Abstract
Meningioma, a tumor of the meninges covering the central nervous system, shows frequent loss of material from human chromosome 22. Homozygous and heterozygous deletions in meningiomas defined a candidate region of >1 Mbp in 22q12.3-q13.1 and directed us to gene cloning in this segment. We characterized a new member of the N-acetylglucosaminyltransferase gene family, the LARGE gene. It occupies >664 kilobases and is one of the largest human genes. The predicted 756-aa N-acetylglucosaminyltransferase encoded by LARGE displays features that are absent in other glycosyltransferases. The human like-acetylglucosaminyltransferase polypeptide is much longer and contains putative coiled-coil domains. We characterized the mouse LARGE ortholog, which encodes a protein 97.75% identical with the human counterpart. Both genes reveal ubiquitous expression as assessed by Northern blot analysis and in situ histochemistry. Chromosomal mapping of the mouse gene reveals that mouse chromosome 8C1 corresponds to human 22q12.3-q13.1. Abnormal glycosylation of proteins and glycosphingolipids has been shown as a mechanism behind an increased potential of tumor formation and/or progression. Human tumors overexpress ganglioside GD3 (NeuAcα2,8NeuAcα2,3Galβ1,4Glc-Cer), which in meningiomas correlates with deletions on chromosome 22. It is the first time that a glycosyltransferase gene is involved in tumor-specific genomic rearrangements. An abnormal function of the human like-acetylglucosaminyltransferase protein may be linked to the development/progression of meningioma by altering the composition of gangliosides and/or by effect(s) on other glycosylated molecules in tumor cells.
Human chromosome 22 is the second smallest autosome and is rich in genes of medical interest. A considerable number of tumors exhibit deletions on this chromosome, suggesting that it contains several cancer-related genes that have not yet been characterized (1). Meningioma is one of the tumors that frequently display total or partial deletions on chromosome 22 (2–4). Previous studies revealed several regions on 22q that were targeted by interstitial homozygous and/or heterozygous tumor-specific deletions, and these regions may therefore harbor tumor suppressor genes. One of the regions was defined by a combination of two interstitial deletions—a homozygous deletion in one tumor and a heterozygous deletion in another (4)—and delineated a segment of >1 Mbp in 22q12.3-q13.1. These findings prompted us to investigate the gene content of this chromosomal region.
Glycosyltransferases (GTs) constitute a heterogeneous group of enzymes that carry out synthesis of glycoprotein and glycosphingolipid sugar chains within different compartments of the Golgi network (5). Analysis of sequences from mammalian GTs cloned so far indicates a family with low sequence conservation but with similar protein structure. They all are between 330–560 amino acids long and share the same type II transmembrane protein structure (Nin/Cout) with four main domains: a short cytoplasmic domain, a targeting/membrane-anchoring domain, a stem region, and a catalytic domain. The latter two are located within the Golgi cisternae (5).
Glycosphingolipids are composed of sphingosine, fatty acid chain, and oligosaccharide head, which constitutes the basis for their diversity. Gangliosides are complex glycosphingolipids containing sialic acid residues in their oligosaccharide head. Gangliosides participate in various cellular processes, and there is evidence for their role in tumorigenesis: e.g., the composition of gangliosides has been shown to change on cellular transformation (6, 7). This process is believed to increase tumorigenicity and/or to affect the metastatic capacity of tumor cells. Some growth factor receptors appears to be regulated by gangliosides, among them ganglioside GD3. Gangliosides may inhibit dimerization of growth factor receptors and thus may modify their actions (8). Melanoma was shown to overexpress gangliosides GD3 and GD2, and clinical trials using antibodies mimicking GD3 or GD2 resulted in inhibition of tumor growth in a number of patients (9, 10). Meningioma also has been studied for its content of gangliosides and has been divided into ganglioside GM3 (NeuAcα2,3Galβ1,4Glc-Cer)- and GD3-rich groups (11). It also has been shown that monosomy 22 in meningiomas correlates with a high GD3 content (12). Moreover, there is a connection between monosomy 22 and an increased aggressiveness of meningioma because deletions on chromosome 22 correlate with signs of tumor recurrence (13).
We report here the cloning of a novel, distinct member of the N-acetylglucosaminyltransferase gene family, the LARGE gene, from a region on human chromosome 22 that previously was shown to be affected by interstitial tumor deletions. An abnormal function of the human like-acetylglucosaminyltransferase (LARGE) protein may be linked to the development/progression of meningioma by altering the composition of gangliosides and/or by effect(s) on other glycosylated molecules in tumor cells.
MATERIALS AND METHODS
Physical Mapping, Analysis of Genomic Sequence, and cDNA Screening.
The contig was constructed and exon amplification was applied to 24 cosmids from the region as described (14–16). Analysis of sequences was performed by using the xgrail2 program (17). Predicted exons were tested by PCR in the following cDNA libraries: fetal brain, fetal muscle, adult muscle, fetal spleen, pancreatic adenocarcinoma, and testis (Stratagene, catalog nos. 936206, 836201, 937209, 937205, 937208, and 939202, respectively) and fetal brain and thyroid (CLONTECH, catalog nos. HL3003a and HL3019a, respectively). DNA fragments were labeled radioactively by PCR (18) or by random priming (19). cDNA screening of human fetal brain (Stratagene, catalog no. 936206), mouse early embryo, and mouse brain (CLONTECH, catalog no. ML3000a) libraries was performed as described (20).
Sequencing and Bioinformatics.
cDNA sequencing was performed by using BigDye-Terminators (Applied Biosystems) (21). Sequencing products were separated by using LongRanger (FMC) on ABI377 (Applied Biosystems). Sequences were assembled by using the gap4 program (22). cDNAs were sequenced with a minimal redundancy of 8.7 reading character per consensus character and at least one sequencing read on each strand. Repeats were filtered out by using repeat masker (http://www.ftp.genome.washington.edu). The blast programs were used on the National Center for Biotechnology Information/National Institutes of Health server (http://www.ncbi.nlm.nih.gov/BLAST). Promoter analysis was done with tssw (http://www.dot.imgen.bcm.tmc.edu:9331/gene-finder/Help/tssw.html) and nntp (http://www-hgc.lbl.gov/projects/promoter.html). Protein sequences were aligned by using clustal (23). psort was used to predict the cellular localization (http://www.psort.nibb.ac.jp:8800/form.html), and tmpred was used for analysis of the protein transmembrane regions (http://www.isrec.isb-sib.ch/software/TMPRED_form.html). Coiled-coil domains were predicted by using coils (24) (http://www.isrec.isb-sib.ch/software/COILS_form.html) and multicoil (25) (http://nightingale.lcs.mit.edu/cgi-bin/multicoil). Secondary protein structures were predicted by using ssp (http://www.dot.imgen.bcm.tmc.edu:9331/pssprediction/pssp.html). A search for prosite patterns (26) was performed by using scanprosite (http://www.expasy.hcuge.ch/sprot/scnpsit1.html).
In Situ Hybridization on Mouse Sections.
Two 45-mer oligonucleotides (5′-AAT GCA GCT TTC GCT GGT CAC ATG TCA ACT CAA TAC AAA CAG CCC, and its complementary sequence) were used as probes and correspond to nucleotides 3,602–3,646 of the mouse Large cDNA. A control probe specific for the cholecystokinin gene was used as described (27). Oligonucleotides were 3′-end-labeled with ATP[α-35S] (28). The specific activities obtained ranged from 1–4 × 109 cpm/μg. Probes were hybridized to mouse sections from day-14.5 and -17.5 embryos and adult brain, according to published procedures (28). Sections were dipped in nuclear track β2 emulsion (Kodak). After exposure at 4°C for 5 weeks, slides were developed and mounted in glycerol-phosphate buffer before microscope analysis (Axiophot, Zeiss).
Fluorescent in Situ Hybridization (FISH) on Mouse Metaphase Chromosomes.
Mouse P1-derived artificial chromosome (PACs) 396N1 and 657P21 from a 129-strain library (RPCI-21, Roswell Park Cancer Institute, Buffalo, NY) were labeled with digoxigenin-11-dUTP by using a DIG-nick system (Boehringer Mannheim). FISH was performed as described (29) on metaphases prepared from BALB/c embryo cultures. Before each hybridization, 100 ng of labeled PAC was preannealed with 20 μg of mouse DNA. The hybridization signal was detected by using the antidigoxigenin-fluorescein antibody (Boehringer Mannheim). Identification of mouse chromosomes was done by consecutive FISH paintings of fluorescein isothiocyanate- or Cy3-labeled mouse chromosomes 6-, 8-, 9-, 10-, 11- and 15-specific probes (Cambio, Cambridge, U.K.) to the same metaphase spreads. Results were analyzed by using a fluorescence microscope (LEITZ-DMRB, Leica, Heidelberg).
RESULTS
Mapping of the Region Deleted in Tumors 11 and 119A and Construction of a Genomic Contig.
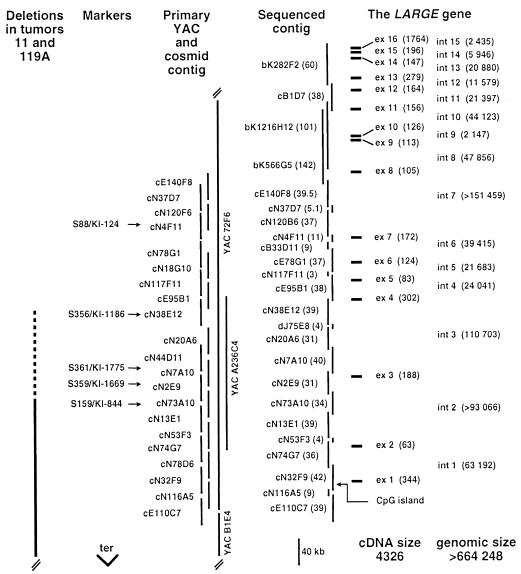
Previous deletion mapping of 170 sporadic meningiomas revealed two cases (11 and 119) with interstitial deletions centered around marker KI-844 (4). We refined here the breakpoint location of the deletions by using the following eight markers: KI-1186, KI-844, KI-106, KI-117, KI-711, W23C, KI-261, and the MB (myoglobin) gene (Fig. 1). Loss of heterozygosity in tumor 11, defining the extent of heterozygous deletion, was observed for the above markers, except KI-1186 at the centromeric side and W23C, KI-261, and MB at the telomeric side of the deletion. The tumor 119 originally was divided into multiple sections, and three of them were analyzed molecularly. On refined analysis, section 119A displayed a homozygous deletion for all above-mentioned markers except for KI-1186 and MB. The overlap between these deletions can be estimated to 1.5 Mbp between markers KI-844 and KI-711 (30). Using the probes previously mapped to this part of chromosome 22 (31, 32), we constructed and present here a physical map over the deleted area, based on a combination of yeast artificial chromosomes and cosmids. Fig. 1 displays the centromeric part of this contig, which was the starting point for systematic gene characterization. This contig also was submitted for sequencing to the Sanger Centre (Hinxton, U.K.).
Figure 1.
Structure of the human LARGE gene. The extent of the deletions in tumors 11 and 119A is shown on the left side. The dashed bar indicates the location of centromeric breakpoints of deletions (4). Markers KI-1186 and KI-844 are retained and deleted, respectively, in both tumors. Cosmids shown in this figure represent only a fraction of all cosmids identified in the course of contig construction. The exact length of cosmid clones and extent of overlap between each cosmid step in the primary contig has not been determined. Clones sequenced from this region are shown in the sequenced contig, which is composed of cosmids, bacterial artificial chromosomes, and one PAC, indicated by prefixes “c”, “b”, and “dJ”, respectively. The vertical bar for each sequenced clone is proportional to the amount of sequence data generated from each clone and is indicated in parentheses and in kilobase pairs, next to the clone name. Clones cE140F8, cN120B6, cN2E9, and bK566G5 are not yet fully sequenced. Accession numbers for sequenced clones are bK282F2, AL008630; cB1D7, Z82173; bK1216H12, AL008715; bK566G5, AL023577; cE140F8, Z82179; cN37D7, Z73421; cN120B6, Z73987 and Z73988; cN4F11, Z69943; cB33D11, AL008640; cE78G1, Z70288; cN117F11, Z97354; cE95B1, Z69042; cN38E12, Z68287; dJ75E8, Z76736; cN20A6, Z69713; cN7A10, Z68324; cN2E9, Z68685, Z68686, and Z68286; cN73A10, Z49866; cN13E1, Z54073; cN53F3, Z77853; cN74G7, Z69715; cN32F9, Z73429; cN116A5, Z69925; and cE110C7, Z68223. “Ter” indicates the direction of the telomere. The human LARGE gene is composed of 16 exons shown by filled rectangles on the right side. The sizes (in base pairs) for each exon (ex) and intron (int) are shown in parentheses.
Cloning of the Human LARGE Gene.
Twenty-four nonoverlapping cosmid clones (eight of which belong to the contig shown in Fig. 1) were analyzed by exon trapping, and 29 putative exons were retrieved. However, on Northern blot analysis and screening of cDNA libraries, we were unable to characterize any cDNA clone (data not shown). As the genomic sequence in 22q12.3-q13.1 became available from the European Molecular Biology Laboratory/GenBank database and no expressed sequence tags matched the genomic sequence, we analyzed the region by using a sequence-based exon-trapping approach (33). We applied the xgrail2 program in the analysis of six sequence contigs, spanning >650 kilobases (kb) in the vicinity of marker KI-844, which resulted in prediction of 209 putative exons. Previous assessment of the accuracy of xgrail predictions indicated that 85% of the exons predicted with “excellent” score truly corresponded to expressed genes (34). We concentrated on 71 exons that were predicted with the highest score. For each excellent exon, we designed a pair of intraexonic primers and tested them by using PCR on a panel of eight human cDNA libraries. Twenty-seven of the predicted exons generated a PCR product of correct size, were further tested on Northern blots, and were used as probes on cDNA screening. A 302-bp exon (no. 4, Fig. 1) predicted from cosmid cE95B1 was the only one to give positive results on Northern blot as it detected an ≈4.5-kb, ubiquitously expressed transcript (results not shown). Screening of the human fetal brain cDNA library resulted in 10 positive cDNA clones, which were end-sequenced. Two clones containing the longest inserts were sequenced fully and resulted in a 4,326-bp consensus [European Molecular Biology Laboratory/GenBank accession no. (acc.) AJ007583]. The longest ORF encoded a protein of 756 amino acids showing, on gapped-blastp search, highest similarity with the human i-β-1,3-N-acetylglucosaminyltransferase (28% identity and 44% similarity; acc. AF029893). This newly cloned gene therefore was named the LARGE (for like-acetylglucosaminyltransferase) gene. One cDNA clone, c4.5, extending over the entire 4,326 bp, was hybridized to a Northern blot and revealed a ubiquitous pattern of expression, highest in heart, brain, and skeletal muscle (Fig. 2E).
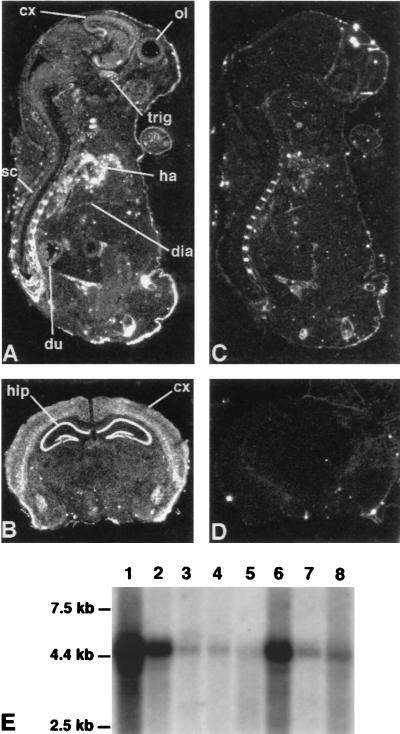
Figure 2.
Expression pattern of the LARGE gene in human and mouse. Dark field autoradiograms illustrate the gene expression in mouse embryo (A) and adult brain (B) by using mRNA in situ hybridization. The antisense probe of the mouse Large cDNA sequence was hybridized to a sagittal section of a day-17.5 mouse embryo (A) and to a coronal section from mouse adult brain (B). Note the ubiquitous pattern of gene expression with a strong signal in heart (ha), central nervous system structures such as cerebral cortex (cx), hippocampus (hip), olfactory lobe (ol), trigerminal ganglion (trig), and spinal cord (sc) as well as in diaphragm (dia) and duodenum (du). As a negative control, the sense probe was hybridized to contiguous sections (C and D). (E) Ubiquitous expression of the LARGE gene in human tissues as an ≈4.5-kb transcript. The entire cDNA was used as probe on Northern blot containing poly(A)+ selected mRNA from human adult tissues (MTN 7760–1, CLONTECH): Lanes: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas.
Comparison between the LARGE cDNA and the genomic sequence (http://www.sanger.ac.uk/HGP/Chr22) revealed that its genomic size is >664 kb, is composed of 16 exons (ranging from 63 to 1,764 bp), and is transcribed in the telomere to centromere direction (Fig. 1). The introns of the LARGE gene range from 2,147 bp to >151 kb. The sequences of all splicing sites of the gene contained the consensus sequence of donor (AG) and acceptor (GT) sites (data not shown; annotated in acc. AJ007583). Exons 1 and 2 are fully untranslated. Exon 3 contains the predicted start of translation, and exon 16 (1,764 bp) contains 1,569 bp of the 3′ untranslated region. The region at the 5′ end of the LARGE gene (within cosmid N32F9; Fig. 1; acc. Z73429) displays characteristics of a CpG island. The 2,795 bp sequence, encompassing the entire exon 1 and stretching 200 bp into the first intron, is GC-rich with 71.7% of C+G nucleotides and has an observed/expected ratio of CpG dinucleotide of 0.83 (35). We therefore conclude that this region is likely to represent the true GC-rich promoter of LARGE, which is consistent with its function as a housekeeping, ubiquitously expressed gene.
We retrieved 31 human expressed sequence tags from the database of expressed sequence tags that corresponded to the human LARGE gene. As expected, all were distributed over the 3′ part of the cDNA with one (acc. H55582) located closest to the 5′ end and starting at position 1,382 in our cDNA sequence. Furthermore, on blastx comparison of the human LARGE protein with the databases, we detected an orthologous gene in cosmid K09C8 (acc. Z68006) from Caenorhabditis elegans. The predicted C. elegans protein is composed of 622 amino acids and displays 33/51% identity/similarity with the human protein (Fig. 3).
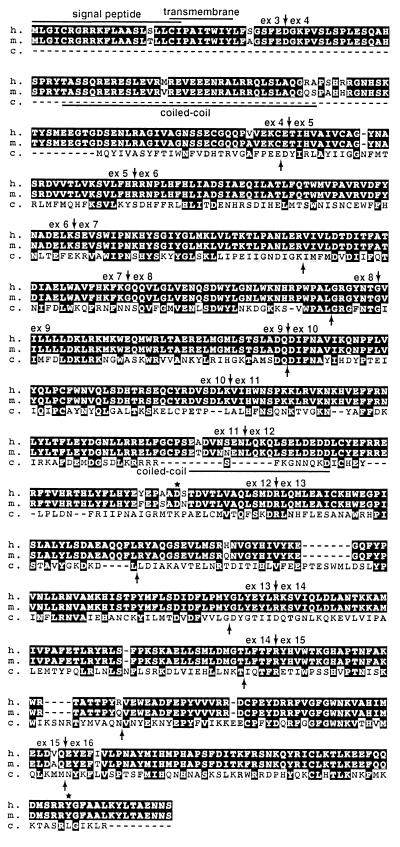
Figure 3.
Alignment of predicted amino acid sequences of the three LARGE genes from human (h.), mouse (m.), and C. elegans (c.). Identity and similarity are indicated by black and gray boxes, respectively. White boxes indicate nonconservative amino acid changes and dashes (−) indicate gaps. The positions of exon/intron borders of the human and C. elegans genes are indicated by vertical arrows above the human sequence and below the C. elegans sequence. Four protein domains of the human LARGE protein predicted by computer-assisted sequence analysis are marked: a signal peptide (amino acids 1–24), a transmembrane domain (amino acids 20–28), and two coiled-coil domains (amino acids 55–90 and 422–441). The portion of the human Large sequence between two asterisks (∗) shows sequence similarity with the part of the human i-β-1,3-N-acetylglucosaminyltransferase (amino acids 91–408, acc. AF029893).
Characterization of the Mouse Large Ortholog Reveals that Human 22q12.3-q13.1 Corresponds to Mouse 8C1.
The human LARGE c4.5 cDNA was used as probe on mouse embryo and brain cDNA libraries, and 30 positive clones were retrieved. Only one of them was derived from the embryo cDNA library. We obtained 3,678 bp of cDNA sequence (acc. AJ006278) by end-sequencing all cDNA clones and further primer-walking. The longest ORF of the mouse gene is 89% identical with the ORF of the human gene and is capable of encoding a 756aa protein, 97.75% identical with its human counterpart. Using mRNA in situ hybridization, we examined the expression pattern of the Large gene in mouse embryos at days 14.5 and 17.5 and in adult mouse brain (Fig. 2 A and B). The mouse Large gene also is expressed ubiquitously and with high levels in heart and diaphragm as well as in the central nervous system, especially in cerebral cortex, hippocampus, and trigerminal ganglion.
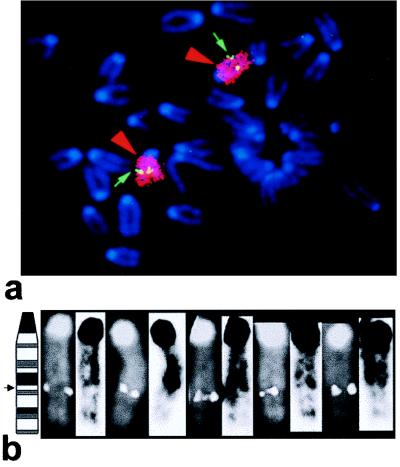
Two probes from mouse Large cDNA were used in screening of the mouse PAC library. One probe located within the translated part of the mouse gene (nucleotides 275–656; acc. AJ006278) and corresponding to exons 3 and 4 of the human gene detected 12 positives PACs. The 450-bp insert of expressed sequence tag clone AA260869 was used as the second probe. It covered the 3′ untranslated part of mouse cDNA (nucleotides 3,240 to the end) and detected three positive PACs. One PAC from each set (657P21 and 396N1) was used for FISH mapping of Large on mouse metaphases. Twenty mouse metaphase spreads were analyzed, and specific FISH signals from PACs 657P21 or 396N1 were detected on all of them. We determined the exact chromosomal localization of these PACs by multistep hybridization with mouse painting probes specific for chromosomes 6, 8, 9, 10, 11, and 15. The localization of both PACs to mouse chromosome 8 was demonstrated by two-color FISH with the PAC and chromosome 8-specific painting probes (Fig. 4a). We also determined the localization of the PAC probe at 8C1 according to 4,6-diamino-2-phenylindole banding (Fig. 4b).
Figure 4.
FISH of PAC 396N1 to mouse metaphase spreads. (a) The green arrow indicates hybridization of PAC 396N1 to mouse chromosome 8 (red arrowhead). (b) PAC 396N1 was localized to chromosome 8C1 by using an inverted 4,6-diamino-2-phenylindole banding pattern.
The Human and Mouse LARGE Proteins Reveal Predicted Coiled-Coil Domains Absent in Other GTs.
Search with blastp and the human and mouse LARGE proteins as query revealed similarity to several GTs. The highest score was noted between the human LARGE protein (amino acids 470–742) and the human i-β-1,3-N-acetylglucosaminyltransferase (amino acids 91–408, acc. AF029893) (Fig. 3). This C-terminal part of the human i-β-1,3-N-acetylglucosaminyltransferase corresponds to its globular, catalytic domain located within the Golgi lumen (5, 36). We therefore can assume that this part of human and mouse LARGE proteins contain a C-terminal catalytic domain because it is the case for other GTs. We further searched the sequence of LARGE proteins for other features that are typical for GTs. psort and tmpred predicted a signal peptide (residues 1–24) and a transmembrane domain (residues 20–28) in the LARGE protein, respectively (Fig. 3). These features are likely to represent the targeting/membrane-anchoring domain found in all GTs (5). As the LARGE proteins have been predicted to contain 756 amino acids, the Golgi-luminal stem region separating the putative catalytic and transmembrane domains is longer (by ≈200–250 amino acids) as compared with other family members (5, 36).
We investigated whether other protein domains could be assigned to the stem region of the LARGE. Using coils and multicoil, we recognized two putative coiled-coil domains (residues 55–90 and 422–441). Analysis of human and mouse proteins displayed identical results. The first domain was predicted with the 1.0 probability score by using coils (MTIDK matrix, with or without weighting and using window 28-option). These results were confirmed by multicoil, which resulted in a 0.973 probability (using window 28-option, 0.921 dimer-probability, and low trimer-probability). The second predicted domain, between amino acids 422–441, only was predicted by using coils with the probability of 0.921 (window 14-option). Coiled-coil domains have been characterized in many proteins. These domains form stable, rod-like structures that mediate protein–protein interactions via formation of two or three α-helices coiled around each other. In a similar way as described above, we analyzed multiple GTs for presence of coiled-coil domains and obtained negative results (e.g., acc. AF029893, X77922, L43494, AF038660, AB003478, U17894, U41514, M97347, and D13789).
DISCUSSION
We report here a novel human gene of extensive genomic size covering a minimum of 664 kb. To date, LARGE is the fifth-largest gene in the human genome, after the dystrophin (2.3 Mbp), DCC (1.4 Mbp), GRM8 (1 Mbp), and utrophin (900 kb) genes [Online Mendelian Inheritance in Man database (http://www.ncbi.nlm.nih.gov/Omim)]. The LARGE gene is composed of 16 exons (4,326 bp cDNA) and has an exon content of <0.66%, which is similar to the exon content of the dystrophin gene (0.6%). The chromosomal segment of 22q containing the LARGE gene is apparently poor in genes. At present, the most efficient method of positional cloning is to exploit the information from the database of expressed sequence tags. However, we obtained no help from scanning this database. This was attributable to the size of the LARGE gene and its position in relation to the area deleted in tumors. We were primarily interested in identifying genes from the deletions. However, this part contains only the 5′-end of LARGE, and, at the time of cloning, no corresponding expressed sequence tags were available.
Chromosomal localization of the mouse Large gene reveals a conservation of synteny between human chromosome 22q12.3-q13.1 and mouse chromosome 8C1. We characterized several additional human genes located at the telomere of 22q as compared with the location of LARGE, e.g., the human ortholog of the chicken Tom1 (target of myb 1) gene. The mouse ortholog of the chicken Tom1 gene also is localized on mouse chromosome 8C1 (E.S., D. Kedra, M.K.-A., A.-C.S.-N., I.F., J. Jacobs, Y. Fu, H.-Q. Pan, B. Roe, S.I., and J.P.D., unpublished work). Seven syntenic groups between 22q and the mouse genome have been reported involving mouse chromosomes 5, 10, 11, 15, and 16 (37). As compared with human chromosome 21, which is of similar size, it is intriguing that human chromosome 22 is divided into many distinct syntenic groups in the mouse genome. The expression pattern of the human and mouse LARGE orthologs is similar. Both genes are expressed ubiquitously, consistent with their function as housekeeping genes. These genes are also evolutionarily well conserved, as we detected an ortholog in C. elegans encoding a polypeptide 33% identical with the human protein.
On computer-assisted sequence analysis, the human and mouse LARGE proteins display features typical for known GTs. We detected the targeting/membrane-anchoring domain present at the N terminus. Furthermore, comparison with the other GTs predicts that the LARGE proteins contain the C-terminal catalytic domain. It is therefore likely that the LARGE protein is a member of the GT family and, more specifically, of the N-acetylglucosaminyltransferase subgroup. These predictions should be verified experimentally by, for example, determination of the normal subcellular localization of the LARGE proteins and examination of their substrate specificity. The human and mouse LARGE proteins display, however, additional features that are absent in the previously characterized GTs. The LARGE polypeptides are longer by ≈200 amino acids as compared with the longest of the known GTs (N-acetylgalactosaminyltransferases, 559 residues, acc. L07780 and U41514). Two coiled-coil domains were detected in the part of the human and mouse LARGE polypeptides between the targeting/membrane-anchoring and catalytic domains. Coiled-coil domains might suggest the existence of a protein(s) that dimerizes with LARGE.
Previously reported tumor-specific deletions (4) have directed us to the cloning of LARGE, which is encompassed by these aberrations. Without additional evidence showing the involvement of LARGE in tumorigenesis, it is not possible to suggest its role as a tumor suppressor. However, this gene may be an attractive object of further investigations with regard to cancer-related questions. Abnormal glycosylation of proteins and glycosphingolipids, especially gangliosides, have been suggested as a mechanism behind increased tumor formation and/or progression potential (6, 7). The screen for LARGE gene mutations in tumors should encompass not only searching for point mutations but also searching for intragenic deletions as well as possible changes in gene expression. Tumor no. 119, which is one of the cases behind this study, has shown a homozygous deletion in only one fraction of tumor cells. Thus, this genetic change is more likely to be related to tumor progression rather than to initial events of tumorigenesis. An analysis of the abnormal function of the LARGE gene should therefore take into account possible tumor heterogeneity.
Acknowledgments
We thank Drs. Darek Kedra and Hans Mehlin for bioinformatics support, Dr. Ulf Eriksson for the mouse embryo cDNA library, Dr. Pam Fredman for critical review of the manuscript, and Mia Nilsson, Joannes Jacobs, Maria Lundin, and Michelle Ekman for excellent technical assistance. This work is supported by the Swedish Cancer Foundation, the Swedish Medical Research Council, the Berth von Kantzow Fond, the Cancer Society in Stockholm, the Karolinska Hospital, and the Karolinska Institutet. Work at the Sanger Centre is supported by the Wellcome Trust. Mapping and sequencing data presented in the “sequenced contig” from Fig. 1 were produced by the Chromosome 22 Mapping and Sequencing groups at the Sanger Centre.
ABBREVIATIONS
- LARGE
human like-acetylglucosaminyltransferase
- FISH
fluorescent in situ hybridization
- acc.
European Molecular Biology Laboratory/GenBank accession number
- GT
glycosyltransferase
- kb
kilobase
- PAC
P1-derived artificial chromosome
Footnotes
References
- 1.Dumanski J P. Neuropathol Appl Neurobiol. 1996;22:412–417. doi: 10.1111/j.1365-2990.1996.tb00912.x. [DOI] [PubMed] [Google Scholar]
- 2.Dumanski J P, Carlbom E, Collins V P, Nordenskjöld M. Proc Natl Acad Sci USA. 1987;84:9275–9279. doi: 10.1073/pnas.84.24.9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seizinger B R, de la Monte S, Atkins L, Gusella J F, Martuza R L. Proc Natl Acad Sci USA. 1987;84:5419–5423. doi: 10.1073/pnas.84.15.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruttledge M H, Xie Y-G, Han F-Y, Peyrard M, Collins V P, Nordenskjöld M, Dumanski J P. Genes Chromosomes Cancer. 1994;10:122–130. doi: 10.1002/gcc.2870100207. [DOI] [PubMed] [Google Scholar]
- 5.Fukuda M, Hindsgaul O. In: Molecular Glycobiology. Hames B D, Glover D M, editors. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 6.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakomori S. Cancer Res. 1996;56:5309–5318. [PubMed] [Google Scholar]
- 8.Yates A J, Saqr H E, Van Brocklyn J. J Neurooncol. 1995;24:65–73. doi: 10.1007/BF01052661. [DOI] [PubMed] [Google Scholar]
- 9.Helling F, Shang A, Calves M, Zhang S, Ren S, Yu R K, Oettgen H F, Livingston P O. Cancer Res. 1994;54:197–203. [PubMed] [Google Scholar]
- 10.Foon K A, Sen G, Hutchins L, Kashala O L, Baral R, Banerjee M, Chakraborty M, Garrison J, Reisfeld R A, Bhattacharya-Chatterjee M. Clin Cancer Res. 1998;4:1117–1124. [PubMed] [Google Scholar]
- 11.Davidsson P, Fredman P, Collins V P, von H H, Mansson J E, Svennerholm L. J Neurochem. 1989;53:705–709. doi: 10.1111/j.1471-4159.1989.tb11761.x. [DOI] [PubMed] [Google Scholar]
- 12.Fredman P, Dumanski J P, Davidsson P, Svennerholm L, Collins V P. J Neurochem. 1990;55:1838–1840. doi: 10.1111/j.1471-4159.1990.tb05765.x. [DOI] [PubMed] [Google Scholar]
- 13.Sanson M, Richard S, Delattre O, Poliwka M, Mikol J, Philippon J, Thomas G. Int J Cancer. 1992;50:391–394. doi: 10.1002/ijc.2910500311. [DOI] [PubMed] [Google Scholar]
- 14.Xie Y-G, Han F-Y, Peyrard M, Ruttledge M H, Fransson I, DeJong P, Collins J, Dunham I, Nordenskjöld M, Dumanski J P. Hum Mol Genet. 1993;2:1361–1368. doi: 10.1093/hmg/2.9.1361. [DOI] [PubMed] [Google Scholar]
- 15.Church D M, Stotler C J, Rutter J L, Murrell J R, Trofatter J A, Buckler A J. Nat Genet. 1994;6:98–105. doi: 10.1038/ng0194-98. [DOI] [PubMed] [Google Scholar]
- 16.Peyrard M, Fransson I, Xie Y-G, Han F-Y, Ruttledge M H, Swahn S, Collins J E, Dunham I, Collins V P, Dumanski J P. Hum Mol Genet. 1994;3:1393–1399. doi: 10.1093/hmg/3.8.1393. [DOI] [PubMed] [Google Scholar]
- 17.Uberbacher E C, Mural R J. Proc Natl Acad Sci USA. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schowalter D B, Sommer S. Anal Biochem. 1989;177:90–94. doi: 10.1016/0003-2697(89)90019-5. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg A P, Vogelstein B. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A laboratory manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Kedra D, Pan H, Seroussi E, Fransson I, Guilbaud C, Collins J E, Dunham I, Blennow E, Roe B, Piehl F, et al. Hum Genet. 1998;103:131–141. doi: 10.1007/s004390050795. [DOI] [PubMed] [Google Scholar]
- 22.Staden R. In: The Staden Package. Griffin A M, Griffin H G, editors. Vol. 25. Totawa, NJ: Humana; 1994. pp. 9–170. [Google Scholar]
- 23.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupas A, Van D M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 25.Wolf E, Kim P S, Berger B. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bucher P, Bairoch A. Ismb. 1994;2:53–61. [PubMed] [Google Scholar]
- 27.Deschenes R J, Lorenz L J, Haun R S, Roos B A, Collier K J, Dixon J E. Proc Natl Acad Sci USA. 1984;81:726–730. doi: 10.1073/pnas.81.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandberg-Nordqvist A-C, von Holst H, Holmin S, Sara V R, Bellander B M, Schalling M. Brain Res Mol Brain Res. 1996;38:285–293. doi: 10.1016/0169-328x(95)00346-t. [DOI] [PubMed] [Google Scholar]
- 29.Fedorova L, Kost-Alimova M, Gizatullin R Z, Alimov A, Zabarovska V I, Szeles A, Protopopov A I, Vorobieva N V, Kashuba V I, Klein G, et al. Eur J Hum Genet. 1997;5:110–116. [PubMed] [Google Scholar]
- 30.Collins J E, Cole C G, Smink L J, Garrett C L, Leversha M A, Soderlund C A, Maslen G L, Everett L A, Rice K M, Coffey A J, et al. Nature (London) 1995;377:367–379. doi: 10.1038/377367a0. [DOI] [PubMed] [Google Scholar]
- 31.Dumanski J P, Geurts van Kessel A H, Ruttledge M, Wladis A, Sugawa N, Collins V P, Nordenskjöld M. Hum Genet. 1990;84:219–222. doi: 10.1007/BF00200562. [DOI] [PubMed] [Google Scholar]
- 32.Ruttledge M H, Xie Y-G, Han F-Y, Giovannini M, Janson M, Fransson I, Werelius B, Delattre O, Thomas G, Evans G, et al. Genomics. 1994;19:52–59. doi: 10.1006/geno.1994.1012. [DOI] [PubMed] [Google Scholar]
- 33.Kedra D, Peyrard M, Fransson I, Collins J E, Dunham I, Roe B A, Dumanski J P. Hum Mol Genet. 1996;5:625–632. doi: 10.1093/hmg/5.5.625. [DOI] [PubMed] [Google Scholar]
- 34.Lopez R, Larsen F, Prydz H. Genomics. 1994;24:133–136. doi: 10.1006/geno.1994.1590. [DOI] [PubMed] [Google Scholar]
- 35.Gardiner-Garden M, Frommer M. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 36.Paulson J C, Colley K J. J Biol Chem. 1989;264:17615–17618. [PubMed] [Google Scholar]
- 37.Debry R W, Seldin M F. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]