Abstract
Methylation of CpG islands is an established transcriptional repressive mechanism and is a feature of silencing in X chromosome inactivation. Housekeeping genes that are subject to X inactivation exhibit differential methylation of their CpG islands such that the inactive alleles are hypermethylated. In this report, we examine two contrasting X-linked genes with CpG islands for regulation by DNA methylation: SYBL1, a housekeeping gene in the Xq pseudoautosomal region, and GPC3, a tissue-specific gene in Xq26 that is implicated in the etiology of the Simpson–Golabi–Behmel overgrowth syndrome. We observed that in vitro methylation of either the SYBL1 or the GPC3 promoter resulted in repression of reporter constructs. In normal contexts, we found that both the Y and inactive X alleles of SYBL1 are repressed and hypermethylated, whereas the active X allele is expressed and unmethylated. Furthermore, the Y and inactive X alleles of SYBL1 were derepressed by treatment with the demethylating agent azadeoxycytidine. GPC3 is also subject to X inactivation, and the active X allele is unmethylated in nonexpressing leukocytes as well as in an expressing cell line, suggesting that methylation is not involved in the tissue-specific repression of this allele. The inactive X allele, however, is hypermethylated in leukocytes, presumably reflecting early X inactivation events that become important for gene dosage in expressing lineages. These and other data suggest that all CpG islands on Xq, including the pseudoautosomal region, are subject to X inactivation-induced methylation. Additionally, methylation of SYBL1 on Yq may derive from a process related to X inactivation that targets large chromatin domains for transcriptional repression.
Mammalian DNA is characterized by the covalent modification of cytosine residues at CpG dinucleotides with methyl groups, resulting in the formation of 5-methylcytosine. This modification is initiated early in development, just after implantation, when a wave of de novo methylation modifies most of the genome. The process is essential for proper embryonic development, because the disruption of the major DNA methyltransferase gene in mice results in death at midgestation (1).
Specific roles for methylation have been suggested for a number of important biological processes, including tumorigenesis, differentiation, X chromosome inactivation, and imprinting (2, 3). In most of these cases, methylation has been associated with transcriptional repression of genes (4, 5). Furthermore, in vitro methylation of genes often results in their transcriptional repression in transfection assays (6). Correspondingly, the demethylation of cells in culture with the drug 5-azacytidine results in the transcriptional activation of many repressed genes (7).
Although the way in which methylation may influence transcription is beginning to be understood (8), the degree to which it is necessary and/or sufficient for repression in various cases remains unknown. To analyze the influence of methylation on transcription further, we have compared the extent and effects of methylation on two genes, each of which is repressed strongly by two distinct mechanisms.
GPC3 lies in Xq26 and has been implicated recently in the etiology of Simpson–Golabi–Behmel overgrowth syndrome (9). It undergoes two types of regulation, X inactivation (see Results), which achieves dosage compensation, and tissue-specific expression, which is limited predominantly to mesodermally derived tissues early in development (9). The second gene, SYBL1, is “pseudoautosomal” (PAR), existing at homologous loci in subtelomeric Xq and Yq (10). This gene codes for a member of the synaptobrevin family, a group of proteins involved in membrane transport. Like GPC3, it is repressed on the inactive X chromosome, but, in sharp contrast, it is expressed in all tissues. It is also the only reported PAR gene that is repressed on the Y chromosome as well as the inactive X (10).
Here, we show that in vitro methylation of multiple CpG dinucleotides in the promoter regions of GPC3 and SYBL1 silences reporter activity in transfection assays, indicating that methylation is sufficient for transcriptional repression. CpG-island methylation also is correlated with in vivo repression for SYBL1, because the gene is activated in either Y or inactive X hybrids by treatment with the demethylating agent 5-aza-2′-deoxycytidine (5aCdr). Thus, methylation is necessary and sufficient to silence SYBL1 expression from both the inactive X and the Y chromosomes. GPC3, a tissue-specific gene, may also be regulated by methylation of the inactive X in expressing tissues, but methylation was not observed on the active X in nonexpressing tissues. We presume that methylation on the inactive X involves most of the CpG islands and occurs early in development without respect to the transcriptional potential of the corresponding active X alleles.
MATERIALS AND METHODS
Cell Culture.
Caco-2 (ATCC no. HTB-37), HeLa (ATCC no. CCL-2), Daudi (ATCC no. CCL-213), and Raji (ATCC no. CCL-86) cells were cultured under standard conditions (American Type Culture Collection). GM1416 (48,XXXX) and GM06318B, hamster–human somatic-cell hybrids containing an active human X chromosome, and THX88 and HY70C4T3, two hamster–human somatic-cell hybrids containing an inactive human X chromosome, were cultured in RPMI medium 1640 supplemented with 10% fetal calf serum. GM06317 and HY853, two hamster–human somatic-cell hybrids containing a human Y chromosome, were cultured in DMEM supplemented with 10% heat-inactivated fetal calf serum, 2% nonessential amino acids, 2% essential amino acids, and 1% MEM vitamins. For reactivation experiments, cultures of THX88 and GM06317 were treated with 5aCdr as described (11).
Plasmid DNA.
A 0.7-kb SmaI–Bsu36I fragment of p1F8–3 (9) containing a GPC3 promoter region conferring complete activation of the gene (residues −585 to +112) was subcloned into pCAT-Basic (Promega; CAT = chloramphenicol acetyltransferase) to yield pGPC3-CAT. The pSYBL-CAT plasmid was constructed by cloning the BamHI–BssHII fragment from positions −233 to +79 into the BamHI–SmaI site of p8CAT0. pSV-βGAL, containing the lacZ gene under the control of the simian virus 40 (SV40) promoter/enhancer, and pCAT-Control, containing the CAT cDNA under the control of the SV40 promoter/enhancer, were purchased from Promega.
Primer Extension.
In vitro transcription experiments were performed by using the HeLa Cell Extract Transcription System (Promega) according to the manufacturer’s protocol. Transcription products were analyzed by primer extension by using a CAT primer according to standard protocols. Reaction products were resolved on a 6% polyacrylamide/8 M urea gel. Sequencing ladders were generated by the dideoxynucleotide chain-termination method under standard conditions.
Sequence Analysis.
GC and CpG content for the two promoters was determined for a 100-bp sliding window across the sequence. GenBank accession numbers are AF003529 for the GPC3 promoter sequence and AJ004799 for the SYBL1 genomic locus.
RNA Preparation.
Total RNA for reverse transcription–PCR (RT-PCR) experiments was prepared under standard conditions according to Sambrook et al. (12). Total RNA from 5aCdr-treated cultures was harvested at different intervals from 2 to 7 days after treatment.
RT-PCR.
RT-PCR was carried out by using the cDNA Cycle Kit (Invitrogen) according to the manufacturer’s instructions. For GPC3 amplification, the oligonucleotides used were GPC3 EX1A (5′-AGGTAGCTGCGAGGAAAC) and GPC3 EX3B (5′-AGGTCACGTCTTGCTCCTC). For Actin gene amplification, ActinA (5′-GGGAAATCGTGCGTGACATT) and ActinB (5′-GGAGTTGAAGGTAGTTTCGTG) were used. For SYBL1 amplification, either SYB-A (5′-CTGGTAGCTCAGCGAGGAGA) and SYB-B (5′-AGTCACATGGATTGCTTTTA) or TBP3 (5′-GAACCTCAAGCTCACTAT) and SYB-B were used. For MIC2 amplification, MIC2A (5′- ACCCAGTGCTGGGGATGACTTT) and MIC2B (5′-CTCTCCATGTCCACCTCCCCT) were used.
Southern Blot Analysis.
Purified, high molecular-weight, genomic DNA was digested, electrophoresed, and transferred to SureBlot Nylon membranes (Oncor) overnight (12). Hybridizations then were carried out according to the manufacturer’s recommendations. For GPC3, the 1.4-kb NheI fragment of p1F8–3 (9) spanning the promoter region and 5′ untranslated region was used as the probe. For SYBL1, the probe was a 1.6-kb EcoRI fragment spanning the promoter region, 5′ untranslated region, and part of the first exon.
In Vitro Methylation.
pGPC3-CAT and pSYBL-CAT were methylated in vitro by using SssI methylase, HhaI methylase, or HpaII methylase according to the manufacturer’s protocol (New England Biolabs). Complete methylation was verified by using HhaI–HpaII digests. Preparations uncut by overnight digestion by HhaI–HpaII were used for subsequent experiments.
Transient Transfections and CAT and β-Galactosidase Assays.
Plasmid (10 μg of pCAT-Control, pGPC3-CAT, or pSYBL-CAT) was cotransfected into HeLa cells with 2 μg of pSV-βGAL with the Calcium Phosphate Profection Kit (Promega) according to the manufacturer’s instructions. After 48 h, total cellular protein extracts were prepared with the Reporter Lysis Buffer (Promega) according to the manufacturer’s instructions. CAT and β-galactosidase assays were carried out according to the manufacturer’s (Promega) instructions.
RESULTS
Differential Expression of SYBL1 and GPC3.
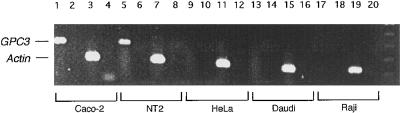
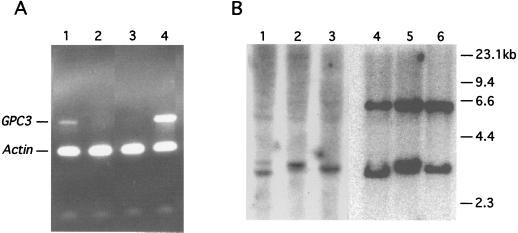
To characterize the regulation of GPC3 and SYBL1, expressing and nonexpressing cell lines have been identified for both genes. SYBL1 has been shown to be expressed ubiquitously, but it undergoes typical X inactivation and is the first PAR gene known to be transcriptionally inactive from a Y chromosome homologue as well (10). To study features of repression on the sex chromosomes, hamster–human somatic-cell hybrids were used to segregate X or Y chromosomes bearing the inactive gene. For GPC3, candidate cell lines were screened for expression by RT-PCR analysis. As shown in Fig. 1, GPC3 expression was detected in both Caco-2 (as in ref. 9) and in NT2 cells. No expression, however, was seen in HeLa cells or in the transformed Burkitt’s lymphoma lines Daudi and Raji.
Figure 1.
RT-PCR analysis of GPC3. GPC3-specific primers or actin gene-specific primers were used to amplify reverse transcribed RNA from the indicated cell lines. The expected amplification products for GPC3 (357 bp) and Actin (247 bp) are indicated.
Sequence Analysis Upstream of GPC3 and SYBL1.
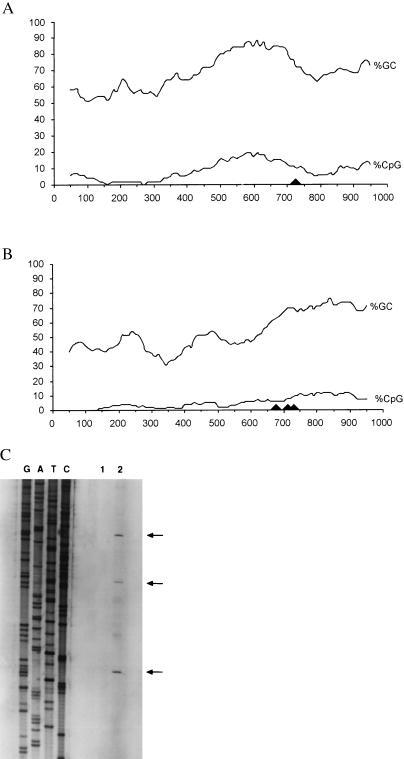
Fig. 2 A and B diagrams the CpG content and the location of the transcription-initiation sites for the two genes. The corresponding sequences of the GPC3 region (13) and the complete sequence of the SYBL1 genomic locus are deposited in GenBank (accession no. AF003529 for GPC3; accession no. AJ004799 for SYBL1).
Figure 2.
CpG content upstream of GPC3 and SYBL1 and determination of the transcription-initiation sites for SYBL1. Percentages of GC and CpG were calculated by using a 100-bp sliding window across the GPC3 (A) and SYBL1 (B) promoter regions. Transcription-initiation sites for the two genes are indicated with arrowheads. (C) Determination of the transcription-initiation sites for SYBL1 by primer extension. The initiation site for GPC3 has been determined (13).
The transcription-initiation site for GPC3 has been identified by 5′ rapid amplification of cDNA ends (RACE) and RNase protection analyses (13) and falls in a near-consensus “initiator” sequence (YYYANTYY, in which Y represents C or T; ref. 14). For SYBL1, determination of the transcription-initiation site was performed by using both primer extension and 5′ RACE. Primer extension analysis shows three transcription-initiation sites (Fig. 2C). These sites were confirmed independently by using 5′ RACE (unpublished results). The occurrence of multiple transcription-initiation sites is not surprising, because there is no TATA-box or initiator element upstream of the SYBL1 coding region.
The putative promoter regions in this study extend 1098 bp upstream of GPC3 and 729 bp upstream of SYBL1. Both are highly GC-rich, with the telltale enrichment of CpG dinucleotides that cues “CpG islands” (15). However, GPC3 shows a markedly higher concentration of CpG across the region (5.6% vs. 3.2% over the length of the entire sequence; compare Fig. 2A with Fig. 2B). Additionally, both genes possess consensus binding sites for several common transcription factors, including Sp1 and AP-2 (ref. 16; M. Strazzullo, M. D’Esposito, and M. D’Urso, unpublished work).
Methylation Is Correlated with Transcriptional Repression of GPC3 and SYBL1.
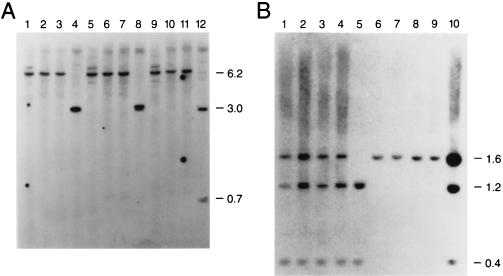
To assess the extent of methylation at both the GPC3 and SYBL1 loci, we studied the susceptibility of the promoter region DNA to methylation-sensitive restriction endonucleases. For GPC3, genomic DNA was extracted from Caco-2 cells, which express GPC3, and Daudi and Raji cells, which do not. All are male cell lines, ensuring analysis of only the active X chromosome. Genomic DNA was restricted with methylation-sensitive endonucleases that have sites in the sequence of the region of high CpG. The sites included one EclXI site, one KspI site, and two BssHII sites. A 1.4-kb NheI fragment spanning the GPC3 CpG island was then used as a probe in Southern blot analysis. As shown in Fig. 3A, DNA from the nonexpressing cell lines Daudi and Raji, as well as from THX88, a hamster–human hybrid cell line containing an inactive human X chromosome, show complete methylation of the upstream promoter region at the four sites analyzed. This methylation produced a 6.2-kb band (Fig. 3A, lanes 1–3, 5–7, and 9–11). In expressing Caco-2 cells (lanes 4, 8, and 12), the same sites are completely unmethylated; the expected restriction fragments produced are of 3.1 kb for KspI and EclXI and 3.0 kb and 0.7 kb for BssHII. These results seem to suggest that the GPC3 promoter must be unmethylated for transcription to occur.
Figure 3.
DNA-methylation analysis of 5′ CpG islands at the GPC3 and SYBL1 loci. Genomic DNA was used in Southern blotting at the GPC3 (A) and SYBL1 (B) loci. (A) Genomic DNA from Raji cells (lanes 1, 5, and 9), Daudi cells (lanes 2, 6, and 10), THX88 (inactive X chromosome-containing hybrid cell line) cells (lanes 3, 7, and 11), and Caco-2 cells (lanes 4, 8, and 12) was restricted with HindIII in combination with KspI (lanes 1–4), EclXI (lanes 5–8), or BssHII (lanes 9–12), and Southern blotting was performed by using a 1.4-kb NheI fragment encompassing the GPC3 CpG island as the probe. (B) This Southern blot is similar to the one shown in A, but it uses the SYBL1 1.6-kb EcoRI fragment as the probe with normal male (lanes 1 and 2) and female DNA (lanes 3 and 4); GM06318B (active X chromosome-containing hybrid cell line) DNA (lane 5); HY70C4T3 and THX88 DNA (lanes 6 and 7); GM06317 and HY853 (Y chromosome-containing hybrid cell lines) DNA (lanes 8 and 9); or GM1416 (48,XXXX human lymphoblastoid cell line) DNA (lane 10). Digestions were performed with EcoRI and the methylation-sensitive enzyme BssHII.
For SYBL1 (Fig. 3B), genomic DNA extracted from normal male and female (lanes 1–4), as well as hybrid cell lines retaining either the active X (lane 5), the inactive X (lanes 6 and 7), the Y chromosome (lanes 8 and 9), or a human lymphoblastoid cell line (48,XXXX; lane 10), was digested with a methylation-insensitive enzyme (EcoRI) and a methylation-sensitive enzyme (BssHII). After hybridization with a 1.6-kb EcoRI probe spanning the entire promoter region, complete demethylation was seen only in the expressing hybrid cells containing active X DNA (1.2-kb and 0.4-kb fragments), whereas complete methylation was apparent in the nonexpressing inactive X and Y chromosome-containing hybrids (1.6-kb fragment). Partial methylation patterns are visible in male, female, and 4X human cells. Confirmatory results were obtained by using other methylation-sensitive enzymes, including MluI and HpaII, which recognize four additional CpG sites (unpublished results).
Methylation Represses Expression of Transfected DNA.
To determine whether methylation is sufficient to silence the promoters of GPC3 and SYBL1, promoter DNA was methylated and then assayed for expression after transient transfection into HeLa cells.
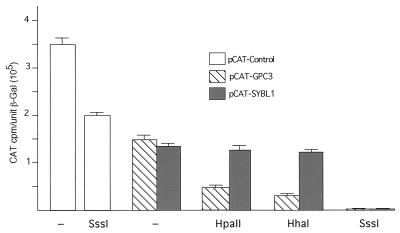
Minimal CAT reporter constructs known to contain the GPC3 (pGPC3-CAT) and SYBL1 (pSYBL-CAT) promoter were methylated in vitro with HhaI, HpaII, or SssI methylase and transiently transfected into HeLa cells. As an internal control for transfection efficiencies, a plasmid containing the lacZ cDNA under the control of the SV40 promoter/enhancer (pSV-βGAL) was cotransfected with each construct, and then CAT activity was normalized to the level of β-galactosidase activity.
As shown in Fig. 4, complete methylation of an SV40 promoter/enhancer-CAT cDNA reporter control plasmid (pCAT-Control) had only a minimal effect on reporter gene activation. Methylation of the minimal GPC3 promoter (pGPC3-CAT), however, markedly reduced transcription by nearly 70% for the site-specific methylases HpaII and HhaI. These enzymes methylate 8 and 10 sites, respectively, in the GPC3 promoter. Complete methylation with SssI methylase virtually abolished reporter gene activity, indicating that methylation is sufficient to silence the GPC3 promoter. The same result was obtained in transfections with decreasing amounts of methylated plasmid DNA, indicating that methylation probably did not act by sequestration or elimination of transcription factors (unpublished results).
Figure 4.
In vitro methylation analysis of the GPC3 and SYBL1 promoters. pCAT-Control, containing the SV40 promoter/enhancer driving the CAT cDNA (open bars), pCAT-GPC3, containing the GPC3 minimal promoter driving the CAT cDNA (hatched bars), or pCAT-SYBL1, containing the SYBL1 minimal promoter driving the CAT cDNA (shaded bars), was methylated in vitro as indicated with HhaI methylase, HpaII methylase, or SssI methylase and transfected transiently into HeLa cells. Resultant CAT activity was normalized to β-galactosidase activity from a cotransfected SV40-βGAL (pSV-βGAL) construct and plotted along the y axis.
For SYBL1, only complete methylation with SssI was able to silence reporter activity (Fig. 4). Methylation with HpaII methylase (three sites) or HhaI methylase (three sites) had little effect on reporter expression, however. This result indicates that methylation is again sufficient to repress reporter activation but only at levels above a certain threshold (see Discussion).
5aCdr Treatment Reactivates SYBL1 but Not GPC3.
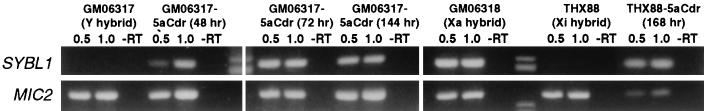
To assess the requirement for methylation to maintain X and Y inactivation of SYBL1, we treated cultures of either an inactive X (THX88) or a Y chromosome-containing (GM06317) somatic-cell hybrid with 5aCdr and analyzed RNA preparations from these cells for the presence or absence of SYBL1 by RT-PCR. As shown in Fig. 5, SYBL1 RNA was not detected in human Y chromosome-containing GM06317 cells before treatment with 5aCdr. Treatment with 5aCdr, however, resulted in the activation of the SYBL1 gene. This activation was detected 48 h, 72 h, and 144 h after the addition of the drug to the culture medium. A similar 5aCdr-induced activation of SYBL1 was observed in inactive X chromosome-containing THX88 cells, which also do not express SYBL1 endogenously. These results indicate that methylation is necessary for the repression of SYBL1 transcription from the inactive X chromosome and from the Y chromosome.
Figure 5.
Activation of SYBL1 expression after 5aCdr treatment of Y and inactive X hybrids. SYBL1 expression and MIC2 expression were examined in hamster–human somatic-cell hybrids by using RT-PCR (as described in Materials and Methods). PCR products were derived from reverse transcriptase reactions that contained 0.5 μg of RNA (0.5), 1.0 μg of RNA (1.0), or 1.0 μg of RNA without reverse transcriptase (−RT). Cell lines and 5aCdr-treated cultures analyzed included GM06317, an untreated hybrid with a Y chromosome, mass cultures of GM06317 isolated at different times after 5aCdr treatment, THX88, an inactive X hybrid (Xi), a mass culture of THX88 after 5aCdr treatment, and GM06318, an active X hybrid (Xa).
In analogous experiments, GPC3 expression was also examined in THX88 hybrids cells cultured in the presence of 5aCdr. No reactivation of expression was observed, however (unpublished results). As GPC3 expression has not been seen in fibroblast-like cells (9), the lack of reactivation may reflect the inability of 5aCdr to reverse tissue-specific forms of regulation that do not depend on methylation (see below).
GPC3 Repression Without Methylation in Peripheral Blood Leukocytes.
Tissue-specific repression of GPC3 apparently does not require promoter methylation, because human male peripheral blood leukocytes, which do not express GPC3 (Fig. 6A), nevertheless contain an unmethylated GPC3 promoter (Fig. 6B; compare the 3-kb band in lanes 1–6 with the 6.2-kb band in lane 4–6). It has been reported that autosomal tissue-specific genes have unmethylated CpG islands in nonexpressing tissues (17); our results suggest that these unmethylated CpG islands also exist in the active X alleles of tissue-specific genes such as GPC3 (see Discussion).
Figure 6.
Expression and DNA methylation analysis of GPC3 in peripheral blood leukocytes. (A) RNA from Caco-2 cells (lane 1), human male peripheral blood leukocytes (lane 2), and human female peripheral blood leukocytes (lane 3) was reverse transcribed and amplified in multiplex by using both GPC3- and actin gene-specific primers. A control PCR with GPC3 and actin cDNA preparations was electrophoresed in lane 4 for comparison. (B) Southern blotting of the GPC3 promoter was carried out as in Fig. 3 by using human male peripheral blood leukocyte DNA (lanes 1–3) or female peripheral blood leukocyte DNA (lanes 4–6).
DISCUSSION
We examined two unusual X-linked genes—SYBL1, a housekeeping gene in XqPAR, and GPC3, a tissue-specific gene in Xq26—for the potential role of methylation in gene regulation.
Methylation and Regulation of GPC3.
Methylation of the large number of CpG dinucleotides in the upstream promoter region of GPC3 is very extensive on the inactive X in hybrids and leukocytes and in two transformed nonexpressing cell lines with a single male-derived X chromosome. All four specific sites that were examined with methylation-sensitive restriction enzymes are methylated fully. Also, when a frequently cutting methylation-sensitive restriction enzyme, HhaI, was used, a high degree of methylation across 10 sites (≈70–90%) was observed in both inactive X chromosome-containing hybrids and the nonexpressing cell lines Daudi and Raji (unpublished results).
Although the human peripheral leukocytes do not express detectable levels of GPC3 mRNA (Fig. 6A), they show little if any methylation of GPC3 on the active X chromosome (Fig. 6B). Thus, although methylation is sufficient to repress the GPC3 promoter in vitro, (Fig. 4A) and the normal inactive X allele is methylated (Figs. 3A and 6B), methylation is not necessary for repression in these cells. This secondary role for methylation in controlling transcription may also be the case in Daudi and Raji cells where methylation may be an effect of transformation and cell culture rather than a primary repressive mechanism (17, 18). Furthermore, treatment of an inactive X hybrid with 5aCdr did not result in GPC3 reactivation.
We suggest that methylation of the GPC3 promoter in nonexpressing cells is the result of an early step in X inactivation in which most CpG islands on the inactive X are methylated without respect to the transcriptional potential of the corresponding active alleles. Support for this view comes from the fact that other X-linked tissue-specific genes also have methylated CpG islands on the inactive X in nonexpressing cells. For example, the androgen receptor is not expressed in peripheral leukocytes (19), yet its 5′ CpG island is unmethylated on the active X and methylated on the inactive X (20) in these cells. In addition, this pattern was also observed (exclusively) when many random clones of X-linked CpG islands were analyzed for methylation (21, 22).
Tissue-specific expression of GPC3 likely requires unique transcription factors. Nonexpressing cell types simply may not make a transcription factor required for GPC3 expression, or they may make a repressing factor that is able to silence GPC3. Lack of a required positive factor is unlikely, however, because in cells in which the endogenous GPC3 gene is repressed fully (i.e., HeLa), transfected reporter DNA is transcribed at a high rate from the unmethylated GPC3 promoter (Fig. 4; ref. 16). As for a repressing protein, the results indicate that any such protein is unable to repress transfected GPC3 DNA constructs, possibly because a critical protein binding site lies outside of the region included in the clones. At present, we have tested greater than 3.3 kb upstream of GPC3 in such analyses and have yet to identify a tissue-specific repressive element (16).
Methylation and Regulation of SYBL1.
Even though SYBL1 is a PAR gene, it is dosage-compensated by X inactivation in the female and by Y inactivation in the male. That methylation is involved in transcriptional repression of this gene was shown by reactivation of the inactive X and Y alleles in hybrids after treatment with 5aCdr. Because SYBL1 is expressed ubiquitously (10), its promoter structure is likely to be simpler than GPC3; the primary control of transcription is that associated with dosage compensation. In fact, the putative promoter region is methylated heavily in the two repressed states examined, on the inactive X and on the Y chromosome (Fig. 3B). Furthermore, a demethylation protocol that resulted in reactivation of at least six other X-linked genes in an inactive X hybrid (23) also produced a clear activation of SYBL1 alleles on inactive X and Y chromosomes in hybrid cells (Fig. 5). Thus, SYBL1 behaves as a typical X-linked gene that is subject to X inactivation, in spite of its XqPAR location. This behavior is in agreement with its evolutionary history, because this gene is part of an ancient region on the X chromosome that is conserved in all mammals (24).
The presence of SYBL1 on the human Y chromosome is unique. It is absent from the Y chromosome of other primates and mammals (24), which implies that SYBL1 had gone through evolutionary elimination from the Y chromosome in these ancestors. We suggest that the apparent recent introduction—or reintroduction—of SYBL1 into the human Y chromosome from the dosage-compensated X may explain the methylation and inactivation of Y linked SYBL1. Alternatively, because SYBL1 is the most proximal XqPAR gene known, it may be close enough to the heterochromatic region of Yq to become susceptible to inactivation by position effect. Perhaps the closely linked IL9R escapes this repression, because it is more distal and lacks a 5′ CpG island.
As predicted, if methylation is sufficient for repression, DNA methylation of the SYBL1 promoter shuts down subsequent expression of the transfected SYBL1 promoter-reporter gene fusion. There is, however, little effect of low or moderate levels of methylation (Fig. 4). This dose-dependent effect of methylation may be related to the relatively lower number of CpG dinucleotides in the promoter region (Fig. 2B). Nan et al. (8) have provided evidence that the density of methylation is itself not as important as the state of particular residues, but it may be that in situations where there is a relatively low number of CpG dinucleotides in a promoter, the overall number of methyl-CpG binding sites for MeCP1 or MeCP2 (25, 26) is more critical.
The comparable methylation of the SYBL1 promoter on the Y chromosome is also notable, because (i) to our knowledge, no other PAR gene has been shown to undergo such inactivation; (ii) the pattern of methylation is qualitatively similar to that on the inactive X chromosome (Fig. 3); and (iii) this methylation is essential for Y chromosome-specific silencing of SYBL1 (Fig. 5). Because there is, as yet, no evidence for a “Y inactivation locus” corresponding to the X inactivation locus (27), it is interesting to speculate that a unique mechanism may be involved in the transcriptional repression of SYBL1 from the Y chromosome. Embryological studies of the relationship between inactive X and Y chromosome-specific silencing of SYBL1 may help to further our understanding of the mechanisms governing mammalian dosage compensation.
Acknowledgments
This work was supported in part by National Institutes of Health Training Grant GM07067 (to R.H.), by Telethon Grant E.526 (to M.D.), by European Community Contract BMH4-CT96-1134 (to M.D.), and by National Institutes of Health Grants HD16659 and GM52463 (to S.M.G. and R.S.H.).
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- 5aCdr
5-aza-2′-deoxycytidine
- PAR
pseudoautosomal
- RT-PCR
reverse transcription–PCR
- SV40
simian virus 40
Footnotes
References
- 1.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 2.Battistuzzi G, D’Urso M, Toniolo D, Persico G M, Luzzatto L. Proc Natl Acad Sci USA. 1985;82:1465–1469. doi: 10.1073/pnas.82.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razin A, Kafri T. Prog Nucleic Acid Res Mol Biol. 1994;48:53–81. doi: 10.1016/s0079-6603(08)60853-3. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 5.Bird A, Tate P, Nan X, Campoy J, Meehan R, Cross S, Tweedie S, Charlton J, Macleod D. J Cell Sci. 1995;19:37–39. doi: 10.1242/jcs.1995.supplement_19.5. [DOI] [PubMed] [Google Scholar]
- 6.Razin A, Cedar H. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu S, Davies P J A. Proc Natl Acad Sci USA. 1997;94:4692–4697. doi: 10.1073/pnas.94.9.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nan X, Compoy J, Bird A. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 9.Pilia G, Hughes-Benzie M, Mackenzie A, Baybayan P, Chen E Y, Huber R, Neri G, Cao A, Forabosco A, Schlessinger D. Nat Genet. 1996;12:241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 10.D’Esposito M, Ciccodicola A, Gianfrancesco F, Esposito T, Flagiello L, Mazzarella R, Schlessinger D, D’Urso M. Nat Genet. 1996;13:227–229. doi: 10.1038/ng0696-227. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki T, Hansen R S, Gartler S M. Mol Cell Biol. 1992;12:3819–3826. doi: 10.1128/mcb.12.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Huber R, Crisponi L, Mazzarella R, Chen C N, Su Y, Chen E, Cao A, Pilia G. Genomics. 1997;45:48–58. doi: 10.1006/geno.1997.4916. [DOI] [PubMed] [Google Scholar]
- 14.Smale S T, Baltimore D. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 15.Bernardi G. Annu Rev Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- 16.Huber R, Schlessinger D, Pilia G. Gene. 1998;214:35–44. doi: 10.1016/s0378-1119(98)00233-9. [DOI] [PubMed] [Google Scholar]
- 17.Antequera F, Boyes J, Bird A. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 18.Jones P A, Wolkowicz M J, Rideout W M, III, Gonzales F A, Marziasz C M, Coetzee G A, Tapscott S J. Proc Natl Acad Sci USA. 1990;87:6117–6121. doi: 10.1073/pnas.87.16.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen J H, Danel L, Cordier G, Saez S, Revillard J P. J Immunol. 1983;131:2767–2771. [PubMed] [Google Scholar]
- 20.Allen R C, Zoghbi H Y, Moseley A B, Rosenblatt H M, Belmont J W. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 21.Norris D P, Brockdorff N, Rastan S. Mamm Genome. 1991;1:78–83. doi: 10.1007/BF02443782. [DOI] [PubMed] [Google Scholar]
- 22.Tribioli C, Tamanini F, Patrossio C, Milanesi L, Villa A, Pergolizzi R, Maestrini E, Rivella S, Bione S, Mancini M, et al. Nucleic Acids Res. 1992;20:727–733. doi: 10.1093/nar/20.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen R S, Canfield T K, Fjeld A D, Gartler S M. Hum Mol Gen. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- 24.D’Esposito M, Matarazzo M R, Ciccodicola A, Strazzullo M, Mazzarella R, Quaderi N A, Fujiwara H, Ko M S, Rowe L B, Ricco A, et al. Hum Mol Genet. 1997;6:1917–1923. doi: 10.1093/hmg/6.11.1917. [DOI] [PubMed] [Google Scholar]
- 25.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 26.Boyes J, Bird A. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 27.Lee J T, Jaenisch R. Curr Opin Genet Dev. 1997;7:274–280. doi: 10.1016/s0959-437x(97)80138-4. [DOI] [PubMed] [Google Scholar]