Abstract
To better understand the molecular basis of posaconazole (POS) resistance in Aspergillus fumigatus, resistant laboratory isolates were selected. Spontaneous mutants arose at a frequency of 1 in 108 and fell into two susceptibility groups, moderately resistant and highly resistant. Azole resistance in A. fumigatus was previously associated with decreased drug accumulation. We therefore analyzed the mutants for changes in levels of transcripts of genes encoding efflux pumps (mdr1 and mdr2) and/or alterations in accumulation of [14C]POS. No changes in either pump expression or drug accumulation were detected. Similarly, there was no change in expression of cyp51A or cyp51B, which encode the presumed target site for POS, cytochrome P450 14α-demethylase. DNA sequencing revealed that each resistant isolate carried a single point mutation in residue 54 of cyp51A. Mutations at the same locus were identified in three clinical A. fumigatus isolates exhibiting reduced POS susceptibility but not in susceptible clinical strains. To verify that these mutations were responsible for the resistance phenotype, we introduced them into the chromosome of a POS-susceptible A. fumigatus strain under the control of the glyceraldehyde phosphate dehydrogenase promoter. The transformants exhibited reductions in susceptibility to POS comparable to those exhibited by the original mutants, confirming that point mutations in the cyp51A gene in A. fumigatus can confer reduced susceptibility to POS.
The last decade has seen a dramatic increase in serious fungal infections. Those primarily at risk are immunocompromised individuals. Up to 80% of human immunodeficiency virus-infected patients are thought to develop a fungal infection at some point in the course of their illness (21). Aspergillus fumigatus, originally viewed as a weak pathogen, is now a major cause of death at cancer centers, with the mortality rate for leukemia patients approaching 90% even with antifungal therapy (12). Neither the current standards, amphotericin B (AMB) and itraconazole (ITZ), nor new therapies, caspofungin (CAS) and voriconazole (VOR), have satisfactorily met the therapeutic needs of patients with aspergillosis. Clinical failures with AMB are common, but correlating such failures with resistance is often difficult (19). Mechanism-based toxicity has limited the usefulness of AMB (12); newer less-toxic formulations are now available but are expensive. Isolates resistant to ITZ have been obtained from both in vivo (5, 7, 8) and in vitro sources (14, 15). VOR-resistant isolates have been generated in the laboratory (16; E. K. Manavathu, I. Baskaran, G. J. Alangaden, and P. H. Chandrasekar, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. J-817, 2001), but there have been no reports of in vivo resistance. Finally, there are insufficient data available on CAS to assess development of resistance.
Posaconazole (POS; SCH56592), a novel triazole currently in phase 3 clinical trials, is active against a broad spectrum of fungal pathogens including Aspergillus and Candida spp. (4, 10, 15). In vitro and in vivo testing has demonstrated that the drug is more effective than either ITZ or AMB against Aspergillus spp. (15, 25, 29) and that it is more effective than fluconazole (FLZ) against Candida spp. (4).
In this study we characterized isolates of A. fumigatus, both clinical and in vitro laboratory-selected mutants, which exhibit a reduced susceptibility to POS. We demonstrate that resistance is caused by mutations in the cyp51A gene, which encodes the target protein for POS.
(Part of this work was presented at the 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, Calif., September 2002.)
MATERIALS AND METHODS
Strains and plasmids.
A. fumigatus strain ND158 (ATCC 201795) was obtained from the Schering-Plough Research Institute (SPRI) culture collection. A pyrG derivative of ND158, ND158-R2, was selected by plating conidia of ND158 on yeast extract-peptone-dextrose (YPD) plates containing 50 μg each of uridine, uracil, and 5-fluroorotic acid/ml. PCR analysis confirmed that ND158-R2 carried a small deletion in the pyrG locus.
Antifungal agents.
POS and VOR were prepared at SPRI (Kenilworth, N.J.). ITZ, FLZ, and AMB powders were obtained from Janssen Pharmaceutica Inc. (Beerse, Belgium), Pfizer Inc. (New York, N.Y.), and Sigma Chemical Co. (St. Louis, Mo.), respectively. All drugs were dissolved in dimethyl sulfoxide.
Azole susceptibility.
The MICs for A. fumigatus strains were determined by the procedures described in National Committee for Clinical Laboratory Standards document M38-P, Reference Method for Broth Dilution Susceptibility Testing of Conidium-Forming Filamentous Fungi (22).
Selection of A. fumigatus mutants.
Conidia from A. fumigatus ND158 were washed from malt extract (ME) agar slants (grown at 29°C) with 5.5 ml of saline (0.9%). The absorbances (at 530 nm) of the conidial preparations were determined. Using a standard curve relating conidial counts to absorbance, we determined that each slant yielded approximately 3 × 108 to 8 × 108 conidia. Eleven separately grown conidial preparations were separately plated on defined synthetic dropout agar plates (Bio 101, Carlsbad, Calif.) supplemented with complete supplement mixture (Bio 101) and POS at 1 μg/ml. Mutants that arose after 5 days of growth at 29°C were replated on fresh plates containing the drug. After 7 days conidia were transferred to ME slants (with and without POS at 0.2 μg/ml), and after an additional 5 days of growth conidia were isolated for MIC determinations.
Measurement of uptake of [14C]POS by A. fumigatus.
Conidia from ME slants were grown for 18 h in 150 ml of YPD at 29°C. Mycelia were collected by filtration, resuspended at 1 g/30 ml in prewarmed 0.9% saline, and dispersed by vortexing with glass beads. Thirty milliliters of this suspension was added to 100 ml of prewarmed YPD containing [14C]POS at 0.02 μg/ml. At various times triplicate 5-ml aliquots were withdrawn, collected on glass fiber filters, and washed with 20 ml of saline containing 2.5 μg of unlabeled POS/ml. The filters were dried and then extracted with 0.75 ml of perchloric acid (30 min at 55°C), and the cell-associated drug was detected by liquid scintillation counting.
Quantitation of transcript levels.
Conidia from ME slants were grown for 18 h in 150 ml of YPD at 29°C. Total RNA was extracted with the RNAeasy minikit (Qiagen Inc., Valencia, Calif.). Real-time PCR analysis was performed as described previously (2). Relative gene expression levels (ΔCT) were calculated by using the 18S rRNA control.
PCR amplification and sequencing.
The cyp51 genes were amplified by PCR in overlapping 600-bp segments from total genomic DNA. Double-stranded DNA sequencing of the PCR products was performed by MWG-Biotech Inc. (High Point, N.C.).
Cloning of cyp51A alleles under the control of the glyceraldehyde phosphate dehydrogenase (gpdA) promoter.
The cyp51A open reading frame was amplified by PCR (oligonucleotides: 5′-GGTACCGATGCTATGGCTTACGGCCTACAT-3′ and 5′-TCTAGATCACTTGGATGTGTTTTTCGA-3′) from strains ND158, MS6 and R7-1 to append flanking KpnI/XbaI restriction sites and cloned into the corresponding sites in pAL3 (30), generating pPAM51 to -53, respectively. The gpdA promoter (nucleotides −679 to +38) (26) from Aspergillus nidulans was amplified by PCR (5′-GGTACCTGCGGAGAGACGGACGGT-3′ and 5′-GGTACCTAAAGGTTCTTGGATGGG-3′) to append flanking KpnI sites and cloned in front of the cyp51A alleles described above, generating pPAM54 (wild-type cyp51A), pPAM55 (encodes an arginine substitution at residue 54) and pPAM56 (encodes a tryptophan substitution at residue 54). All sequences were verified by double-stranded-DNA sequencing. Plasmids were introduced into strain ND158-R2 by the method of Tilburn et al. (27). Correct integration of the plasmid in the transformants was verified by isolating chromosomal DNA and screening for a PCR product of the appropriate size with an oligonucleotide pair that annealed to the gpdA promoter and the cyp51A gene.
RESULTS
Isolation of strains displaying reduced susceptibility to POS.
Five of 11 independently grown conidial suspensions from A. fumigatus ND158, plated on solid media containing POS at 1 μg/ml (approximately 30 times the MIC), yielded colonies. The overall frequency of resistance was approximately 1 per 108 conidia plated. A single colony from each independent plating experiment was analyzed further. The five mutants fell into two groups based on POS MICs: four exhibited moderate levels of resistance, and one exhibited high-level resistance (Table 1). The mutants also exhibited decreased susceptibility to ITZ, but there were no changes in susceptibility to either VOR or AMB.
TABLE 1.
Correlation between amino acid substitutions in codon 54 of cyp51A and decreases in azole susceptibility in clinical isolates and spontaneous mutants of A. fumigatus
| Strain | Source of strain | MIC (μg/ml)
|
CYP51A residue 54 | |||
|---|---|---|---|---|---|---|
| POS | ITZ | VOR | AMB | |||
| ND158 | Wild type | 0.03 | 0.12 | 0.25 | 1 | Glycine |
| MS6 | Laboratory mutant | 1 | >16 | 0.12 | 1 | Arginine |
| R1-1 | Laboratory mutant | 1 | >16 | 0.12 | 1 | Arginine |
| R4-1 | Laboratory mutant | 1 | >16 | 0.25 | 1 | Glutamate |
| R6-1 | Laboratory mutant | 1 | >16 | 0.25 | 1 | Glutamate |
| R7-1 | Laboratory mutant | >8 | >16 | 0.25 | 1 | Tryptophan |
| ND215 | Clinical isolate | 0.03 | 0.12 | 0.25 | 2 | Glycine |
| ND216 | Clinical isolate | 0.03 | 0.12 | 0.25 | 2 | Glycine |
| ND267 | Clinical isolate | 0.03 | 0.25 | 0.5 | 2 | Glycine |
| ND223 | Clinical isolate | 1 | >16 | 0.12 | 1 | Arginine |
| ND202 | Clinical isolate | >8 | >16 | 0.12 | 2 | Tryptophan |
| ND229 | Clinical isolate | >8 | >16 | 0.25 | 1 | Tryptophan |
Expression levels of cyp51 and efflux pump genes.
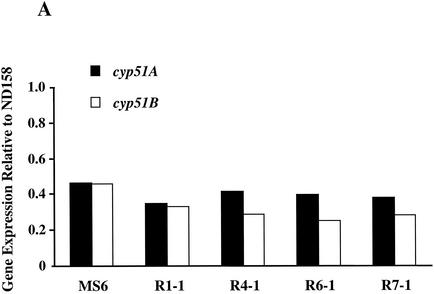
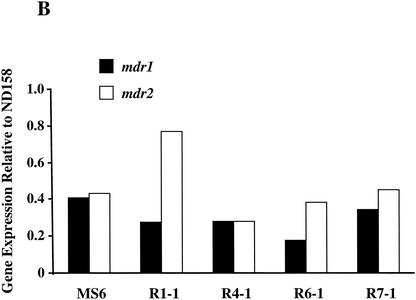
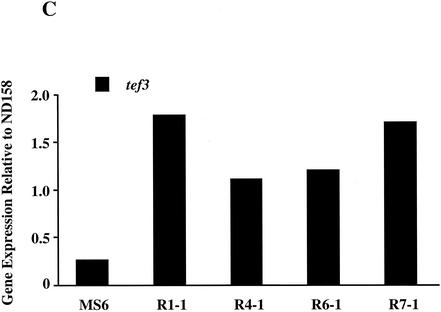
Azole resistance in fungi has previously been associated with increased expression of genes encoding efflux pumps and/or cyp51, which encodes the target protein for POS. A. fumigatus has two cyp51 genes, cyp51A and cyp51B (9, 18). To date only two genes in A. fumigatus encoding multiple-drug resistance-like transporters, mdr1 and mdr2, have been described (28). Levels of transcripts of cyp51A, cyp51B, mdr1, and mdr2 in 18-h-old mycelia from all five mutants were quantified by real-time PCR. As a control, the expression levels of tef3, which encodes peptide elongation factor 3, were also measured. The expression levels of the test genes in all five mutants fluctuated less than twofold compared to those in the azole-sensitive parental strain, ND158 (Fig. 1).
FIG. 1.
Measurement of the expression levels of the cyp51 and mdr genes in A. fumigatus mutants. For each target gene the expression levels were quantified by real-time PCR and then compared to the expression level of the same gene in the azole-susceptible parental strain, ND158. Expression levels of a control gene, tef3, were also determined. Differences in expression levels between the two strains are plotted for cyp51A and cyp51B (A), mdr1 and mdr2 (B), and tef3 (C). The data presented are the averages of two independent determinations. Deviations from the mean were less than 10%, and therefore error bars are not shown.
Uptake of [14C]POS by mutants.
To determine if an as yet uncharacterized efflux pump was responsible for the reduction in POS susceptibility, we monitored the accumulation of the radiolabeled drug in 18-h-old mycelia in the parental strain, ND158, and in strains MS6 and R7-1. In ND158 [14C]POS rapidly accumulated over the first 5 min, after which accumulation plateaued (data not shown). MS6 and R7-1 exhibited kinetics similar to those of ND158 (data not shown).
Nucleotide sequences of the cyp51 genes.
Azole resistance in both yeasts and molds has been associated with amino acid substitutions in the cytochrome P450 14α-demethylase (13). The cyp51A and cyp51B genes from the wild-type parent and the five mutants were sequenced. The sequences from the parental strain, ND158, matched the sequences deposited in the GenBank (9, 18). There were no missense mutations in the cyp51B gene in any of the mutants. In contrast, we identified single nucleotide substitutions in codon 54 of cyp51A in the mutant strains (Table 1). In the four isolates exhibiting moderate levels of POS resistance glycine 54 was mutated to either glutamate or arginine. In the mutant with a high level of POS resistance glycine 54 was mutated to tryptophan.
Sequencing the cyp51 genes from selected clinical isolates.
The cyp51 genes from several clinical isolates were also sequenced (Table 1). ND215 and ND216 are POS-susceptible isolates from different patients. ND267 and ND223 are susceptible and moderately resistant sequential isolates, respectively, from a single patient. ND202 and ND229 are sequential isolates from the same patient and exhibited high-level POS resistance. There were no missense mutations in cyp51B in any of these isolates. Also, there were no missense mutations in cyp51A in any of the susceptible isolates. However, we detected single nucleotide substitutions in codon 54 of the cyp51A genes from isolates for which the POS MICs were elevated. The substitutions of either arginine or tryptophan for glycine were again associated with moderate and high levels of resistance, respectively (Table 1).
Expression of cyp51A alleles in an azole-sensitive A. fumigatus strain.
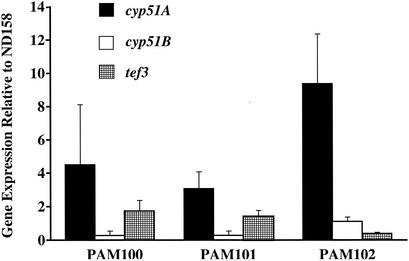
To determine if the cyp51A mutations were directly responsible for the azole resistance, the cyp51A genes from ND158, MS6, and R7-1 were cloned in front of the constitutive gpdA promoter on pAL3 and the resultant plasmids were used to transform strain ND158-R2 to uracil prototrophy. The transformants were subsequently screened for changes in POS susceptibility. Approximately 30% of the transformants derived from plasmids carrying the mutant cyp51A alleles exhibited increases in POS resistance comparable to those exhibited by the strains from which the cyp51A alleles were originally cloned (Table 2). For the transformants for which the POS MIC did not change, it is possible that either the plasmid underwent rearrangement during integration or that it integrated into a region of the chromosome that did not permit efficient expression of the cloned genes (3). For none of the 45 transformants obtained by using the plasmid carrying the wild-type cyp51A did the POS MICs exhibit any changes. However, we confirmed by PCR analysis (see Materials and Methods) that in approximately 50% of the strains the cyp51A gene was linked to the gpdA promoter. To rule out the possibility that the elevated MICs seen in the transformants derived from the mutant cyp51A alleles were simply due to overexpression of these particular alleles, perhaps resulting from multiple plasmid insertions, cyp51A transcript levels in representative transformants (PAM100 to -102; Table 2) were measured. All three transformants expressed cyp51A at comparable levels. Compared to those in the parental strain, ND158-R2, the cyp51A transcript levels in the transformants were elevated approximately three- to ninefold (Fig. 2). In contrast, expression levels of cyp51B and tef3 fluctuated less than threefold.
TABLE 2.
Effect of expressing cyp51A alleles carrying substitutions in codon 54 on POS susceptibility in A. fumigatus ND158-R2
| Strain | CYP51A residue 54 | Plasmid-encoded CYP51A residue 54 | POS MIC (μg/ml) |
|---|---|---|---|
| ND158-R2 | Glycine | 0.03 | |
| PAM100a | Glycine | Glycine | 0.03 |
| MS6 | Arginine | 1 | |
| PAM101a | Glycine | Arginine | 0.5 |
| R7-1 | Tryptophan | >8 | |
| PAM102a | Glycine | Tryptophan | >8 |
Transformants derived from transforming ND158-R2 with plasmids carrying the indicated cyp51A alleles.
FIG. 2.
Measurement of the expression levels of the cyp51 genes in A. fumigatus transformants. cyp51A alleles with point mutations in codon 54 were transformed into a wild-type A. fumigatus strain. The expression levels of cyp51A and cyp51B in selected transformants exhibiting reduced susceptibility to POS were quantified by real-time PCR. The differences in expression levels between the transformants and the azole-susceptible parental strain, ND158, are shown. Expression levels of a control gene, tef3, were also determined. The data presented are the averages of three independent determinations.
DISCUSSION
Azole resistance in fungi, particularly in Candida albicans, is well documented (most recently reviewed in reference 13). Resistance most often results from either energy-dependent drug efflux or alterations in either the expression level or sequence of the target enzyme, 14α-demethylase. In contrast, azole resistance in Aspergillus has received less attention. A. fumigatus isolates resistant to either ITZ or VOR have been selected in the laboratory. The resistance frequencies for both drugs were the same as that for POS measured in this study (14, 16). For a number of the laboratory-generated VOR-resistant isolates, mutations were identified in two positions in cyp51A (Manavathu et al., 41st ICAAC). Interestingly, one of the sites is analogous to the site of a mutation causing FLZ resistance in C. albicans. Similarly, two ITZ-resistant A. fumigatus isolates from a patient receiving ITZ therapy exhibited reduced susceptibility to ITZ in a cell-free assay measuring 14α-demethylase activity (8). Plasmid-mediated overexpression of cyp51A resulted in ITZ resistance in A. fumigatus (24), but there are no reports of either in vitro or in vivo resistance being ascribed to overexpression of the cyp51 genes. Finally, in vivo (8) and spontaneous in vitro (14) isolates exhibited reduced accumulation of ITZ. However, no particular pump has been identified as being responsible for efflux.
In this study we demonstrated that mutations in cyp51A, specifically in codon 54, are associated with POS resistance in A. fumigatus. Depending on the amino acid substitution the strains were either moderately or highly resistant. To confirm that the mutant alleles were responsible for the resistance phenotype, they were cloned into an integrative vector and introduced into a sensitive strain. The resultant transformants exhibited decreases in POS susceptibility comparable to those exhibited by the strains from which the cyp51A alleles were originally cloned. We further verified that the resistance phenotype was specifically dependent on expression of cloned mutant alleles by deleting the promoter upstream of the cyp51A alleles and repeating the transformations (data not shown). In the absence of a promoter driving cyp51A expression none of the transformants exhibited any changes in POS susceptibility.
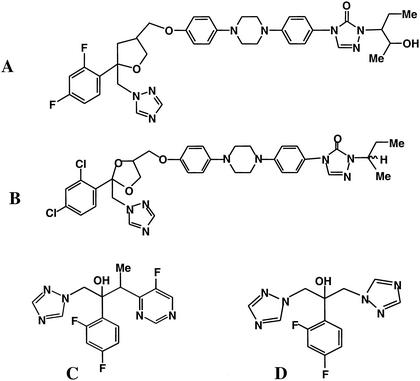
As might be expected from the structural similarity between POS and ITZ (Fig. 3), the POS-resistant mutants exhibited cross-resistance to ITZ. However, there was no cross-resistance to VOR. In two recent studies the reciprocal observation was made: A. fumigatus isolates, for which the VOR MICs ranged from 4 to 16 μg/ml, remained susceptible to POS (15; Manavathu et al., 41st ICAAC). These findings differ from those for C. albicans, where FLZ-resistant strains consistently exhibited cross-resistance to ITZ and VOR (6, 20, 23). Whether these apparent differences between C. albicans and A. fumigatus are due to the operation of different mechanisms of resistance or differences in the way the azoles bind the 14α-demethylase (see below) remains to be determined.
FIG. 3.
Structures of POS (A), ITZ (B), VOR (C), and FLZ (D).
The finding that the substitutions that result in azole resistance are located close to the N terminus of the 14α-demethylase was unusual. To date, the closest mutation resulting in azole resistance was at codon 105 in C. albicans (17). The first 43 amino acids of the 14α-demethylase from C. albicans are predicted to form a membrane-spanning domain that anchors the protein to the cytoplasmic face of the endoplasmic reticulum (1). Deleting this anchor as far as (but not including) glycine 65, which is analogous to glycine 54 in A. fumigatus, generated a soluble protein which retained the ability to bind FLZ (11). The N terminus of the A. fumigatus protein has a similar hydrophobicity profile and has significant homology with the C. albicans protein, suggesting that it may perform a similar role. However, in contrast to what was found for the C. albicans protein, this study demonstrated that mutations in the region immediately after the hydrophobic domain resulted in POS resistance. It is therefore possible that FLZ and POS interact with different regions of the 14α-demethylase protein, a finding that may explain why most FLZ-resistant C. albicans strains with defined mutations in ERG11 (the C. albicans orthologue of cyp51A) show no changes in susceptibility to POS (D. Sanglard, F. Ischer, and J. Bille, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-11, 1997).
Acknowledgments
We thank Dayna Daubaras (SPRI) and Gregory May (University of Texas) for helpful comments and suggestions and Todd Black (SPRI) for critically reading the manuscript.
REFERENCES
- 1.Boscott, P. E., and G. H. Grant. 1994. Modeling cytochrome P450 14α demethylase (Candida albicans) from P450cam. J. Mol. Graphics 12:185-192. [DOI] [PubMed] [Google Scholar]
- 2.Brieland, J., D. Essig, C. Jackson, D. Frank, D. Loebenberg, F. Menzel, B. Arnold, B. DiDomenico, and R. Hare. 2001. Comparison of pathogenesis and host immune responses to Candida glabrata and Candida albicans in systemically infected immunocompetent mice. Infect. Immun. 69:5046-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buxton, F. P., D. I. Gwynne, and R. W. Davies. 1985. Transformation of Aspergillus niger using the argB gene of Aspergillus nidulans. Gene 37:201-214. [DOI] [PubMed] [Google Scholar]
- 4.Cacciapuoti, A., D. Loebenberg, E. Corcoran, F. Menzel, Jr., E. L. Moss, Jr., C. Norris, M. Michalski, K. Raynor, J. Halpern, C. Mendrick, B. Arnold, B. Antonacci, R. Parmegiani, T. Yarosh-Tomaine, G. H. Miller, and R. S. Hare. 2000. In vitro and in vivo activities of SCH 56592 (posaconazole), a new triazole antifungal agent, against Aspergillus and Candida. Antimicrob. Agents Chemother. 44:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chryssanthou, E. 1997. In vitro susceptibility of respiratory isolates of Aspergillus species to itraconazole and amphotericin B. Acquired resistance to itraconazole. Scand. J. Infect. Dis. 29:509-512. [DOI] [PubMed] [Google Scholar]
- 6.Cuenca-Estrella, M., T. M. Diaz-Guerra, E. Mellado, A. Monzon, and J. L. Rodriguez-Tudela. 1999. Comparative in vitro activity of voriconazole and itraconazole against fluconazole-susceptible and fluconazole-resistant clinical isolates of Candida species from Spain. Eur. J. Clin. Microbiol. Infect. Dis. 18:432-435. [DOI] [PubMed] [Google Scholar]
- 7.Denning, D. W., S. A. Radford, K. L. Oakley, L. Hall, E. M. Johnson, and D. W. Warnock. 1997. Correlation between in-vitro susceptibility testing to itraconazole and in-vivo outcome of Aspergillus fumigatus infection. J. Antimicrob. Chemother. 40:401-414. [DOI] [PubMed] [Google Scholar]
- 8.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. W. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edlind, T. D., K. W. Henry, K. A. Metera, and S. K. Katiyar. 2001. Aspergillus fumigatus CYP51 sequence: potential basis for fluconazole resistance. Med. Mycol. 39:299-302. [DOI] [PubMed] [Google Scholar]
- 10.Galgiani, J. N., and M. L. Lewis. 1997. In vitro studies of activities of the antifungal triazoles SCH56592 and itraconazole against Candida albicans, Cryptococcus neoformans and other pathogenic yeasts. Antimicrob. Agents Chemother. 41:180-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb, D. C., D. E. Kelly, K. Venkateswarlu, N. J. Manning, H. F. Bligh, W. H. Schunck, and S. L. Kelly. 1999. Generation of a complete, soluble, and catalytically active sterol 14 alpha-demethylase-reductase complex. Biochemistry 38:8733-8738. [DOI] [PubMed] [Google Scholar]
- 12.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupetti, A., R. Danesi, M. Campa, M. Del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 14.Manavathu, E. K., J. A. Vazquez, and P. H. Chandrasekar. 1999. Reduced susceptibility in laboratory-selected mutants of Aspergillus fumigatus to itraconazole due to decreased intracellular accumulation of the antifungal agent. Int. J. Antimicrob. Agents 12:213-219. [DOI] [PubMed] [Google Scholar]
- 15.Manavathu, E. K., J. L. Cutright, D. Loebenberg, and P. H. Chandrasekar. 2000. A comparative study of the in vitro susceptibilities of clinical and laboratory-selected resistant isolates of Aspergillus spp. to amphotericin B, itraconazole, voriconazole and posaconazole (SCH 56592). J. Antimicrob. Chemother. 46:229-234. [DOI] [PubMed] [Google Scholar]
- 16.Manavathu, E. K., O. C. Abraham, and P. H. Chandrasekar. 2001. Isolation and in vitro susceptibility of voriconazole-resistant laboratory isolates of Aspergillus fumigatus. Clin. Microbiol. Infect. 7:130-137. [DOI] [PubMed] [Google Scholar]
- 17.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. S. C. Ramaekers, F. C. Odds, and H. V. Bossche. 1999. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 18.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2001. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore, C. B., N. Sayers, J. Mosquera, D. Slaven, and D. W. Denning. 2000. Antifungal drug resistance in Aspergillus. J. Infect. 41:203-220. [DOI] [PubMed] [Google Scholar]
- 20.Müller, F. M., M. Weig, J. Peter, and J. T. Walsh. 2000. Azole cross-resistance to ketoconazole, fluconazole, itraconazole and voriconazole in clinical Candida albicans isolates from HIV-infected children with oropharyngeal candidosis. J. Antimicrob. Chemother. 46:338-340. [DOI] [PubMed] [Google Scholar]
- 21.Musial, C. E., F. R. Cockerill, and G. D. Roberts. 1988. Fungal infections of the immunocompromised host: clinical and laboratory aspects. Clin. Microbiol. Rev. 1:349-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 1998. Reference method for broth dilution antifungal susceptibility testing of conidium-forming filamentous fungi. Proposed standard M38-P. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.Nguyen, M. H., and C. Y. Yu. 1998. Voriconazole against fluconazole-susceptible and resistant Candida isolates: in-vitro efficacy compared with that of itraconazole and ketoconazole. J. Antimicrob. Chemother. 42:253-256. [DOI] [PubMed] [Google Scholar]
- 24.Osherov. N., D. P. Kontoyiannis, A. Romans, and G. S. May. 2001. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14α-demethylase gene, pdmA. J. Antimicrob. Chemother. 48:75-81. [DOI] [PubMed] [Google Scholar]
- 25.Pfaller, M. A., S. A. Messer, R. J. Hollis, R. N. Jones, and the SENTRY Participants Group. 2002. Antifungal activities of posaconazole, ravuconazole, and voriconazole compared to those of itraconazole and amphotericin B against 239 clinical isolates of Aspergillus spp. and other filamentous fungi: report from the SENTRY Antimicrobial Surveillance Program, 2000. Antimicrob. Agents Chemother. 46:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Punt, P. J., M. A. Dingemanse, A. Kuyvenhoven, R. D. Soede, P. H. Pouwels, and C. A. van den Hondel. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101-109. [DOI] [PubMed] [Google Scholar]
- 27.Tilburn, J., C. Scazzocchio, G. G. Taylor, J. H. Zabicky-Zissman, R. A. Lockington, and R. W. Davies. 1983. Transformation by integration in Aspergillus nidulans. Gene 26:205-221. [DOI] [PubMed] [Google Scholar]
- 28.Tobin, M. B., R. B. Peery, and P. L. Skatrud. 1997. Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene 200:11-23. [DOI] [PubMed] [Google Scholar]
- 29.Uchida, K., N. Yokota, and H. Yamaguchi. 2001. In vitro antifungal activity of posaconazole against various pathogenic fungi. Int. J. Antimicrob. Agents 18:167-172. [DOI] [PubMed] [Google Scholar]
- 30.Waring, R. B., G. S. May, and R. N. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79:119-130. [DOI] [PubMed] [Google Scholar]