig. 1).
FIG. 2.
Procedure for selecting mefloquine resistance in P. chabaudi. Clone AS(15CQ) was passaged through 24 successive groups of mice on the days indicated (x axis). Mefloquine was administered to the mice as four daily doses starting on the day of parasite inoculation. The doses (y axis) were increased gradually during the course of the experiment.
Tests for mefloquine response.
Parasites were tested for their susceptibility to mefloquine by comparing their response to the drug with that of sensitive control parasites. For this test, six mice were each inoculated with 106 parasites (day 0). Four mice were treated with mefloquine at 5 mg kg−1 at 3 h after inoculation and at the same time on the following 3 days, the remaining two mice being undrugged. Blood smears were taken every 2 days from day 6 onward, stained with Giemsa stain, and examined microscopically for parasites. Each clone was tested for mefloquine response in this way on at least two occasions.
Genetic crosses.
Two crosses were made between AJ and AS (15MF/3) by using the method previously described (5). Briefly, this procedure was as follows. Each clone was first grown separately in mice. When the parasitemia had reached ca. 30%, equal volumes of infected blood (0.5 ml) were mixed and injected intraperitoneally into a splenectomized rat to promote gametocyte production (11). Mosquitoes (Anopheles stephensi) were subsequently allowed to feed on the rat to permit cross-fertilization of gametes of each clone to occur. The mosquitoes were examined for the presence of oocysts after 7 days and, when sporozoites were subsequently present in the salivary glands, the mosquitoes were permitted to feed on mice in order to transmit the infection. The blood forms which developed in these animals, the progeny of the cross, were then cloned by dilution into mice and characterized for their drug response and polymorphic genetic markers distinguishing AS(15MF/3) and AJ (5). A total of 16 independent recombinant parasite clones, identified by their unique combinations of the parent markers, were chosen for further study.
As controls, mosquitoes were also fed on each parent clone alone and fed on new mice on the same day as those fed on the mixed parasites. AJ was successfully transmitted, but AS(15MF/3) failed to establish an infection in these animals.
Preparation of chromosome blocks, and PFGE.
Infected mice exhibiting high parasitemias (≥50%) at the trophozoite stage were exsanguinated into heparinized phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 8.8]), and the blood was passed down a CF11 cellulose powder column to remove host leukocytes (23). Parasites were liberated from host cells by saponin lysis and then used to make agarose blocks for pulsed-field gel electrophoresis (PFGE) by using a standard procedure (25). Chromosomes were separated by PFGE (44) by using a CHEF DR II (Bio-Rad) apparatus as follows: 140 V and a 120-s pulse time for 24 h; 130 V and a 300-s pulse time for 24 h; 140 V and a 180-s pulse time for 24 h.
Genome markers.
Genome markers for all 14 P. chabaudi chromosomes that distinguished AS from AJ were used to characterize parent and cross-progeny clones. Details of the methods used to characterize all markers except msp1, ama1, pccg10, and pfs28 are given by Carlton et al. (5; see also http//www.ncbi.nlm.nih.gov/Malaria/Rodent/ctabgenmarker.html).
Sequence data for P. chabaudi msp1 (28), ama1 (15), pccg10 (P. Hunt, unpublished data) and pfs28 (45) were used to develop PCR-based polymorphic markers; msp1 of clone AJ and ama1 of clones AS and AJ were provided by S. Cheesman and R. Carter (University of Edinburgh). Msp1 products gave a restriction fragment length polymorphism (RFLP) and a nested primer-specific product of different lengths; ama1 gave a HindIII RFLP; pccg10 had a size polymorphism and an ApoI RFLP; and pfs28 gave a BspTI RFLP (data not shown).
DNA and RNA extraction, Southern and Northern blotting, and probe hybridization.
DNA was extracted from parasites by incubating them overnight with lysis solution (10 mM Tris [pH 8.0], 50 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], proteinase K [1 mg/ml]) at 42°C. After phenol extraction, DNA was precipitated by using propan-2-ol and NH4 acetate. RNA was extracted from saponin-lysed infected blood cells by using an RNA isolator (Genosys). Parasite DNA separated by electrophoresis was transferred onto Hybond N+ membrane (Amersham) according to the manufacturer's instructions. DNA probes were radiolabeled by random hexamer priming (Boehringer Mannheim High Prime radiolabeling kit), and hybridization and washing were performed by standard procedures (43). The intensity of radioactive signals was measured with a PhosphorImager and quantitated with ImageQuant software (Molecular Dynamics).
Sequencing of pcmdr1.
The pcmdr1 genes from AS(15CQ), AS(15MF/3), and progeny clones 440/5 and 1016/11 were sequenced from overlapping PCR products generated from the following pairs of PCR primers: pcmdr1-45 (5′-AATTTGTGTCGTTATCCACAC-3′) and pcmdr1-30 (5′-CGGATGTTAATTTAGAACCTGG-3′), pcmdr1-29 (5′-GTTTATGCATGGGTACTGTTAC-3′) and pcmdr1-18 (5′-CCAATTTTTGATCTCCACC-3′), pcmdr1-40 (5′-GTCAAATTTCATTATGGTACTAG-3′) and pcmdr1-14 (5′-TTTTGCTTTCAACCTTTTCTCC-3′), pcmdr1-26 (5′-GATTGAGTACTATTCGATATGC-3′) and pcmdr1-11 (5′-CAATTTTTCCATTCGTACTCTTGC-3′), pcmdr1-13 (5′-AATTGGAGAAAAGGTTGAAAGC-3′) and pcmdr1-49 (5′-ATTGGTTCTTGGTTAACTATCG-3′), pcmdr1-9 (5′-CTAATGCAACCTCTATGAACG-3′) and pcmdr1-10 (5′-GTTTTTGACCACCTGATAAGC-3′), and pcmdr1-05 (5′-GTAGGACCTTATGGAAAAAGC-3′) and pcmdr1-51 (5′-TATTTTCCTGGGAATAACATCC-3′). These primers are based upon the AS pcmdr1 sequence (P. Hunt et al., unpublished data). PCR products were either sequenced directly using appropriate primers or cloned by using TOPO TA cloning (Invitrogen) prior to sequencing of plasmid DNA. After being cloned, isolated transformants were used to generate plasmid DNA by using QIAprep spin miniprep kits (Qiagen). In both cases, sequencing used ABI Prism BigDye Terminator (Applied Biosystems) cycle sequencing ready reaction kits.
RESULTS
Selection for mefloquine resistance.
Three related cloned lines of P. chabaudi, derived sequentially from a drug-sensitive clone denoted AS(sens), were involved in the mefloquine resistance selection experiment (Fig. 1). These were a pyrimethamine-resistant clone denoted AS(0CQ), a clone exhibiting low-level chloroquine-resistance denoted AS(3CQ), and a more highly chloroquine-resistant clone denoted AS(15CQ). Attempts to establish stable mefloquine resistance in AS(0CQ) and AS(3CQ) were unsuccessful (data not shown). However, AS(15CQ) responded well to the selection procedure, and a line resistant to four daily doses of 30 mg of mefloquine kg (mouse body weight)−1, AS(30MF), was selected after 24 passages (Fig. 2).
The stability of the resistance was assessed following mosquito transmission and multiple blood passage in the absence of the drug. Two lines of AS(30MF) were established that were transmitted through mosquitoes on separate occasions (Fig. 1 and 2). Both lines appeared to have lost some degree of resistance, from resistant to four daily doses of mefloquine at 30 mg kg−1 to resistant to four doses of 15 mg kg−1 (data not shown). These two lines were then designated AS(15MF/1) and AS(15MF/2). After 15 passages in the absence of drug, AS(15MF/1) had maintained its level of resistance, and a clone was established from these parasites that was denoted AS(15MF/3) (Fig. 1). The resistance of AS(15MF/2) was unstable, and studies on these parasites were discontinued.
Karyotype of mefloquine-resistant lines.
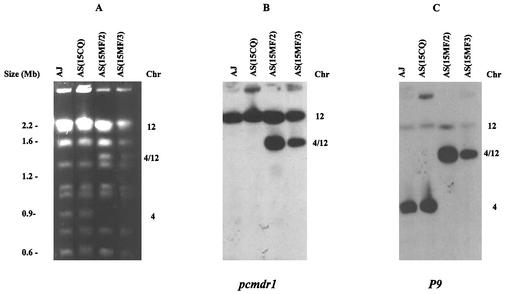
To investigate whether any chromosomal rearrangements had occurred during mefloquine selection, intact chromosomes from AS(15CQ), AS(15MF/2), and AS(15MF/3) were separated by PFGE (Fig. 3). The karyotype of AS(15CQ) was the same as that of the AS clone from which it had originally been isolated and was similar to that of the unrelated clone AJ (Fig. 3A). However, chromosome separations of the two AS(15MF) lines showed karyotypic changes. Chromosome 4 (0.9 Mb) appeared not to be present and, instead, a 1.3-Mb chromosome was seen between chromosome 7 and chromosomes 8 and 9 (which migrate as a doublet on PFGE gels) (Fig. 3A).
FIG. 3.
Duplication and translocation of the pcmdr1 gene in a mefloquine-resistant mutant of P. chabaudi. (A) Chromosomes, separated by PFGE, of AJ, AS(15CQ), AS(15MF/2), and AS(15MF/3). Chromosomes 1 and 2, 8 and 9, and 12 to 14 were run as doublets under the PFGE conditions used here; note the absence of the chromosome 4 band and the presence of a novel 4/12 band in AS(15MF/2) and AS(15MF/3). (B) Southern blot of panel A hybridized with a pcmdr1 probe. Note the positive hybridization to chromosome 12 in AJ and AS(15CQ) and to 12 and 4/12 in AS(15MF/2) and AS(15MF/3). (C) Southern blot of panel A hybridized with a chromosome 4-specific probe P9. Note the hybridization only to chromosome 4.
Since amplification of pfmdr1 has been associated with mefloquine-resistance in P. falciparum (14), we investigated whether amplification of pcmdr1 had occurred in our mefloquine-selected P. chabaudi lines. Southern blots of PFGE gels of AS(15CQ) and the two AS(15MF) lines were hybridized at high stringency with a pcmdr1 probe (Fig. 3B). In AS(15CQ), hybridization occurred to chromosome 12. In the two AS(15MF) lines, the probe hybridized not only to chromosome 12 but also to the novel 1.3-Mb chromosome. This indicated that during selection of the mefloquine-resistant parasites there had been a duplication of an ∼400-kb segment of chromosome 12 containing the pcmdr1 gene and that the duplicated region had translocated to chromosome 4, producing a novel-sized chromosome. We refer to this chromosome as number 4/12.
Characterization of the translocation in AS(15MF/3).
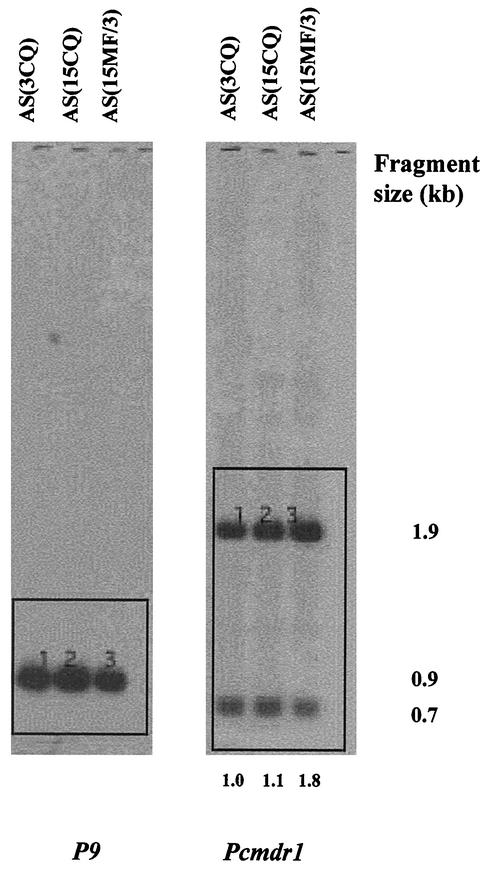
In order to characterize the translocated portion of chromosome 12 in more detail, we first investigated whether there was any evidence for amplification of the pcmdr1 gene in this region. DraI-digested DNA from AS(3CQ), AS(15CQ), and AS(15MF/3) was electrophoresed, blotted, and probed with pcmdr1 (Fig. 4). Two fragments (0.7 and 1.9 kb) were seen, indicating that DraI cuts once within the probe. PhosphorImager analysis of the intensity of the signal from the 1.9-kb bands revealed that AS(15MF/3) had a pcmdr1 copy number of approximately two compared to AS(3CQ) and AS(15CQ). This was shown by comparing the intensities of hybridization of pcmdr1 with those of the single-copy chromosome 4 gene P9 in each respective parasite (Fig. 4). Thus, although duplication of pcmdr1 and transposition of the duplicated form had occurred in AS(15MF/3), the increase in size of chromosome 4/12 compared to 4 in AS(3CQ) and AS(15CQ) was not due to further pcmdr1 gene amplification on the translocated element.
FIG. 4.
Demonstration that the size exhibited by chromosome 4/12 is not due to tandem duplication of pcmdr1. Southern blot of DraI digests of AS(3CQ), AS(15CQ), and AS(15MF/3) genomic DNA hybridized with pcmdr1 and P9 probes. The ratios of the 1.9-kb pcmdr1 band intensities to P9 intensities, quantified by using PhosphorImager software, is shown under each lane, with the P9 band intensities of each respective parasite (labeled 1, 2, and 3) as standards for comparison. These values are normalized to AS(3CQ), which is given a value of 1.0.
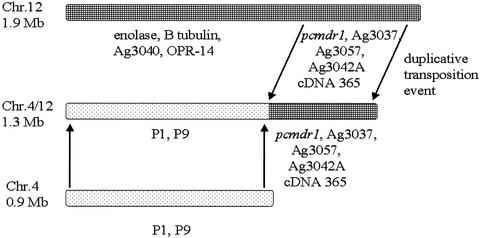
A partial linkage map of the translocation was constructed from an analysis of the hybridization pattern of chromosome 4- and 12-specific probes to PFGE blots. Two chromosome 4 markers (P1 and P9) mapped to chromosome 4/12, four chromosome 12-specific markers (enolase, β-tubulin, Ag3040, and OPR-14) mapped to chromosome 12, and four chromosome-12 specific probes (Ag3037, Ag3057, Ag3042A, and cDNA365) mapped to both chromosomes 12 and 4/12 (Fig. 3C). Figure 5 shows a schematic map of these findings.
FIG. 5.
Partial chromosome map of AS(15MF/3) showing some of the genes identified as duplicated and translocated from chromosome 12 to chromosome 4/12. Note that the order of the genes is unknown at present.
Sequence of pcmdr1.
We sought to investigate whether there were sequence polymorphisms in either copy of pcmdr1 of AS(15MF/3) which could be correlated with mefloquine response. Primers based on the gene sequence of pcmdr1 from AS(sens) were used first to amplify overlapping gene fragments from each of the AS clones used in the work. The PCR products from AS(15MF/3) were sequenced in two ways to ensure that the sequences of both gene copies were represented. First, the PCR products of this parasite were cloned, and five clones selected for sequencing. Second, the products were sequenced, and the fluorescence electropherograms were then inspected for evidence of two pcmdr1 sequences being present in this parasite.
The sequences of pcmdr1 from AS(sens), AS(15CQ) and AS(15MF/3) were identical (Table 1). There were no differences among any of the five cloned sequences from AS(15MF/3), and no evidence for mixed alleles was seen in the electropherograms. It could be concluded that no mutations had occurred in either copy of the gene in this parasite during mefloquine selection. The pcmdr1 sequence of clone AJ was also obtained and was found to differ from that of the AS clones at six positions, four within the coding region, all of which were synonymous differences, i.e., they did not result in altered amino acids (Table 1). These sequence data have been submitted to the DDB/EMBL/GenBank databases under accession number AY123625.
TABLE 1.
Synonymous nucleotide differences in the P. chabaudi pcmdr1 gene in clones AS(sens), AS(15CQ), AS(15MF/3), and AJa
| Position in coding sequence | Position in genomic sequence | AS(sens), AS(15CQ), or AS(15MF/3) (codon) | AJ (codon) | Amino acid |
|---|---|---|---|---|
| 72 | 286 | C(ACC) | T(ACT) | T |
| 3423 | 3637 | A(TTA) | G(TTG) | L |
| 3516 | 3730 | T(TAT) | C(TAC) | Y |
| 3681 | 3895 | T(GGT) | C(GGC) | G |
| 3′-untranslated region | 4508 | G | A | |
| 4516-7 | AT | TA |
Full sequence data are in DDBJ/EMBL/GenBank databases under accession number AY123625.
Table 2 compares the amino acids in the pcmdr1 sites with those in the equivalent polymorphic sites in the P. falciparum gene.
TABLE 2.
P. chabaudi pcmdr1 and P. falciparum pfmdr1 polymorphisms in cloned lines and their sensitivity to mefloquinea
| Strain | Mefloquine response | Pgh-1 amino acid at position:
|
||||
|---|---|---|---|---|---|---|
| 86 | 184 | 1034 | 1042 | 1246 | ||
| P. falciparum | ||||||
| K1 | Sensitive | Y | Y | S | N | D |
| 7G8 | Sensitive | N | F | C | D | Y |
| HB3 | Sensitive | N | F | S | D | D |
| 3D7 | Low resistance | N | Y | S | N | D |
| D10 | Resistant | N | Y | S | N | D |
| Dd2 | Resistant | Y | Y | S | N | D |
| P. chabaudi | ||||||
| AJ | Sensitive | N | Y | S | N | D |
| AS(sens) | Sensitive | N | Y | S | N | D |
| AS(15CQ) | Sensitive | N | Y | S | N | D |
| AS(15MF/3) | Resistant | N | Y | S | N | D |
| 440/5 | Resistant | N | Y | S | N | D |
P. falciparum data are from references 18, 20, and 38.
Expression of pcmdr1 and other genes in the mefloquine-resistant clone AS(15MF/3).
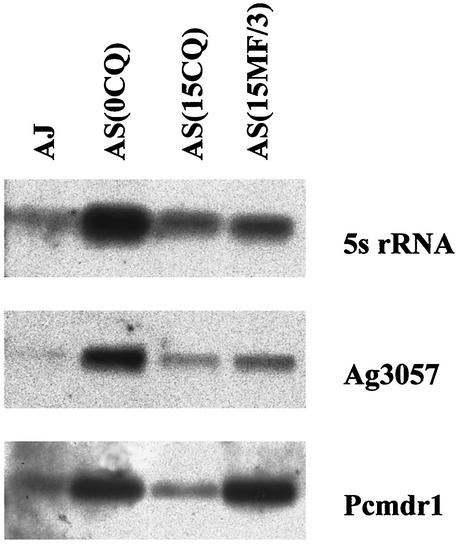
Expression of pcmdr1, Ag3057, and 5S rRNA genes at the level of transciption was examined. Northern blots of total RNA extracted from AS(0CQ), AS(15CQ), AS(15MF/3), and AJ were hybridized with appropriate probes for these genes. PhosphorImager analysis was used to quantify the ratio of intensities of signal from each clone. The results are shown in Fig. 6 and Table 3.
FIG. 6.
Expression of pcmdr1 in P. chabaudi clones demonstrated by Northern hybridization. After gel fractionation and transfer onto membranes, RNA from AJ, AS (0CQ), AS (15CQ), and (AS 15MF/3) was hybridized with 5S rRNA probe (top) and subsequently with the ag3057 (middle) and pcmdr1 (lower) probes. The intensities of the bands were estimated by using a PhosphorImager and ImageQuant software (Table 3).
TABLE 3.
Levels of pcmdr1 RNA expression in AJ, AS(0CQ), AS(15CQ), and AS(15MF/3)a
| Probe | Clone | Intensity relative to AS(15CQ) | Intensity relative to 5S rRNA and AS(15CQ) |
|---|---|---|---|
| Pcmdr1 | AJ | 0.8 | 1.2 |
| AS(0CQ) | 2.7 | 1.2 | |
| AS(15CQ) | 1.0 | 1.0 | |
| AS(15MF/3) | 3.3 | 2.9 | |
| 5S rRNA | AJ | 0.7 | 1.0 |
| AS(0CQ) | 2.3 | 1.0 | |
| AS(15CQ) | 1.0 | 1.0 | |
| AS(15MF/3) | 1.1 | 1.0 | |
| Ag3057 | AJ | 0.7 | 1.2 |
| AS(0CQ) | 2.7 | 1.2 | |
| AS(15CQ) | 1.0 | 1.0 | |
| AS(15MF/3) | 1.4 | 1.2 |
The intensities of expression of pcmdr1, 5S rRNA, and ag3057 are shown relative to their expression in AS(15CQ). Compensation for the differences in total RNA loading is made by dividing these relative intensities by the corresponding 5S rRNA intensities.
The intensities of the bands obtained with hybridization of 5S rRNA in Fig. 6 were used to provide a comparative measure of the amount of RNA in each track. The band intensities were then normalized to the values of AS(15CQ) on the same blot (Table 3). This showed that AS(15MF/3) had between two and three times as much expression of pcmdr1 as did AS(15CQ), AS(0CQ), or AJ. Expression of Ag3057 was similar in each clone. pcmdr1 was the only one of the genes examined that showed significantly increased expression in AS(15MF/3).
Analysis of crosses between AS(15MF/3) and AJ.
Two crosses were made between AS(15MF/3) and AJ by infecting splenectomised rats with a mixture of the two clones and transmitting the parasite mixture through mosquitoes into mice (see Materials and Methods). The parent lines alone were subjected to the same procedures at the same time.
AJ alone was transmitted successfully through mosquitoes into mice, but AS(15MF/3) failed to infect mice from sporozoites, despite having produced oocysts in mosquitoes. Sporozoites in mosquitoes infected from the mixed clones were successfully transmitted into mice. These parasites, denoted as the progeny of the cross, were examined for parental clone markers and then cloned into further mice.
(i) Progeny genotypes.
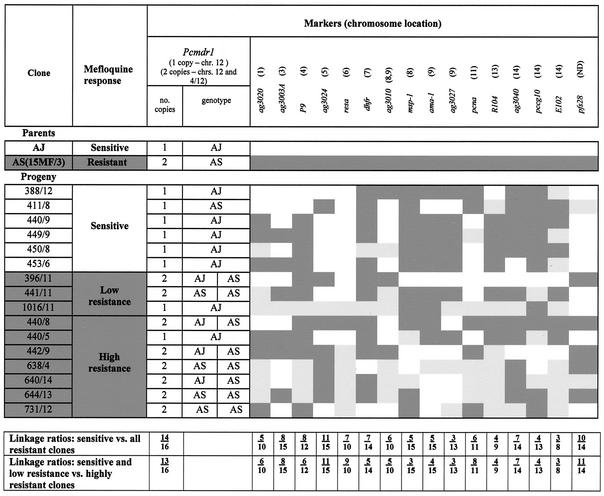
The uncloned progeny were found to possess markers from both AS(15MF/3) and AJ (data not shown). A total of 47 clones were obtained from these parasites, which were characterized for each parental marker. Seven possessed pure parent AJ markers. The remaining 40 were recombinant forms, with combinations of AJ and AS(15MF/3) alleles which could be allocated into 16 groups, each characterized by a unique combination of markers. A representative clone of each of these groups was chosen for detailed characterization (Fig. 7).
FIG. 7.
Characteristics of AS(15MF/3), AJ, and 16 progeny clones from the cross showing the inheritance of mefloquine response, pcmdr1 alleles and duplication, and 16 chromosome-specific markers. A dark shaded background represents the inheritance of characters of AS(15MF/3), and a white background indicated AJ. Light shading indicates that the inheritance was not determined. Linkage ratios are ratios of the number of progeny clones showing linkage of drug resistance or sensitivity with each respective AS(15MF/3) or AJ allele to the total number of progeny analyzed.
(ii) Mefloquine responses.
Each progeny clone was typed at least twice for its mefloquine response. After mefloquine treatment, AS(15MF/3) and seven progeny clones underwent recrudescence by day 7 after parasite inoculation and were classified as resistant (Fig. 7). AJ and six progeny clones underwent recrudescence later than day 13 and were classified as mefloquine sensitive. The remaining three progeny clones were mefloquine resistant, but at a level intermediate between each parent clone, with recrudescence occurring on day 9; these clones were classified as having low resistance (Fig. 7). These drug responses were consistent in each of at least two tests carried out on each clone.
(iii) Karyotypes.
The progeny clones possessed karyotypes which, with the exception of minor size variations in some chromosomes, were similar to either AS(15MF/3) or AJ (Fig. 8).
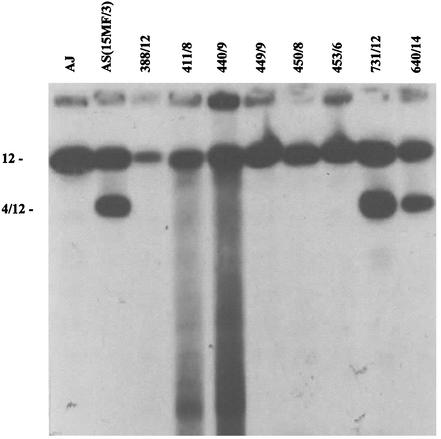
FIG. 8.
Southern blot of PFGE gel of chromosomes of parents and progeny clones, hybridized with the pcmdr1 probe. See Fig. 7 for progeny clone characters. Note the hybridization to chromosomes 12 and 4/12 in AS(15MF/3) and the progeny clones 731/12 and 640/14.
(iv) pcmdr1 duplication and sequencing.
The results of pcmdr1 typing are shown in Fig. 7 and 8. Each of the six mefloquine-sensitive clones possessed just a single copy of pcmdr1, on chromosome 12. Six of the seven clones exhibiting high resistance and two with low resistance possessed two copies of pcmdr1 located on chromosomes 12 and 4/12 as seen in AS(15MF/3). Clone (440/5), resistant to mefloquine at the same level as parent AS (15MF/3), and clone 1016/11, characterized by low resistance, had only a single copy of pcmdr1 (on chromosome 12).
By using an EcoRI RFLP, it was possible to determine the inheritance of each pcmdr1 allele among the progeny clones (Fig. 7). All of the progeny inheriting a single copy of the gene possessed the AJ-type allele, with the exception of sensitive clone 411/8, which had the AS-type allele. Among the progeny inheriting two pfmdr1 copies, four clones had inherited both AS-type alleles, like parent clone AS(15MF/3), and four had one AS-type allele and one AJ-type allele, presumably due to random assortment of chromosomes 12 and 4/12 during meiosis of heterozygous forms in the mosquito.
(v) Expression of pcmdr1 among the progeny clones.
mRNA levels were studied in progeny clones by Northern blot analysis by using the pcmdr1 probe. Clones containing two copies of pcmdr1 possessed increased levels of gene expression compared to those with single copies. Clones 440/5 and 1016/11, which possessed only a single copy of pcmdr1 but were resistant to high and low levels of mefloquine, respectively, did not show increased expression of the gene (data not shown).
Linkage of resistance with chromosome-specific markers.
Linkage ratios of pcmdr1 duplication and of 16 other chromosome-specific markers to mefloquine susceptibility were calculated. The linkage ratio for each marker is the ratio of the number of progeny clones showing linkage of drug resistance or sensitivity with each respective parent marker to the total number of progeny analyzed. The results are shown in Fig. 7. When clones exhibiting low and high resistance were classified together as a single resistant category, the linkage ratio for pcmdr1 duplication and resistance was 14/16; when the low-resistance clones were classified with the sensitive group, it was 13/16. No clear associations were seen between the inheritance of other markers and response to mefloquine, with the possible exception of RESA which had a linkage ratio of 9/10 when low-resistance and sensitive clones were considered as a single category.
DISCUSSION
The principal finding of the present study is that, although duplication of the pcmdr1 gene has an important rôle in the resistance of the mutant P. chabaudi clone to mefloquine, it is not the only gene involved. In the cross between resistant and sensitive clones, all progeny that contained the duplicated gene were resistant. However, two progeny clones exhibited resistance in the absence of duplicated pcmdr1, one of which was characterized by low resistance and another of which was characterized by high resistance. This showed that other genes are primary determinants of the resistance. Our results are consistent with the fact that some, but not all, studies on P. falciparum show an association between pfmdr1 amplification and resistance to mefloquine.
There have been surprisingly few attempts to select mefloquine-resistant mutants of malaria parasites in the laboratory. The only previous report of experimental selection of mefloquine resistance in rodent malaria parasites is in Plasmodium berghei and Plasmodium yoelii (32), which was carried out on uncloned material. Parasites exhibiting high resistance were obtained after some 30 passages of sensitive forms under increasing drug pressure, but they reverted to sensitivity once the drug pressure was removed. In P. falciparum, a mefloquine-resistant mutant denoted W2Mef was derived successfully from the chloroquine-resistant clone W2 by prolonged mefloquine treatment of cultured forms (29). The resistance obtained was stable in the absence of the drug. The P. chabaudi AS(15MF/3) clone which we have obtained by continuous and increasing mefloquine pressure gave rise initially to a highly resistant line, but after mosquito transmission on two occasions the resistance became slightly lower. However, this resistance proved stable and contrasted with the instability seen in other derivatives of our selection procedure.
It was noteworthy that the selection of stable mefloquine-resistance reported here in P. chabaudi was successfully achieved only from a parasite clone already resistant to chloroquine and pyrimethamine and not from one sensitive to these drugs. Other attempts to produce stable chloroquine or mefloquine resistance from drug-sensitive rodent species have also failed, e.g., in Plasmodium vinckei (33). In P. falciparum, also, the only mefloquine-resistant mutants obtained by selection have originated from parasites already resistant to both chloroquine and pyrimethamine (14, 29). Rathod et al. (37) found similarly that resistance to atovaquone and 5-fluoroorotate could be selected only from a clone resistant to other drugs. These authors considered that such clones had acquired ARMD (accelerated resistance to multiple drugs); the genetic basis of this phenotype was unknown but could include mechanisms such as acquisition of a general mutator phenotype or “bursts of mutagenic activity that involve genetic rearrangements” (37).
The most striking finding of our study was that pcmdr1, the P. chabaudi orthologue of the pfmdr1 gene of P. falciparum, had undergone duplication in the mefloquine-resistant AS(15MF/3) clone and that the second copy had translocated, along with other genes, in a 400-kb portion of chromosome 12 to chromosome 4. The mRNA level of pcmdr1 in AS(15MF/3) was approximately twice that of the mefloquine sensitive clone from which it was derived. These results lend support to earlier claims that amplification of the pfmdr1 gene in P. falciparum and of its orthologue in P. berghei plays a role in resistance to mefloquine (14, 21, 31, 34, 51). The work of Cowman et al. (14) on P. falciparum is most relevant to our own results, because it involved the selection of a stable mefloquine-resistant clone (W2Mef) from a sensitive clone (W2), in contrast to the unstable resistance obtained in the P. berghei work (32). P. falciparum W2Mef possessed an increased copy number of pfmdr1, an increase in the size of chromosome 5 containing the gene, and increased pfmdr1 expression compared to W2 (50). When the mefloquine resistance of W2mef was increased in two further selection experiments (14, 31), there were further increases in the copy number and expression of pfmdr1. In related work (2), the mefloquine resistance of W2mef became lower after selection for chloroquine resistance, and this was accompanied by a decrease in the size of chromosome 5 and of the copy number of pfmdr1.
Amplification of a gene whose product is the target of a chemotherapeutic agent is a well-established mechanism by which microorganisms can develop resistance. This has recently been demonstrated for the dhfr gene in experimentally produced resistance to pyrimethamine in P. falciparum (46). Similarly, in selecting a pyrimethamine-resistant mutant of P. chabaudi, the pcdhfr gene underwent a duplication event that involved a partial chromosome duplication after continuous low pressure of the drug on the parasites (12). In this instance, the duplicated portion of the chromosome did not become translocated to another chromosome. Subsequent drug selection on this P. chabaudi line resulted in a point mutation (S106N) in one copy of the gene and loss of the second copy (13). Similar results have been reported for two pyrimethamine-resistant lines selected from P. berghei (47). In both of these studies, it appeared that different methods of selection had given rise to different types of resistance mechanism; selection with low drug doses had selected for parasites with gene duplication and consequent increased gene expression, whereas selection with higher doses had selected for parasites with functional gene mutations (13). Point mutations in dhfr appear to be the most common cause of resistance to higher levels of this drug in both P. chabaudi (10, 13) and in field populations of P. falciparum (24, 36).
Our crossing work has demonstrated that duplication and overexpression of the pcmdr1 gene correlate with mefloquine resistance. No progeny clones that were mefloquine sensitive contained the duplicated and/or overexpressed gene. We do not know whether the translocation event by which the duplicated portion of chromosome 12 containing pcmdr1 became attached to chromosome 4 was important in establishing the resistance. The duplication was maintained by AS(15MF/3) in the absence of the drug. The translocation event may allow stable inheritance of the character by enabling the duplicated portion to replicate during mitosis along with the chromosome to which it became attached. The 4/12 chromosome was also seen in the uncloned AS(15MF/2) line immediately after drug selection (Fig. 3), but this parasite line subsequently became sensitive after its prolonged passage in the absence of the drug. It seems probable that immediately after selection this line still contained a minority of drug-sensitive parasites without the duplication that outgrew the resistant forms during passages without drug. It would have been of obvious interest to have examined the karyotype of AS(15MF/2) at this later stage, but samples of this line were not preserved.
Sequence differences between alleles of pfmdr1 have been reported to correlate with variations in mefloquine sensitivity in some studies on P. falciparum (17, 31, 35, 38), but no clear pattern has emerged (Table 2). The progeny of the 3D7/HB3 cross of P. falciparum showed cosegregation of low mefloquine resistance with a pfmdr1 allele containing 184Y and 1042N (18). The transfection work of Reed et al. (38) produced evidence that an allele of pfmdr1 with 1034S, 1042N, and 1246D caused an increase in mefloquine resistance when it replaced the allele with 1034C, 1042D, and 1246Y present in the sensitive recipient clone; the reciprocal transfection experiment confirmed the association of each respective allele with a mefloquine response. We found here no sequence differences between the pcmdr1 alleles of AS(sens), AS(15CQ), or AS(15MF/3), showing that no mutations in the gene had occurred during selection for resistance (Table 2). Also, the level of resistance in each progeny clone derived from the cross with a single copy of pfmdr1 showed no correlation with its inherited parental allele (AS or AJ).
The appearance among the progeny of the cross of three clones exhibiting levels of mefloquine resistance intermediate between the parent clones provides further evidence that more than one gene determined the resistance of AS(15MF/3). Two of these clones contained the duplicated pcmdr1, and just one (1016/11) had only a single copy. In addition, one progeny clone with a high level of resistance (440/5) possessed only a single copy of pcmdr1. Duplication of the gene was thus not an essential condition for the manifestation of resistance. The role of mdr1 in resistance to mefloquine and related drugs could therefore be a secondary rather than a primary one, perhaps representing secondary adaptations that enhance parasite fitness in the presence of mutations in other genes, as suggested in other microbial systems (26, 52). We do not yet know the identity of the other genes involved in mefloquine resistance in P. chabaudi. None of the genetic markers distinguishing the parent clones cosegregated with the resistance character among the progeny. Identification of the other loci involved in the mefloquine resistance will require further linkage analysis when more markers are available. In this regard, many new polymorphic genetic markers, e.g., amplified fragment length polymorphisms (22), are becoming available for this species, which means that more powerful linkage analysis of crosses will be achievable than has been possible hitherto.
The work reported here is the first in which a mefloquine-resistant mutant of a malaria parasite obtained by drug selection has been used in a cross to analyze the genetic basis of the resistance. We have demonstrated that pcmdr1 amplification is associated with mefloquine resistance, but that at least one other gene must be involved, findings that are consistent with field and other laboratory studies on P. falciparum. P. chabaudi has the important advantage that drug-resistant mutants can be selected from sensitive clones in the laboratory more readily than in P. falciparum and also that crosses can be performed much more easily (7). Furthermore, the identification of genes linked to markers of resistance phenotypes will be helped by the availability of considerable genome sequence data for P. chabaudi (http://www.sanger.ac.uk/Projects/P_chabaudi/). Such an approach will allow the identification of genes involved in the drug resistance of malaria parasites to progress rapidly.
Acknowledgments
We thank Margaret Mooney and Richard Fawcett for technical assistance and the ICAPB Animal House staff for their excellent animal husbandry.
This work was supported by the UK Medical Research Council. P.V.L.C. acknowledges the support of Portugal's Fundaçao para a Ciencia e Tecnologia and the Centro de Malaria e Outras Doenças Tropicais, Lisbon, Portugal, for providing personal research funds.
REFERENCES
- 1.Babiker, H. A., S. J. Pringle, A. Abdel-Mushin, M. Mackinnon, P. Hunt, and D. Walliker. 2001. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine-resistance transporter gene pfcrt and the multidrug-resistance gene pfmdr1. J. Infect. Dis. 183:1535-1538. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, D., S. Foote, D. Galatis, D. Kemp, and A. F. Cowman. 1992. Selection for high level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J. 11:3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basco, L., J. Le Bras, Z. Rhoades, and C. Wilson. 1995. Analysis of Pfmdr1 and drug susceptibility in fresh isolates of Plasmodium falciparum from Subsaharan Africa. Mol. Biochem. Parasitol. 74:157-166. [DOI] [PubMed] [Google Scholar]
- 4.Beale, G. H., R. Carter, and D. Walliker. 1978. Genetics, p. 213-245. In R. Killick-Kendrick and W. Peters (ed.), Rodent malaria. Academic Press, Inc., New York, N.Y.
- 5.Carlton, J., M. Mackinnon, and D. Walliker. 1998. A chloroquine resistance locus in the rodent malaria parasite Plasmodium chabaudi. Mol. Biochem. Parasitol. 93:57-72. [DOI] [PubMed] [Google Scholar]
- 6.Carlton, J. M. R., R. Vinkenoog, A. P. Waters, and D. Walliker. 1998. Gene synteny in species of Plasmodium. Mol. Biochem. Parasitol. 93:285-294. [DOI] [PubMed] [Google Scholar]
- 7.Carlton, J. M., K. Hayton, P. V. Cravo, and D. Walliker. 2001. Of mice and malaria mutants: unravelling the genetics of drug resistance using rodent malaria models. Trends. Parasitol. 17:236-242. [DOI] [PubMed] [Google Scholar]
- 8.Carter, R., and D. Walliker. 1975. New observations on the malaria parasites of rodents of the Central African Republic: Plasmodium vinckei petteri subsp. nov. and Plasmodium chabaudi Landau, 1965. Ann. Trop. Med. Parasitol. 69:187-196. [DOI] [PubMed] [Google Scholar]
- 9.Chaiyaroj, S. C., A. Buranakiti, P. Angkasekwinai, S. Looareesuwan, and A. F. Cowman. 1999. Analysis of mefloquine resistance and amplification of pfmdr1 in multidrug-resistant Plasmodium falciparum isolates from Thailand. Am. J. Trop. Med. Hyg. 61:780-783. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, Q., and A. Saul. 1994. The dihydrofolate reductase domain of rodent malarias: point mutations and pyrimethamine resistance. Mol. Biochem. Parasitol. 65:361-363. [DOI] [PubMed] [Google Scholar]
- 11.Cornelissen, A. W., and D. Walliker. 1985. Gametocyte development of Plasmodium chabaudi in mice and rats: evidence for host induction of gametocytogenesis. Zentbl. Parasitenkd. 171:297-303. [DOI] [PubMed] [Google Scholar]
- 12.Cowman, A. F., and A. M. Lew. 1989. Antifolate drug selection results in duplication and rearrangement of chromosome 7 in Plasmodium chabaudi. Mol. Cell. Biol. 9:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowman, A. F., and A. M. Lew. 1990. Chromosomal rearrangements and point mutations in the DHFR-TS gene of Plasmodium chabaudi under antifolate selection. Mol. Biochem. Parasitol. 42:21-29. [DOI] [PubMed] [Google Scholar]
- 14.Cowman, A., D. Galatis, and J. Thompson. 1994. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the Pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc. Natl. Acad. Sci. USA 91:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crewther, P. E., M. L. S. M. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djimde, A., O. K. Doumbo, R. W. Steketee, and C. V. Plowe. 2001. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet 358:890-891. [DOI] [PubMed] [Google Scholar]
- 17.Duraisingh, M. T., P. Jones, L. Sambou, L. Von Seidlein, M. Pinder, and D. C. Warhurst. 2000. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 108:13-23. [DOI] [PubMed] [Google Scholar]
- 18.Duraisingh, M. T., C. Roper, D. Walliker, and D. C. Warhurst. 2000. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol. Microbiol. 36:955-961. [DOI] [PubMed] [Google Scholar]
- 19.Fidock, D. A., T. Nomura, A. T. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. B. Ursos, A. B. S. Sidhu, B. Naudé, K. W. Deitsch, X.-Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein Pfcrt and evidence for their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foote, S. J., D. E. Kyle, R. K. Martin, A. M. Oduola, K. Forsyth, D. J. Kemp, and A. F. Cowman. 1990. Several alleles of the multidrug resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:255-258. [DOI] [PubMed] [Google Scholar]
- 21.Gervais, G. W., K. Trujillo, B. L. Robinson, W. Peters, and A. E. Serrano. 1999. Plasmodium berghei: identification of an mdr-like gene associated with drug resistance. Exp. Parasitol. 91:86-92. [DOI] [PubMed] [Google Scholar]
- 22.Grech, K., A. Martinelli, S. Pathirana, D. Walliker, P. Hunt, and R. Carter. 2002. Numerous robust genetic markers for Plasmodium chabaudi by the method of amplified fragment length polymorphism. Mol. Biochem. Parasitol. 123:95-104. [DOI] [PubMed]
- 23.Homewood, C. A., and K. D. Neame. 1976. A comparison of methods used for the removal of white cells from malaria-infected blood. Ann. Trop. Med. Parasitol. 70:249-251. [DOI] [PubMed] [Google Scholar]
- 24.Hyde, J. E. 1990. The dihydrofolate reductase-thymidylate synthetase gene in the drug-resistance of malaria parasites. Pharmacol. Ther. 48:45-59. [DOI] [PubMed] [Google Scholar]
- 25.Kemp, D. J., L. M. Corcoran, R. L. Coppel, H. D. Stahl, A. E. Bianco, G. V. Brown, and R. F. Anders. 1985. Size variation in chromosomes from independent cultured isolates of Plasmodium falciparum. Nature 315:347-350. [DOI] [PubMed] [Google Scholar]
- 26.Levin, B. R., V. Perrot, and N. Walker. 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154:985-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim, A. S., D. Galatis, and A. F. Cowman. 1996. Plasmodium falciparum: amplification and overexpression of pfmdr1 is not necessary for increased mefloquine resistance. Exp. Parasitol. 83:295-303. [DOI] [PubMed] [Google Scholar]
- 28.McKean, P. G., K. O'Dea, and K. N. Brown. 1993. Nucleotide sequence analysis and epitope mapping of the merozoite surface protein 1 from Plasmodium chabaudi chabaudi AS. Mol. Biochem. Parasitol. 62:199-210. [DOI] [PubMed] [Google Scholar]
- 29.Oduola, A. M. J., W. K. Milhous, N. F. Weatherly, J. H. Bowdre, and R. E. Desjardins. 1988. Plasmodium falciparum: induction of resistance to mefloquine in cloned strains by continuous drug exposure in vitro. Exp. Parasitol. 67:354-360. [DOI] [PubMed] [Google Scholar]
- 30.Padua, R. A. 1981. Plasmodium chabaudi: genetics of resistance to chloroquine. Exp. Parasitol. 52:419-426. [DOI] [PubMed] [Google Scholar]
- 31.Peel, S., P. Bright, B. Yount, J. Handy, and R. Baric. 1994. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homologue (Pfmdr1) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 51:648-658. [DOI] [PubMed] [Google Scholar]
- 32.Peters, W., J. Portus, and B. L. Robinson. 1977. The chemotherapy of rodent malaria. XXVIII. The development of resistance to mefloquine (WR 142,490). Ann. Trop. Med. Parasitol. 71:419-427. [DOI] [PubMed] [Google Scholar]
- 33.Powers, K. G., R. L. Jacobs, W. C. Good, and L. C. Koontz. 1969. Plasmodium vinckei: production of chloroquine-resistant strain. Exp. Parasitol. 26:193-202. [DOI] [PubMed] [Google Scholar]
- 34.Price, R., G. Robinson, A. Brockman, A. Cowman, and S. Krishna. 1997. Assessment of Pfmdr1 gene copy number by tandem competitive polymerase chain reaction. Mol. Biochem. Parasitol. 85:161-169. [DOI] [PubMed] [Google Scholar]
- 35.Price, R. N., C. Cassar, A. Brockman, M. Duraisingh, M. Van Vugt, N. J. White, F. Nosten, and S. Krishna. 1999. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob. Agents Chemother. 43:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quaye, I., and C. H. Sibley. 2002. Molecular data on Plasmodium falciparum chloroquine and antifolate resistance: a public health tool. Trends Parasitol. 18:184-186. [Google Scholar]
- 37.Rathod, P. K., T. McErlean, and P.-C. Lee. 1997. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 94:9389-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 39.Riffkin, C. D., R. Chung, D. M. Wall, J. R. Zalcberg, A. F. Cowman, M. Foley, and L. Tilley. 1996. Modulation of the function of human MDR1 P-glycoprotein by the antimalarial drug mefloquine. Biochem. Pharmacol. 52:1545-1552. [DOI] [PubMed] [Google Scholar]
- 40.Ritchie, G. I., M. Mungthin, J. E. Gree, P. G. Bray, S. R. Hawley, and S. A. Ward. 1996. In vitro selection of halofantrine resistance in Plasmodium falciparum is not associated with increased expression of Pgh1. Mol. Biochem. Parasitol. 83:35-46. [DOI] [PubMed] [Google Scholar]
- 41.Rosario, V. E. 1976. Genetics of chloroquine resistance in malaria parasites. Nature 261:585-586. [DOI] [PubMed] [Google Scholar]
- 42.Rubio, J., and A. F. Cowman. 1996. The ATP-binding cassette (ABC) gene family of Plasmodium falciparum. Parasitol. Today 2:135-140. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbour Laboratory, Cold Spring Harbor, N.Y.
- 44.Schwartz, D. C., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed-field gradient gel electrophoresis. Cell 37:67-75. [DOI] [PubMed] [Google Scholar]
- 45.Taylor, D., N. Cloonan, V. Mann, Q. Cheng, and A. Saul. 2000. Sequence diversity in rodent malaria of the Pfs28 ookinete surface antigen homologs. Mol. Biochem. Parasitol. 110:429-434. [DOI] [PubMed] [Google Scholar]
- 46.Thaithong, S., L. C. Ranford-Cartwright, N. Siripoon, P. Harnyuttanakorn, N. S. Kanchanakhan, A. Seugorn, et al. 2001. Plasmodium falciparum: gene mutations and amplification of dihydrofolate reductase genes in parasites grown in vitro in presence of pyrimethamine. Exp. Parasitol. 98:59-70. [DOI] [PubMed] [Google Scholar]
- 47.van Dijk, M. R., G. A. McConkey, R. Vinkenoog, A. P. Waters, and C. J. Janse. 1994. Mechanisms of pyrimethamine resistance in two different strains of Plasmodium berghei. Mol. Biochem. Parasitol. 68:167-171. [DOI] [PubMed] [Google Scholar]
- 48.Walliker, D., R. Carter, and A. Sanderson. 1975. Genetic studies on Plasmodium chabaudi: recombination between enzyme markers. Parasitology 66:309-320. [DOI] [PubMed] [Google Scholar]
- 49.White, N. J. 1994. Mefloquine. BMJ 308:286-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, C. M., A. E. Serrano, A. Wasley, M. P. Bogenschutz, A. H. Shankar, and D. F. Wirth. 1989. Amplification of a gene related to mammalian mdr genes in drug-resistant Plasmodium falciparum. Science 244:1184-1186. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, C., S. Volkman, S. Thaithong, R. Martin, D. Kyle, W. Milhous, and D. Wirth. 1993. Amplification of Pfmdr1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol. Biochem. Parasitol. 57:151-160. [DOI] [PubMed] [Google Scholar]
- 52.Wilson, M., J. De Risi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]