Abstract
Signal transduction by two-component systems involves phosphorylation and thereby activation of the response regulator by the cognate histidine kinase. Bifunctional histidine kinases have two opposing activities: depending on the environmental stimuli they either promote phosphorylation or stimulate the rapid dephosphorylation of the response regulator. To determine the mechanism of this switch, we analyzed the domain organization of the bifunctional histidine kinase NtrB. Based on sequence alignments with other histidine kinases and a deletion analysis, we defined three separate subdomains of the transmitter module, the H domain (amino acids 123–221), the N domain (amino acids 221–269), and the G domain (amino acids 269–349). The transmitter module, when separately expressed, exhibited a constitutive positive phenotype. In contrast, in the absence of the G domain, the H domain exhibits a constitutive negative phenotype. This negative regulatory activity of the H domain is inhibited by the G domain. The G domain could be physically uncoupled; when coexpressed with the H–N fragment, the constitutive positive phenotype of the transmitter was restored. We demonstrate, in vitro, that the constitutive negative phenotype of the fragments lacking the G domain is caused by stimulation of dephosphorylation of the response regulator NtrC-P. Based on our analysis, we suggest that the function of the sensor domain is to control the interaction of the H and G domains. If these subdomains interact, NtrB acts as a positive regulator; if they cannot interact, NtrB acts as a negative regulator.
The protein family of the two-component systems allows the cell to adapt to a variety of different environmental conditions. Members of this family sense different environmental stimuli and, in response, change the expression of target genes or the activity of a target enzyme (1, 2). The basic mechanism of signal transmission by two-component systems is a phosphorylation cascade that involves aspartate and histidine residues (3–5). A classical two-component system consists of two proteins, the histidine kinase and the response regulator. Two-component systems consist of conserved modules involved in reactions common to all two-component systems (the phosphotransfer reactions) and variable domains involved in reactions unique for a particular two-component system. The first reaction in the signaling cascade is the autophosphorylation of the highly conserved histidine of the transmitter module. This reaction is under the control of the sensor domain dependent on the conditions of the environment. The phosphate group is subsequently transferred to an aspartate residue of the conserved N-terminal receiver module of the response regulator. This induces activation of the output domain and thus activation of the response reactions.
Adaptation to changing environmental conditions requires that the adaptive response be efficient in both directions; that is, the response reactions cannot only be switched on on demand, but also must be switched off immediately under different conditions. To some extent, the instability of the aspartylphosphate (the “autophosphatase activity”) of the response regulators contributes to a transient signal (4, 6). However, for most two-component systems, additional proteins have been identified that stimulate the fast dephosphorylation of the response regulator under conditions where the system has to be shut off. In many instances, this negative regulatory function resides in the histidine kinase itself. Depending on the environmental stimulus, these bifunctional histidine kinases either autophosphorylate to promote the phosphorylation of the response regulator or they adopt a different conformation to stimulate the dephosphorylation of the response regulator (3, 6, 7).
One example of a bifunctional histidine kinase is NtrB, the histidine kinase for nitrogen regulation (8–10). Under nitrogen-limited conditions, NtrB promotes the phosphorylation of the response regulator NtrC. Phosphorylated NtrC is active as a transcriptional activator for the Ntr regulon. Under nitrogen-excess conditions, NtrB has the opposite activity and promotes the dephosphorylation and thus inactivation of NtrC-P. Genetic and biochemical data have established that the PII protein (product of glnB) and uridylyltransferase (UTase, product of glnD) are involved in signaling the nitrogen status of the cell to NtrB (8, 11, 12). UTase uridylylates PII under nitrogen-limited conditions and deuridylylates PII under nitrogen-excess conditions. It is believed that interaction of deuridylylated PII with the sensor domain of NtrB induces the switch to the negative regulatory function in NtrB, leading to the dephosphorylation of NtrC-P (12). In the absence of PII or in the presence of PII-UMP, NtrB acts as an autokinase.
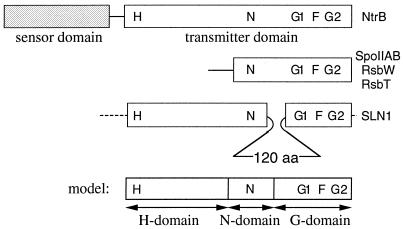
To determine how the bifunctional histidine kinase switches between its two opposing activities, we analyzed the domain organization of the transmitter module of NtrB. This module contains all conserved motifs characteristic for a transmitter of two-component systems (1)—that is, the H motif carrying the active-site histidine, the N motif of unknown function, and the glycine-rich G1 and G2 motifs believed to be important for ATP-binding (Fig. 1). We found that the transmitter of NtrB can be divided into three subdomains [designated the H, N, and G domains (Fig. 1)] and provide evidence that the negative regulatory function of NtrB resides in the H domain and that the switch between the positive and negative regulatory activities is based on a change in the interaction between the H and the G domain.
Figure 1.
Domain organization of the transmitter module of SLN1, SpoIIAB and homologues, and NtrB. Conserved motifs (1) are indicated. The deduced model for the transmitter of NtrB is shown at the bottom.
MATERIALS AND METHODS
Strains and Plasmids.
RB9132 (ΔntrB) is derived from YMC10 (thi endA1 hsdR17 ΔlacU169 hutCk supE44) (8). Plasmids pB1, pBSHN1, pBHN1, and pBHNG1 are all derivatives of the vector pTrc99B (13). By using PCR with pgln62 as the template, ntrB fragments were amplified such that they were flanked by an NcoI restriction site at the 5′ end and a stop codon followed by an EcoRI restriction site at the 3′ end. The sequence of the primer for fragments starting with Met-1 was 5′-CGCTAGCCATGGCAACAGGCACGC-3′ and starting at Val-81 was 5′-CGGGATCCCATGGTGACGCTGGTCATCG-3′; for fragments ending with Gln-269, 5′-GTGAATTCTTATTGAAACGCGGTGCGGG-3′ and for fragments ending with Lys-349, 5′-GTGAATTCGTTGCATAAACGTCACC-3′. Fragments were digested with NcoI and EcoRI and ligated to the large NcoI/EcoRI fragment of pTrc99B.
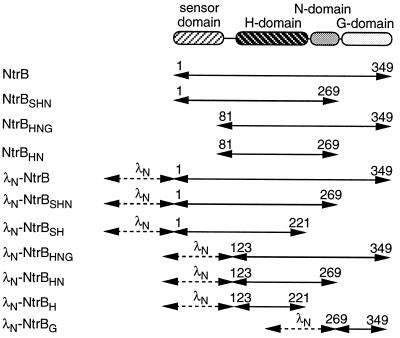
Plasmid pJH391 (14) was used for the construction of fusions between DNA encoding the N-terminal domain of λ repressor (base pairs 1–396 of λcI ind1) and fragments of ntrB. By using PCR with pgln62 as the template, ntrB fragments were amplified such that they were flanked by an XhoI restriction site at the 5′ end and a stop codon followed by an BamHI restriction site at the 3′ end. The sequences of the primers were, for ntrB fragments starting at Met-1, 5′-ACGTCTCGAGAATGGCAACAGGCACGCAGCC-3′; at Gln-123, 5′-ACGTCTCGAGACAGCACGCCCAGCAGGTTGC-3′; at Gln-269, 5′-ACGTCTCGAGACAACTGACCTTACACGGCA-3′; for fragments ending at Ile-221, 5′-GTGGATCCTTAAATCAACCGCACGTTGTCC-3′; at Gln-269, 5′-GTGGATCCTCATTGAAACGCGGTGCGGGT-3′; and at Lys-349, 5′-GTGGATCCTTATTTCCTGATAGGCAGGTA-3′. Amplified fragments were cut with XhoI and BamHI and ligated to the large SalI/BamHI fragment of pJH391. The inserts carried the following fragments of ntrB: entire ntrB (base pairs 1–1050) in pFB1; ntrB (base pairs 1–663) encoding amino acids 1–221 of NtrB in pFBSH1; ntrB (base pairs 1–807) encoding amino acids 1–269 of NtrB in pFBSHN1; ntrB (base pairs 367–663) encoding amino acids 123–221 of NtrB in pFBH1; ntrB (base pairs 367–807) encoding amino acids 123–269 of NtrB in pFBHN1; ntrB (base pairs 367-1050) encoding amino acids 123–349 of NtrB in pFBHNG1; ntrB (base pairs 805-1050) encoding amino acids 269–349 of NtrB in pFBG1. For the in vivo complementation analysis in Fig. 7, a NheI/EcoRI fragment of pFBHN1 containing the coding region of λN-NtrBHN (including the lacUV5 promoter) was blunt-ended by using the Klenow Fragment of DNA polymerase I (Promega) and cloned into the ScaI site of pIq. pIq carries lacIq and is a derivative of pACYC177. pFBHNG287 was constructed like plasmid pFBHNG1 by using pgln62–287 as a template. pgln62–287 carries a mutation in ntrB (A860G), causing the replacement of Asp-287 by a glycine residue.
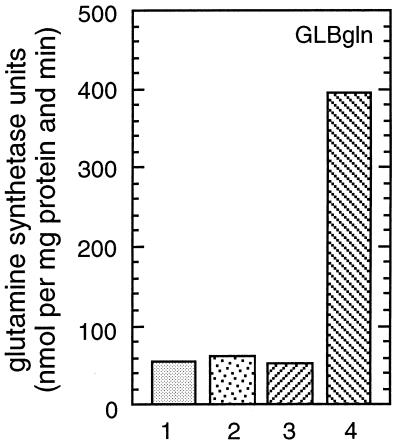
Figure 7.
Coexpression of λN-NtrBHN and λN-NtrBG. RB9132 carrying the following plasmids was grown in GLBgln containing 10 μM IPTG. Column 1, pIq and pJH391 (vectors); column 2, pIq and pFBG1 (λN-NtrBG); column 3, pIq-HN (λN-NtrBHN) and pJH391; and column 4, pIq-HN and pFBG1 (λN-NtrBHN and λN-NtrBG).
The plasmids encoding the Trx–NtrB fusion proteins are derivatives of pET32a (Novagen). ntrB fragments encoding the different NtrB domains were amplified by using PCR and ligated to the NcoI/BamHI-digested large fragment of pET32a, resulting in the plasmids pET-SHN1, which encodes TrxSHN (amino acids 125–349) and pET-HN1, which encodes TrxHN (amino acids 125–269). The primer for the fusion point at Met1 was the same as for amplification of entire ntrB in pB1. For the fusion point at Ala-125 we used 5′-AGCTACCCATGGCCCAGCAGGTTGCTGCC-3′. For fragments ending at Gln-269, we used the primer for amplification of the ntrB fragment in pFBSHN1. The plasmid pET-HN139 encoding TrxHNH139N was constructed by using the QuikChange kit (Stratagene) and the mutagenesis primer 5′-AGTGCGCGGCCTGGCCAATGAGATTAAAAATCC-3′. All DNA fragments obtained with PCR were verified by using DNA sequencing.
Western Blotting.
Cells were grown at 30°C in GLBgln (8) supplemented with ampicillin, kanamycin, and isopropyl β-d-thiogalactoside (IPTG) to an OD of 0.5 at 578 nm and harvested by using centrifugation. Cell lysis was performed by using SDS and chloroform. Proteins were separated by SDS/PAGE and transferred onto a poly(vinylidene) difluoride (PVDF) membrane (Pall). Fifty micrograms of total cellular protein was applied per lane. NtrB and fragments were detected by using purified rabbit polyclonal antibodies against NtrB. A goat anti-rabbit IgG alkaline phosphatase conjugate and a chromogenic substrate (Tropix, Bedford, MA) were used for visualization of NtrB and fragments therefrom.
Enzyme Assays.
For glutamine synthetase assays cells were grown at 30°C to an OD at 578 nm of 0.5 and analyzed as described (15).
Protein Purification.
The Trx–NtrB fusion proteins were purified from 1.5 liters of IPTG-induced culture. Cells were harvested by using centrifugation and resuspended in buffer A (50 mM Tris, pH 7.5/50 mM KCl/2 mM MgCl2/20% glycerol). Cells were disrupted in a French pressure cell and the extract was clarified by using centrifugation. The supernatant was incubated on ice with 8 ml of Ni2+-agarose (Qiagen) for 30 min. The resin was washed with 1 liter of buffer A plus 20 mM imidazole and packed in a column. Protein was eluted with buffer A plus 250 mM imidazole and dialyzed against buffer A plus 0.1 mM EDTA. NtrB (16) and NtrC (17) were purified as described.
Dephosphorylation Assays.
Radioactively labeled NtrC-P was prepared as described (4). The dephosphorylation activity of the NtrB fragments on purified 32P-labeled NtrC-P was analyzed in buffer B (50 mM Tris⋅HCl, pH 7.5/50 mM KCl/10 mM MgCl2/10% glycerol) at 37°C as described (3, 4).
RESULTS
The Transmitter Module of Histidine Kinases: Indications for Subdomains.
To identify the boundaries for potential subdomains in the transmitter module of NtrB, we aligned its sequence with other histidine kinases, focusing on SLN1, a histidine kinase in yeast (18) and the serine protein kinases SpoIIAB, RsbW, and RsbT (19, 20) from Bacillus subtilis. In SLN1, the transmitter module is interrupted by a 120-aa insert positioned between the N and the G1 motifs (indicated by the loop in Fig. 1). SpoIIAB and its homologues RsbW and RsbT are homologous to the C-terminal part of the transmitter module and contain the N, G1, and G2 motifs but lack the region containing the active-site histidine (Fig. 1). This indicates that the C-terminal and N-terminal parts of the transmitter module may be able to fold independently of each other. Moreover, the region containing the N motif is part of SpoIIAB (which lacks entirely the region containing the H-motif) but also is separated from the region containing the G1 and G2 motif by the 120-aa insert in SLN1. Therefore, this segment may constitute a separate subdomain.
Based on this analysis, our strategy was to divide the transmitter module of NtrB into three subdomains (Fig. 1) designated the H domain (carrying the active site histidine), the N domain (carrying the conserved N motif), and the G domain (carrying the ATP-binding motifs G1 and G2). To test this model, we performed a deletion analysis of NtrB.
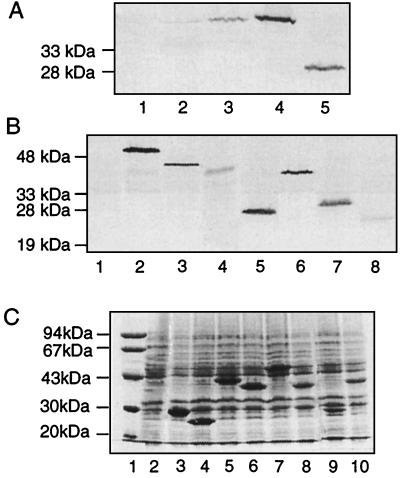
We cloned different fragments of NtrB (NtrBHNG, NtrBSHN, and NtrBHN; Fig. 2) under the control of the ptrc promoter. The plasmids were derived from pBR322 and carried in addition lacIq. Expression of these fragments was analyzed under different inducing conditions. On Coomassie-stained SDS/PAGE gels, expression of NtrB and fragments was not detectable after induction with 1 mM IPTG (not shown). Using Western analysis under low inducing conditions (0–10 μM IPTG), NtrB was clearly detectable when carried on pB1 under the control of the ptrc promoter (Fig. 3A, lane 4). Less NtrB was detected when ntrB was carried on plasmid pgln62 under the control of its own promoter (Fig. 3A, lane 3). Expression was lowest for chromosomal ntrB even under nitrogen-limited conditions (Fig. 3A, lane 2). Fragments of NtrB were much less stable. Only NtrBSHN was detectable on induction with 10 μM IPTG (Fig. 3A, lane 5). We could not detect the transmitter domain (NtrBHNG) or NtrBHN under low inducing conditions (not shown).
Figure 2.
Schematic representation of the derivatives of NtrB used in this study. The numbers indicate the boundaries of the NtrB fragments.
Figure 3.
Expression of NtrB and derivatives. (A) Total proteins of cells harboring the indicated proteins were analyzed by using Western blot analysis. Lane 1, RB9132 (ΔntrB) carrying pTrc99B (vector); lane 2, YMC10 (NtrB+) grown in Ggln; lane 3, RB9132 carrying pgln62 (NtrB); lane 4, RB9132 carrying pB1 (NtrB); and lane 5, RB9132 carrying pBSHN1 (NtrBSHN). With the exception of YMC10, cells were grown under nitrogen-excess conditions (GLBgln). Expression of NtrBSHN was induced with 10 μM IPTG (lane 5); all other cells were grown in the absence of IPTG. (B) Western blot analysis of hybrid proteins containing the N-terminal domain of λ repressor. Cells (RB9132) were grown under nitrogen-excess conditions (GLBgln). Expression was induced with 10 μM IPTG except for λN-NtrBSH (50 μM IPTG). Lane 1, pJH391; lane 2, pFB1 (λN-NtrB); lane 3, pFBSHN1 (λN-NtrBSHN); lane 4, pFBSH1 (λN-NtrBSH); lane 5, pFBS1 (λN-NtrBS); lane 6, pFBHNG1 (λN-NtrBHNG); lane 7, pFBHN1 (λN-NtrBHN); and lane 8, pFBG1 (λN-NtrBG). (C) Overexpression of hybrid proteins containing the N-terminal domain of λ repressor. Crude cell extracts of RB9132 carrying the indicated plasmids were subjected to electrophoresis on an SDS/PAGE gel (12%) and visualized by staining with Coomassie blue. Expression was induced by the addition of 1 mM IPTG. Lane 1, molecular weight markers; lane 2, pJH391 (vector); lane 3, pFBHN1 (λN-NtrBHN); lane 4, pFBG1 (λN-NtrBG); lane 5, pFBSHN1 (λN-NtrBSHN); lane 6, pFBHNG1 (λN-NtrBHNG); lane 7, pFB1 (λN-NtrB); lane 8, pFBSH1 (λN-NtrBSH); and lane 9, pFBH1 (λN-NtrBH); and lane 10, pFBHNG287 (λN-NtrBHNG287).
To stabilize the NtrB fragments, we fused them to the N-terminal DNA-binding domain of λ repressor (Fig. 2B). This domain also serves as a reporter domain for dimerization in vivo (14). All fusion proteins were readily detected on Coomassie-stained polyacrylamide gels after overproduction in the presence of 1 mM IPTG (Fig. 3C) and migrated at their expected molecular weights. With the exception of λN–NtrBH (not shown) all fragments were detected on immunoblots when induced with 10 μM IPTG. (Fig. 3B).
The Transmitter Module of NtrB Is a Constitutive Positive Regulator.
To analyze the activity of the NtrB derivatives in vivo, we measured the amount of glutamine synthetase formed in an ntrB deletion strain carrying plasmids encoding the different NtrB derivatives. Expression of glnA, the structural gene for glutamine synthetase, strongly depends on the amount of NtrC phosphorylated in the cell and thus is a direct measure of the activity of the NtrB derivatives (8, 21, 22).
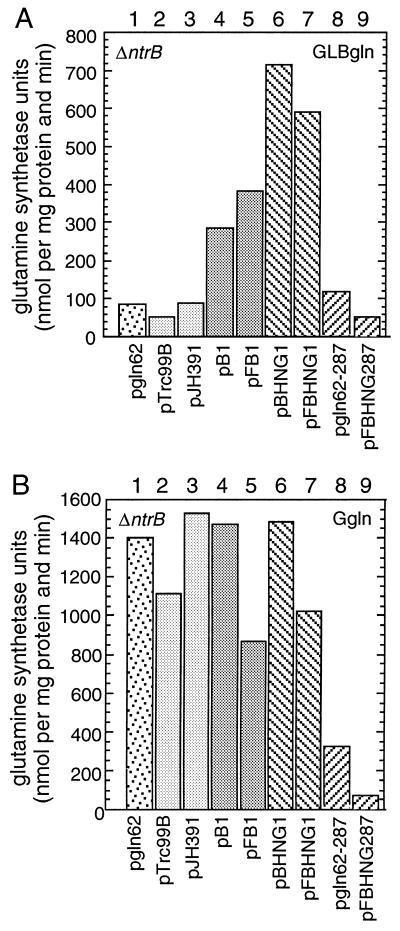
As shown in Fig. 4 constructs carrying the transmitter module but lacking the sensor domain (NtrBHNG and λN-NtrBHNG) were analyzed together with wild-type NtrB and λN-NtrB. Wild-type regulation of glutamine synthetase synthesis, i.e., low levels of glutamine synthetase under nitrogen-excess conditions (GLBgln) and high levels of glutamine synthetase under nitrogen-limited conditions (Ggln) were observed for RB9132 carrying ntrB on plasmid pgln62 (Fig. 4, lane 1). In accordance with previous results (8), apparent wild-type regulation also was observed in a strain carrying no functional NtrB (Fig. 4, lanes 2 and 3). It has been demonstrated that this is caused by the phosphorylation of NtrC by acetyl phosphate (23, 24). Acetyl phosphate accumulates under nitrogen-limited conditions but not under nitrogen-excess conditions in a strain lacking ntrB. In contrast, NtrB carried on pB1 and λN-NtrB carried on pFB1 lead to elevated levels of glutamine synthetase under nitrogen-excess conditions (Fig. 4, lanes 4 and 5). This is best explained by the rather high intracellular concentration of these proteins (Fig. 3). Presumably, the signal for nitrogen-excess conditions, PII, is titrated out by excess NtrB leading to the observed phenotype.
Figure 4.
Expression of glnA in RB9132 (ΔntrB) carrying NtrB derivatives containing the transmitter module. (A) Cells grown in GLBgln carried the following plasmids. Column 1, pgln62 (NtrB); column 2, pTrc99B (vector); column 3, pJH391 (vector); column 4, pB1 (NtrB); column 5, pFB1 (λN-NtrB); column 6, pBHNG1 (NtrBHNG); column 7, pFBHNG1 (λN-NtrBHNG); column 8, pgln62–287 (NtrBD287G); and column 9, pFBHNG287 (λN-NtrBHNG287). Cells containing pFB1, pFBHNG1 and pFBHNG287 (columns 5, 7 and 9) were grown in the presence of 10 μM IPTG, cells containing pBHNG1 (column 6) in the presence of 50 μM IPTG. (B) Columns as in A, but cells were grown in Ggln medium.
Glutamine synthetase expression was strongest in RB9132 containing the derivatives lacking the sensor domain, NtrBHNG or the fusion protein λN-NtrBHNG (Fig. 4, lanes 6 and 7). This phenotype was independent of the intracellular concentration of the transmitter because expression of λN-NtrBHNG was high (Fig. 3), whereas NtrBHNG was not detectable by Western analysis under the conditions of the assay. Our data are in accordance with previous results. For several histidine kinases, e.g., for PhoR, FixL, and NarX (25–27), it has been demonstrated that the transmitter module separately expressed acts as a constitutive positive regulator. Kamberov et al. (28) demonstrated in vitro that an NtrB derivative lacking the sensor domain behaves as a constitutive positive regulator and independent of PII. Accordingly, the constitutive positive phenotype of NtrBHNG and λN-NtrBHNG was as strong in a strain that carried (in addition a transposon in glnD) the structural gene for UTase (not shown).
To demonstrate that the transmitter module has retained the active site both for the positive and negative regulatory function, we analyzed a mutant of NtrB, NtrBD287G. In NtrBD287G, Asp-287 within the G1 motif is changed to a glycine residue leading to a constitutive negative phenotype (Fig. 4, lane 8). We introduced this mutation into λN-NtrBHNG. As shown in Fig. 4 (lane 9), λN-NtrBHNG287 was fully active as a constitutive negative regulator of glnA expression. Therefore the constitutive positive phenotype of the transmitter module is the result of the inability of this fragment to sense nitrogen-excess conditions rather than the loss of the active site for the negative regulatory function of NtrB. This demonstrates that in wild-type NtrB, the sensor domain is required to induce the negative regulatory function but not to induce the positive regulatory function of NtrB.
The Negative Regulatory Function of NtrB Resides in the H Domain.
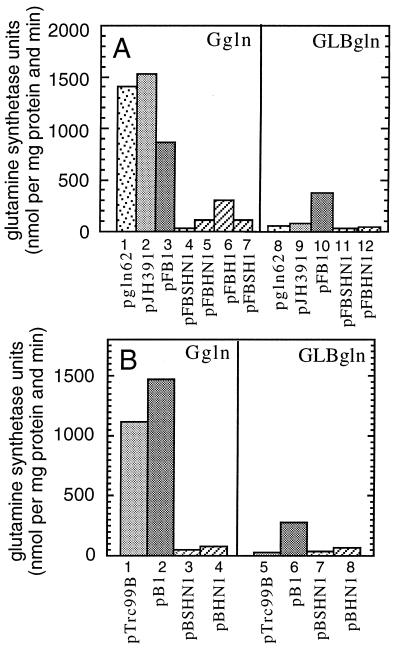
Next we examined the properties of the NtrB fragments lacking the G domain (Fig. 5). As shown in Fig. 5A, deletion of the G domain results in constitutive negative regulators of glnA expression (Fig. 5A, lanes 4–7, 11, and 12 and Fig. 5B, lanes 3, 4, 7, and 8). This is clearly seen when cells are grown in Ggln medium. Here, glutamine synthetase levels are high not only in strains carrying wild-type ntrB but also in strains lacking a functional ntrB because of the slow phosphorylation of NtrC by acetyl phosphate (Fig. 5A, lanes 1–3 and Fig. 5B, lanes 1 and 2). In contrast, glutamine synthetase levels are low in strains carrying NtrBSHN, NtrBHN, λN-NtrBSHN, or λN-NtrBHN. An analysis of the derivatives in a glnB deletion strain revealed that the phenotype was independent of the presence or absence of the PII protein (not shown), which is expected because the constitutive negative phenotype does not require the presence of the sensor domain. Moreover, the NtrB derivatives also lacking the N domain (λN-NtrBH and λN-NtrBSH; Fig. 5A, lanes 6 and 7) retained the constitutive negative phenotype. Therefore, all determinants necessary for the negative regulatory function of NtrB are located within the H domain. Obviously, the sensor domain is only required to induce the phosphatase activity in the H domain if the transmitter module is intact and carries the G domain.
Figure 5.
Expression of glnA in RB9132 (ΔntrB) carrying different NtrB derivatives lacking the G domain. Cells were grown in Ggln or GLBgln as indicated. (A) Analysis of NtrB-derivatives fused to the N-terminal domain of λ repressor. Cells contained the following plasmids. Columns 1 and 8, pgln62 (NtrB); columns 2 and 9, pJH391 (vector); columns 3 and 10, pFB1 (λN-NtrB); columns 4 and 11, pFBSHN1 (λN-NtrBSHN); columns 5 and 12, pFBHN1 (λN-NtrBHN); column 6, pFBH1 (λN-NtrBH); column 7, pFBSH1 (λN-NtrBSH). (B) Analysis of NtrB derivatives cloned under the control of the trc promoter. Cells contained the following plasmids. Columns 1 and 5, pTrc99B (vector); columns 2 and 6, pB1 (NtrB); columns 3 and 7, pBSHN1 (NtrBSHN); columns 4 and 8, pBHN1 (NtrBHN). All cultures were grown in the presence of 10 μM IPTG with the exception of cells carrying pgln62 (no IPTG), cells carrying pFBSH1 (50 μM IPTG), and cells carrying pFBH1 (100 μM IPTG).
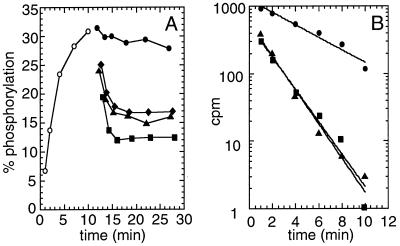
To verify that the constitutive negative phenotype of the NtrB derivatives lacking the G domain is the result of the ability to stimulate the fast dephosphorylation of NtrC-P we purified fusion proteins between TrxA (thioredoxin) and the S–H–N or H–N fragments of NtrB. The fusion proteins carried, in addition, six histidine residues for easy purification using Ni2+ chelate affinity chromatography. In TrxHNH139N the active-site histidine was changed to an asparagine residue. This mutation converts NtrB into a constitutive negative regulator able to stimulate the fast dephosphorylation of NtrC-P in vitro (28, 29). In Fig. 6A, NtrC was phosphorylated with wild-type NtrB until steady-state levels were reached. Addition of TrxSHN, TrxHNH139N, or TrxHN to the reaction mixture shifted the equilibrium to a lower steady-state level, consistent with a faster rate of dephosphorylation of NtrC-P. To confirm this result, we analyzed the rate of dephosphorylation of NtrC-P in the presence of TrxSHN or TrxHNH139N quantitatively by using purified radioactively labeled NtrC-32P (Fig. 6B). Consistent with earlier results, the apparent first-order rate constant for dephosphorylation of NtrC-P in the absence of these proteins was 0.24 min−1 (t1/2 = 3.25 min). Addition of TrxSHN or TrxHNH139N clearly stimulated the rate of dephosphorylation, with t1/2 = 1.25 min for TrxSHN and t1/2 = 1.2 min for TrxHNH139N.
Figure 6.
Dephosphorylation of NtrC-P by NtrB fragments lacking the G domain. (A) NtrC (2 μM) was incubated with 200 nM NtrB in buffer B. After 10 min, the reaction product was split and to 60 μl of product, 6 μl of the following samples were added: buffer B containing 1 mg/ml BSA (•), TrxHN (♦), TrxSHN (▴), and TrxHNH139N (■) to a final concentration of 1 μM. Samples (10 μl) were withdrawn at the indicated times, spotted on glass fiber filters and, analyzed as described (3, 4). (B) Dephosphorylation of purified NtrC-32P in the presence of buffer B containing 1 mg/ml BSA (•), 1 μM TrxSHN (▴), and 1 μM TrxHNH139N (■).
Coexpression of the H–N Fragment and the G Domain.
To probe the activity of the G domain, we coexpressed λN-NtrBG and λN-NtrBHN in strain RB9132. For this experiment, λN-NtrBG was expressed from a pBR322-derived plasmid and λN-NtrBHN was expressed from a pACYC-derived vector that also carried lacIq. Glutamine synthetase levels were analyzed under nitrogen-excess conditions. As shown in Fig. 7, the level of glutamine synthetase increased ≈8-fold under nitrogen-excess conditions, that is, the negative regulatory activity of the H domain was inhibited in the presence of the G domain, and the phenotype of the transmitter module was restored. We conclude that conditions under which the H–N fragment and the G domain are able to interact induce the positive regulatory function of NtrB. In contrast, conditions in which the H domain is “free” result in the rapid dephosphorylation of NtrC-P. Apparently the G domain is able to fold independently into an active domain and phosphorylate the histidine residue of the H domain in trans, and the phosphorylated H–N fragment is able to pass its phosphate group on to NtrC.
DISCUSSION
In this work we show that the transmitter module of NtrB consists of separate subdomains. We show that the mode of interaction between these subdomains determines whether the transmitter acts as a constitutive positive or as a constitutive negative regulator. The ultimate target for regulation of the two opposing activities is clearly the H domain (amino acids 123–221), which constitutes the N-terminal subdomain of the transmitter module. This domain carries both the active site for the positive regulatory function (the conserved histidine for the phosphotransfer to NtrC) and the determinants for the negative regulatory function of NtrB, as demonstrated by the constitutive negative phenotype of λN-NtrBH.
The H domain acts as a constitutive negative regulator in the absence of the G domain. We show in vitro, that this is in fact the result of the ability to stimulate the fast dephosphorylation of NtrC-P, as has been shown for wild-type NtrB (3, 28). As for wild-type NtrB (28, 29), the active-site histidine was not required for dephosphorylation of NtrC-P because TrxHNH139N was active in stimulating the rapid dephosphorylation of NtrC-P. In contrast to wild-type NtrB (6), the fragments lacking the G domain did not require PII and ATP to stimulate the rapid dephosphorylation of NtrC-P (not shown).
The negative regulatory activity of the H domain was inhibited by the G domain (Fig. 7). We infer that the H domain acts as a negative regulator under conditions in which it cannot interact with the G domain, that is, when not phosphorylated at the active-site histidine. Interaction with the G domain—presumably phosphorylation of the active-site histidine—induces the positive regulatory activity of the H domain.
The function of the N domain (amino acids 222–269) for regulation of NtrB is still ambiguous. This domain appears to stabilize the H domain (Fig. 3B) and possibly represents a hinge domain necessary for the proper alignment of the H and G domains.
Previously it has been shown that phosphorylation of the active-site histidine occurs in trans within a dimer (30–32), where the histidine of one subunit is phosphorylated by the other subunit. Therefore the function of the sensor domain could be to change the relative orientation of the transmitter domains toward each other such that in one conformation, the G domain of one subunit would be able to interact with the H domain of the other subunit, leading to phosphorylation of the active-site histidine. In the other conformation, the interaction between G and H domains would be prevented, thereby inducing the negative regulatory activity of the H domain. Such a mechanism is reminiscent of the mechanism of signal transduction suggested for Tar (33, 34). It is believed that a conformational change of a coiled-coil structure within the linker region affects the relative orientation of the cytoplasmic domains of Tar and thereby affects the activity of these domains. Interestingly, chimeric sensor proteins consisting of the receptor domain of the chemoreceptors Tar or Trg and the transmitter module of the histidine kinase EnvZ are able to transmit signals in response to chemoattractants (35, 36). This observation suggests a common principle in signal transduction by chemoreceptors and histidine kinases.
There are several indications that the domain organization of the transmitter module of NtrB applies for other bifunctional histidine kinases as well. We have found for PhoR (the histidine kinase for phosphate regulation) that deletion of the G domain (amino acids 339–431) results in a constitutive negative regulator for the expression of alkaline phosphatase (unpublished data). Recently Park and Inouye demonstrated (37) that the transmitter module of EnvZ can be split into two fragments designated subdomain A and subdomain B. Because of the homology of NtrB and EnvZ, subdomain B comprises the N and the G domain and part of the H domain in our nomenclature and subdomain A comprises ≈2/3 of the H domain. Park and Inouye demonstrate in vitro that subdomain B is able to phosphorylate the fragment carrying the active-site histidine, subdomain A, in trans. However, the authors could not detect phosphatase activity in subdomain A. The lack of the negative regulatory function of subdomain A may be the result of the fact that in subdomain A the C-terminal third of the H domain (as defined here) is deleted. In our hands further N-terminal and C-terminal truncations of the H domain of NtrB resulted in the loss of the negative regulatory activity.
Acknowledgments
We thank B. Magasanik and J. Hu for bacterial strains and plasmids and I. Mettke for expert technical assistance. We are indebted to H. Bujard, whose generous support allowed us to perform this study at the University of Konstanz. This work was carried out in the laboratory of W. Boos and supported by a grant of the Deutsche Forschungsgemeinschaft to H. Bujard and V.W.; G.K. was supported by a fellowship from the Studienstiftung des Deutschen Volkes.
ABBREVIATION
- IPTG
isopropyl β-d-thiogalactoside
References
- 1.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 2.Stock J B, Surette M G, Levit M, Park P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 25–51. [Google Scholar]
- 3.Ninfa A J, Magasanik B. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss V, Magasanik B. Proc Natl Acad Sci USA. 1988;85:8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appleby J L, Parkinson J S, Bourret R B. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 6.Keener J, Kustu S. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tokishita S, Yamada H, Aiba H, Mizuno T. J Biochem (Tokyo) 1990;108:488–493. doi: 10.1093/oxfordjournals.jbchem.a123226. [DOI] [PubMed] [Google Scholar]
- 8.Bueno R, Pahel G, Magasanik B. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E G. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 67–88. [Google Scholar]
- 10.Reitzer L J. In: Escherichia coli and Salmonella. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 391–407. [Google Scholar]
- 11.Atkinson M R, Kamberov E S, Weiss R L, Ninfa A J. J Biol Chem. 1994;269:28288–93. [PubMed] [Google Scholar]
- 12.Kamberov E S, Atkinson M R, Ninfa A J. J Biol Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 13.Amann E, Ochs B, Abel K J. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 14.Hu J C, O’Shea E K, Kim P S, Sauer R T. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 15.Pahel G, Zelenetz A D, Tyler B M. J Bacteriol. 1978;133:139–148. doi: 10.1128/jb.133.1.139-148.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reitzer L J, Magasanik B. Proc Natl Acad Sci USA. 1983;80:5554–5558. doi: 10.1073/pnas.80.18.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiedler U, Weiss V. EMBO J. 1995;14:3696–3705. doi: 10.1002/j.1460-2075.1995.tb00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ota I M, Varshavsky A. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 19.Min K T, Hilditch C M, Diederich B, Errington J, Yudkin M D. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 20.Kang C M, Brody M S, Akbar S, Yang X, Price C W. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atkinson M R, Ninfa A J. J Bacteriol. 1992;174:4538–4548. doi: 10.1128/jb.174.14.4538-4548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiau S P, Schneider B L, Gu W, Reitzer L J. J Bacteriol. 1992;174:179–185. doi: 10.1128/jb.174.1.179-185.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng J, Atkinson M R, McCleary W, Stock J B, Wanner B L, Ninfa A J. J Bacteriol. 1992;174:6061–6070. doi: 10.1128/jb.174.19.6061-6070.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCleary W R, Stock J B. J Biol Chem. 1994;269:31567–72. [PubMed] [Google Scholar]
- 25.Yamada M, Makino K, Shinagawa H, Nakata A. Mol Gen Genet. 1990;220:366–372. doi: 10.1007/BF00391740. [DOI] [PubMed] [Google Scholar]
- 26.Gilles-Gonzalez M A, Ditta G S, Helinski D R. Nature (London) 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 27.Cavicchioli R, Chiang R C, Kalman L V, Gunsalus R P. Mol Microbiol. 1996;21:901–911. doi: 10.1046/j.1365-2958.1996.491422.x. [DOI] [PubMed] [Google Scholar]
- 28.Kamberov E S, Atkinson M R, Chandran P, Ninfa A J. J Biol Chem. 1994;269:28294–9. [PubMed] [Google Scholar]
- 29.Atkinson M R, Ninfa A J. J Bacteriol. 1993;175:7016–7023. doi: 10.1128/jb.175.21.7016-7023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Inouye M. Proc Natl Acad Sci USA. 1991;88:11057–61. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ninfa E G, Atkinson M R, Kamberov E S, Ninfa A J. J Bacteriol. 1993;175:7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson R V, Bourret R B, Simon M I. Mol Microbiol. 1993;8:435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 33.Surette M G, Stock J B. J Biol Chem. 1996;271:17966–73. doi: 10.1074/jbc.271.30.17966. [DOI] [PubMed] [Google Scholar]
- 34.Cochran A G, Kim P S. Science. 1996;271:1113–1116. doi: 10.1126/science.271.5252.1113. [DOI] [PubMed] [Google Scholar]
- 35.Utsumi R, Brissette R E, Rampersaud A, Forst S A, Oosawa K, Inouye M. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 36.Baumgartner J W, Kim C, Brissette R E, Inouye M, Park C, Hazelbauer G L. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park H, Saha S K, Inouye M. Proc Natl Acad Sci USA. 1998;95:6728–6732. doi: 10.1073/pnas.95.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]