Over the last decade, the underlying genetic bases of several neurodegenerative disorders, including Huntington disease (HD), Friedreich ataxia, hereditary spastic paraplegia, and rare familial forms of Parkinson disease (PD), Alzheimer disease (AD), and amyotrophic lateral sclerosis (ALS), have been identified. However, the etiologies of sporadic AD, PD, and ALS, which are among the most common neurodegenerative diseases, are still unclear, as are the pathogenic mechanisms giving rise to the various, and often highly stereotypical, clinical features of these diseases. Despite the differential clinical features of the various neurodegenerative disorders, the fact that neurons are highly dependent on oxidative energy metabolism has suggested a unified pathogenetic mechanism of neurodegeneration, based on an underlying dysfunction in mitochondrial energy metabolism.
Mitochondria are the seat of a number of important cellular functions, including essential pathways of intermediate metabolism, amino acid biosynthesis, fatty acid oxidation, steroid metabolism, and apoptosis. Of key importance to our discussion here is the role of mitochondria in oxidative energy metabolism. Oxidative phosphorylation (OXPHOS) generates most of the cell’s ATP, and any impairment of the organelle’s ability to produce energy can have catastrophic consequences, not only due to the primary loss of ATP, but also due to indirect impairment of “downstream” functions, such as the maintenance of organellar and cellular calcium homeostasis. Moreover, deficient mitochondrial metabolism may generate reactive oxygen species (ROS) that can wreak havoc in the cell. It is for these reasons that mitochondrial dysfunction is such an attractive candidate for an “executioner’s” role in neuronal degeneration.
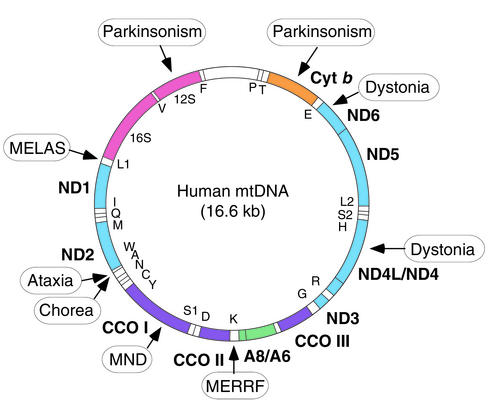
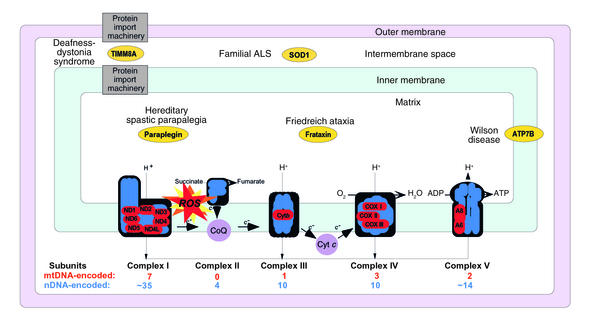
The mitochondrion is the only organelle in the cell, aside from the nucleus, that contains its own genome and genetic machinery. The human mitochondrial genome (1) is a tiny 16.6-kb circle of double-stranded mitochondrial DNA (mtDNA) (Figure 1). It encodes 13 polypeptides, all of which are components of the respiratory chain/OXPHOS system, plus 24 genes, specifying two ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs), that are required to synthesize the 13 polypeptides. Obviously, an organelle as complex as a mitochondrion requires more than 37 gene products; in fact, about 850 polypeptides, all encoded by nuclear DNA (nDNA), are required to build and maintain a functioning organelle. These proteins are synthesized in the cytoplasm and are imported into the organelle, where they are partitioned into the mitochondrion’s four main compartments — the outer mitochondrial membrane (OMM), the inner mitochondrial membrane (IMM), the intermembrane space (IMS), and the matrix, located in the interior (the organelle’s “cytoplasm”). Of the 850 proteins, approximately 75 are structural components of the respiratory complexes (Figure 2) and at least another 20 are required to assemble and maintain them in working order. The five complexes of the respiratory chain/OXPHOS system — complexes I (NADH ubiquinone oxidoreductase), II (succinate ubiquinone oxidoreductase), III (ubiquinone–cytochrome c reductase), IV (cytochrome c oxidase), and V (ATP synthase) — are all located in the IMM. There are also two electron carriers, ubiquinone (also called coenzyme Q), located in the IMM, and cytochrome c, located in the IMS.
Figure 1.
Map of the human mitochondrial genome (1). Polypeptide-coding genes (boldface) are outside the circle and specify seven subunits of NADH dehydrogenase–coenzyme Q oxidoreductase (ND), one subunit of coenzyme Q–cytochrome c oxidoreductase (Cyt b), three subunits of cytochrome c oxidase (CCO), and two subunits of ATP synthase (A) (see also Figure 2). Protein synthesis genes (12S and 16S rRNAs, and 22 tRNAs [one-letter code]) are inside the circle. Mutations in mtDNA associated with MELAS and MERRF, and mutations with features of neurodegenerative disorders, such as ataxia, chorea, dystonia, motor neuron disease (MND), and parkinsonism, are boxed. A complete list of pathogenic mtDNA mutations may be found in ref. 64.
Figure 2.
Schematic representation of the mitochondrion with its electron transport chain (ETC). The ETC is the principal source of ROS in the cell. In addition to mutations in mtDNA- and nDNA-encoded components of the ETC, a number of mutant mitochondrial proteins that do not belong to the ETC have been associated with neurodegenerative disorders. The intramitochondrial localization and the clinical phenotypes associated with mutations of some of these proteins (in ovals) are indicated.
Besides the fact that it operates under dual genetic control, four other features unique to the behavior of this organelle are important in understanding mitochondrial function in neurodegeneration. First, as opposed to the nucleus, in which there are two sets of chromosomes, there are thousands of mtDNAs in each cell, with approximately five mtDNAs per organelle. Second, organellar division and mtDNA replication operate independently of the cell cycle, both in dividing cells (such as glia) and in postmitotic nondividing cells (such as neurons). Third, upon cell division, the mitochondria (and their mtDNAs) are partitioned randomly between the daughter cells (mitotic segregation). Finally, the number of organelles varies among cells, depending in large part on the metabolic requirements of that cell. Thus, skin fibroblasts contain a few hundred mitochondria, whereas neurons may contain thousands and cardiomyocytes tens of thousands of organelles. Taken together, these features highlight the fact that mitochondria obey the laws of population genetics, not mendelian genetics.
Mitochondrial diseases
Mitochondria and their DNA are inherited exclusively from the mother. Thus, pathogenic mutations in mtDNA can cause maternally inherited syndromes. Since these mutations arise initially in only one mtDNA, the population of mtDNAs that was originally homoplasmic (i.e., only one mtDNA genotype) becomes heteroplasmic (two or more coexisting mtDNA genotypes). Obviously, the ratio of normal to mutated mtDNAs in a heteroplasmic population, and their spatial and temporal distributions, will be critical in determining whether and when a subpopulation of mutant mtDNAs will have overt phenotypic consequences. In this regard, it is important to note that most mitochondrial diseases due to maternally inherited mutations in mtDNA are recessive, that is, a very high amount of mutated mtDNA must be present (typically more than 70% of the total population of mtDNAs) in order to cause overt dysfunction. Since mtDNA encodes subunits of the respiratory complexes, pathogenic mutations in mtDNA cause diseases arising from problems in OXPHOS. On the other hand, mutations in nucleus-encoded proteins that are targeted to mitochondria, or that affect organellar function indirectly, can affect almost any aspect of cellular function, not just oxidative energy metabolism.
Neurodegenerative diseases associated with mutations in mitochondrial genes
Diseases associated with mtDNA mutations are typically heterogeneous and are often multisystemic. Since heart, skeletal muscle, and brain are among the most energy-dependent tissues of the body, it is not surprising that many mitochondrial disorders are encephalo-cardiomyopathies. For example, mutations in the tRNALeu(UUR) gene cause mitochondrial encephalomyopathy with lactic acidosis and strokelike episodes (MELAS), mutations in tRNALys cause myoclonus epilepsy with ragged-red fibers (MERRF), and large-scale spontaneous deletions cause Kearns-Sayre syndrome (KSS) and progressive external ophthalmoplegia (PEO) (2). While these diseases are clinically distinct, they share a number of features found in many (but by no means all) mitochondrial diseases, including lactic acidosis, massive mitochondrial proliferation in muscle (resulting in ragged-red fibers [RRFs]), and cytochrome c oxidase (CCO) deficiency. In muscle biopsies, the mutated mtDNAs accumulate preferentially in the RRFs, and RRFs are typically negative for CCO activity.
On the other hand, there are some mtDNA mutations that tend to cause rather selective neuronal degeneration (Figure 1 and Table 1). This situation applies, for example, to mutations in the mtDNA genes coding for subunits of complexes I and V. Mutations in complex I subunits have been associated specifically with Leber hereditary optic neuropathy (LHON), a subacute degeneration of the optic nerve causing bilateral visual failure and blindness. Remarkably, LHON affects males far more than females. Equally remarkably, many patients are homoplasmic for the mutation (i.e., the mutation affects all mtDNAs), which is present in all tissues, and yet the disease is essentially confined to the optic nerve. Complex I activity in cells from these patients ranges from extremely mild to severe. Mutations in complex I genes are also associated with an “LHON-plus” syndrome, characterized pathologically by basal ganglia degeneration and dystonia, and with a maternally inherited form of levodopa-responsive parkinsonism.
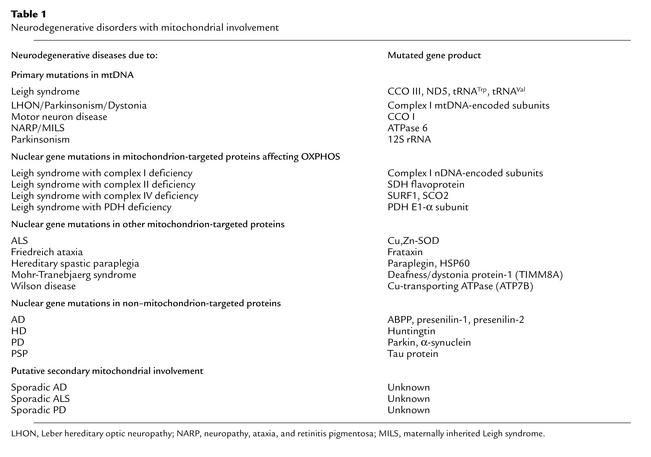
Table 1.
Neurodegenerative disorders with mitochondrial involvement
Mutations in the mtDNA-encoded subunit 6 of complex V (ATPase 6) are associated with a syndrome characterized clinically by neuropathy, ataxia, and retinitis pigmentosa (NARP) at mutation loads of 70–90% in brain, and with a form of subacute necrotizing encephalomyelopathy known as Leigh syndrome (maternally inherited Leigh syndrome [MILS]) at mutation loads above 90%. These mutations cause varying degrees of impairment of mitochondrial ATP synthesis. Other mutations in mtDNA that cause Leigh syndrome (LS) have been found in ND5, tRNATrp, tRNAVal, and CCO III; patients with the latter two were atypical, in that the disease was of relatively late onset.
Neurodegenerative diseases associated with mutations in nuclear gene products that are targeted to mitochondria
Mutations in a small but growing subset of the 850-odd nDNA-encoded gene products that are present in mitochondria have been implicated in neurodegenerative disease (Table 1). While some, such as LS, are essentially the mendelian counterpart of diseases previously associated with mutations in mtDNA, the others are sui generis and have highly diverse genetic origins.
Leigh syndrome
LS is a fatal neurodegenerative condition pathologically characterized by subacute symmetrical necrotic lesions in the subcortical regions of the CNS, including thalamus, basal ganglia, brainstem, and spinal cord, accompanied by demyelination, gliosis, and vascular proliferation in affected areas. Onset is most frequently in infancy or early childhood but may sometimes be in adult life. Symptoms include motor and mental regression, dystonia, ataxia, and abnormal breathing. Death generally occurs within 2 years after onset. It is now clear that LS results from impaired mitochondrial energy metabolism, which can arise from a variety of causes. As noted above, LS can be caused by maternally inherited mutations, but inheritance of LS can also be autosomal-recessive or X-linked. In the latter group are mutations in subunits of the pyruvate dehydrogenase complex, mutations in nuclear-encoded respiratory chain subunits (mainly of complex I but sometimes also of complex II), and mutations in proteins involved in respiratory chain assembly.
CCO deficiency is one of the most common causes of autosomal-recessive LS, but interestingly, all the mutations described to date have been in genes that are required for CCO assembly. Only one sporadic mutation causing LS was found in a CCO structural subunit, in mtDNA-encoded CCO III. CCO-deficient LS is most commonly due to mutations in SURF1, a mitochondrially targeted CCO assembly protein of unknown function, and in SCO2, a mitochondrially targeted protein required for insertion of copper into the CCO holoprotein.
Friedreich ataxia
Friedreich ataxia (FRDA) is the most common of the hereditary ataxias. It is defined clinically by progressive limb and gait ataxia, axonal sensory neuropathy, absent tendon reflexes, and pyramidal signs. FRDA, which is autosomal-recessive, results from mutations in FRDA, which encodes frataxin, a mitochondrially targeted iron-storage protein (3, 4). Trinucleotide repeat (GAA) expansions in intron 1, sometimes in combination with point mutations on the other allele, reduce frataxin expression, resulting in decreased activities of holoproteins containing iron-sulfur groups, including complexes I, II, and III, and aconitase, a Krebs cycle enzyme. Notably, frataxin is particularly abundant in the cerebellum and spinal cord. Mitochondrial enzyme deficiencies and late-stage iron accumulation were also found in a frataxin knockout mouse (5). While the pathogenesis of FRDA is still unclear, one possibility is that the presence of unbound (free) reactive iron, via the Fenton reaction, generates free radicals within the mitochondria, leading to oxidative damage and inactivation of mitochondrial enzymes. For unknown reasons, mitochondrial superoxide dismutase (Mn-SOD), a key antioxidant protein, is deficient in cultured cells from FRDA patients, making them even more prone to oxidative damage. The role of oxidative damage in the pathogenesis of FRDA is also supported by the finding that idebenone, an antioxidant similar to ubiquinone, can reduce myocardial hypertrophy and also decrease markers of oxidative stress in FRDA patients (6, 7).
Hereditary spastic paraplegia
Hereditary spastic paraplegia is a progressive disorder resulting in paraparesis, with onset in childhood or early adulthood. Upper motor neurons are involved selectively, and ataxia and retinitis pigmentosa are also common. Autosomal-dominant, X-linked, and autosomal-recessive forms have been described. Patients with a recessive form of hereditary spastic paraplegia harbor mutations in SPG7, which encodes paraplegin (8), a mitochondrially targeted protein with strong similarity to yeast metalloproteases. Although the underlying pathogenic mechanism is still unclear, mutations in paraplegin cause OXPHOS dysfunction with CCO deficiency and mitochondrial proliferation (i.e., RRFs) in muscle (8). Recently, an autosomal-dominant form of hereditary spastic paraplegia (SPG13) has been attributed to mutations in the mitochondrial import chaperonin HSP60 (9).
Deafness-dystonia syndrome
Deafness-dystonia syndrome, also called Mohr-Tranebjaerg syndrome, is an X-linked recessive disorder characterized by progressive sensorineural deafness, dystonia, cortical blindness, and psychiatric illness. It results from mutations in TIMM8A, which encodes deafness/dystonia protein-1 (DDP1) (10), a component of the mitochondrial protein import machinery located in the IMS. In fibroblasts from patients, mitochondrial protein import is impaired (although not in a global fashion), probably because the DDP1 mutation impairs assembly of TIMM8A into a functional subcomplex of the import machinery (11).
Wilson disease
Wilson disease is an early-onset autosomal-recessive disease characterized by movement disorders (e.g., parkinsonism and dystonia), psychiatric symptoms, and liver failure. The defect results from mutations in ATP7B, which encodes a copper-transporting P-type ATPase and results in copper accumulation in kidney, liver, and the basal ganglia of the brain. ATP7B exists in two isoforms, a 159-kDa form that localizes to the trans-Golgi network, and a 140-kDa form that localizes to mitochondria (12). The mitochondrial localization of the 140-kDa isoform (presumably it is in the inner membrane, but this has not yet been demonstrated) suggests that it might play a role in the functions of copper-dependent enzymes, such as CCO. In fact, mitochondria in affected tissues have morphological abnormalities, as well as deficiency of liver mitochondrial enzymes, especially complex I and aconitase. Thus, oxidative damage mediated by mitochondrial copper accumulation may play a role in the pathogenesis of Wilson disease. Since ATP7B is present in multiple subcellular compartments, the role of mitochondria in the pathogenesis of the disease may be secondary rather than primary, and the issue of mitochondrial involvement in Wilson disease remains controversial.
Mitochondrial dysfunction in disorders associated with mutations in nonmitochondrial proteins, and in neurodegenerative disorders with unknown causes
The most problematic and contentious connections between mitochondrial dysfunction and neurodegeneration involve those disorders that result either from primary mutations in gene products not targeted to mitochondria or from still unknown causes (Table 1). Because, in toto, these disorders account for the vast majority of neurodegenerative disease, the possibility of a unifying principle based on mitochondrial dysfunction as a primary cause of neuronal cell death (13) has become not only attractive, but even seductive. The key question, of course, is whether the data supporting this hypothesis are persuasive. We summarize here the evidence for and against such connections.
Alzheimer disease
AD is the most common form of dementia in the elderly. Approximately 5% of AD cases are familial (FAD) with autosomal-dominant transmission; these result mainly from mutations in the amyloid β precursor protein (ABPP) or in the ABPP processing proteins presenilin-1 (PS1) and presenilin-2 (PS2). Most patients with AD have sporadic cases (SAD) without a known genetic defect. The neuropathology in both FAD and SAD is characterized by neuronal loss and by the presence of amyloid plaques and neurofibrillary tangles. Aggregated β-amyloid peptide (Aβ), a 40– to 42–amino acid subfragment of the much larger ABPP, is considered by many to be the principal culprit in the pathogenesis of AD, but the mechanism by which Aβ40-42 causes neurodegeneration is highly controversial. It is not clear whether plaques and tangles are a cause or an effect, and it has even been proposed that the accumulation of Aβ40-42 is a protective response to oxidative damage caused by mitochondrial dysfunction (14).
Although the findings of mutated forms of APP or presenilins in FAD point toward a pathogenetic role for Aβ40-42, the lack of a clear-cut mechanism for pathogenesis has caused hypotheses involving mitochondrial dysfunction to flourish. In fact, there are several disparate lines of evidence pointing to mitochondrial involvement in the disease. For example, it has long been known that patients with trisomy 21 (Down syndrome) develop the neuropathology of AD, presumably because of an increased dosage of the ABPP gene, which resides on chromosome 21. Not only did cultured astrocytes from Down syndrome patients show increased intracellular levels of Aβ42 associated with impaired mitochondrial membrane potential, but the converse was also true, namely, that impairment of mitochondrial OXPHOS in normal astrocytes resulted in alterations of ABPP processing and elevated levels of Aβ42 (15). These data are consistent with the finding that Aβ can inhibit respiration when added to isolated mitochondria (16).
Several groups have found either reduced CCO activity (17) or reduced levels of immunohistochemically detectable CCO polypeptides (18, 19) in AD brains. At this point, there is no conclusive evidence that mtDNA mutations are higher in AD patients than in normal elderly individuals, or that any particular mtDNA mutation is pathogenic in AD. Therefore, it is more likely that the respiratory chain defects observed in AD brains derive from nuclear gene defects or from acquired damage to respiratory chain components.
Amyotrophic lateral sclerosis
ALS is a neurodegenerative disorder affecting cortical motor neurons and the anterior horn cells of the spinal cord. The disease generally starts in the fourth or fifth decade of life and progresses rapidly, leading to paralysis and premature death. While most cases are sporadic and of unknown etiology, about 15–20% are familial (FALS); of these, about 10% are caused by dominant mutations in SOD1, encoding Cu,Zn-superoxide dismutase (Cu,Zn-SOD). Consistent with the dominant nature of the disorder, mutations in SOD1 do not decrease Cu,Zn-SOD activity significantly, implying that the mutations confer some as-yet unknown toxic “gain of function” on the protein.
Mutant mice carrying the common G93A mutation on a transgene develop severe motor neuron disease reminiscent of ALS, and in addition, the motor neurons develop massive mitochondrial degeneration. In SOD1G93A mice, the motor symptoms were preceded by a transient explosive increase in vacuoles derived from degenerating mitochondria in the motor neurons (20). These mice also had reduced respiratory chain function (21, 22), and, when fed creatine, a key component of the mitochondrial energy buffering and transfer system, their motor performance improved, as did survival (23).
Interestingly, some patients with sporadic ALS (SALS) accumulate abnormal mitochondria in anterior horn cells (24). Reductions in respiratory chain activity have been observed in spinal cords from SALS patients (25, 26), and this reduction appeared to correlate with a concomitant decline in total mtDNA in the tissue, implying that total numbers of organelles were reduced in the spinal cords of these patients (26).
It had long been thought that Cu,Zn-SOD was confined to the cytosol, and that only Mn-SOD (SOD2) was present in mitochondria. Recently, however, it has been determined that SOD1 is also located in the IMS of mitochondria. Thus, SOD activity in mitochondria is compartmentalized within the organelle itself, with SOD1 in the IMS and SOD2 in the matrix. This observation now provides a stronger rationale to explain some of the mitochondrion-related pathology in ALS, as mutant SOD1 could damage mitochondria directly. Transgenic mice expressing the G93A mutation in SOD1 show early activation of the mitochondrion-dependent apoptotic pathway (see below), with release of cytochrome c to the cytosol and translocation of the proapoptotic protein Bax to mitochondria, and similar effects have been observed in the spinal cords of SALS patients. Furthermore, mouse neuroblastoma cells expressing mutant SOD1 had impaired mitochondrial calcium buffering capacity, leading to elevated cytoplasmic calcium, a potential stimulus for apoptosis. Another proposed mitochondrion-related mechanism is based on the finding that cytosolic chaperones, such as HSP70, block the import of mutant SOD1, but not wild-type SOD1, into mitochondria (27). In this scenario, the mutant SOD1 forms aggregates with these HSPs and at the same time titrates them out of the cytosol, thereby killing the cell. Finally, mutant SOD1, but not wild-type SOD1, has been found to interact with at least four proteins, one of which — lysyl-tRNA synthetase — has an isoform that is targeted to mitochondria (28).
Taken together, these findings suggest that mitochondrial dysfunction might play a role in the pathogenesis of both sporadic and familial ALS (29). The mechanisms, however, appear to be complex and may very well involve not only OXPHOS deficiency, but also apoptosis and oxidative damage, as culprits. Notably, treatment of SOD1-mutant mice with the antibiotic minocycline, which inhibits mitochondrial permeability transition-mediated cytochrome c release, extended survival (30). Recent data, however, have failed to support the hypothesis that the gain of function of mutant Cu,Zn-SOD is an increase in copper-mediated free radical damage (31). Specifically, mice that lacked dismutase activity still developed the features of ALS, implying that the gain of function is not mediated by an increase in ROS that results from elevated or altered SOD1 activity.
Huntington disease
HD is a chronic autosomal-dominant disease characterized by choreoathetotic movements and progressive cognitive and emotional disturbances. Pathologically, there is selective degeneration of striatal neurons, with marked atrophy of the putamen and caudate nucleus. HD is a trinucleotide repeat disorder, with expansions of a CAG repeat near the N-terminus of huntingtin, the product of the IT15 gene on chromosome 4. The mutated protein accumulates as nuclear inclusions in striatum and cortex.
Nevertheless, a defect in energy metabolism has been proposed as a potential pathogenic mechanism underlying HD, based on evidence obtained both in vivo and at autopsy. MRI spectroscopy in occipital cortex and basal ganglia of HD patients showed elevated lactate (32), as well as a reduced ratio of phosphocreatine to inorganic phosphate (PCr/Pi) in muscle (33). Animals treated with malonate, which inhibits respiratory chain complex II, develop pathological lesions in the striatum that closely resemble those of HD (34). Defects in complexes II and III, and in aconitase, have been described in postmortem HD brains, especially in the basal ganglia. In transgenic mouse models of HD, similar defects have been observed by some groups, but not by others. Lymphoblasts from HD patients showed increased cyanide-induced depolarization of mitochondria, as well as a greater vulnerability to staurosporin, an inducer of mitochondrion-mediated apoptosis. Recently, it was demonstrated by electron microscopy that mutant huntingtin localizes on neuronal mitochondrial membranes. Furthermore, it was shown that lymphoblast mitochondria from patients with HD have a lower membrane potential and a tendency to become depolarized at lower calcium levels than control mitochondria. Brain mitochondria from transgenic mice expressing mutant huntingtin showed similar mitochondrial defects (35).
Despite evidence in support of mitochondrial involvement in the pathogenesis of HD, the mechanism by which mutated huntingtin could cause bioenergetic dysfunction is still unknown, but some hypotheses have been formulated. Mutant huntingtin is known to bind to, and presumably downregulate, the activity of several polyglutamine-containing transcription factors that modulate nuclear gene transcription, such as Sp1, p53, TATA-binding protein, and cAMP-responsive element–binding (CREB) protein. These factors are thought to be involved in regulating the transcription of proteins involved in mitochondrial energy metabolism and in mitochondrion-initiated apoptosis. For example, mitochondrial dysfunction was shown to be associated with increased activated CREB in cultured cells (36). Therefore, by blocking the activity of critical transcription factors, mutant huntingtin might cause mitochondrial dysfunction. This hypothesis is partially weakened by the recent observation that, in a transgenic mouse model of HD, polyglutamine-containing transcription factors did not seem to be critically depleted in the nuclei of brain cells (37).
The issue of cause and effect has been highlighted by the recent finding that the expanded polyglutamine stretch in mutated huntingtin prevents it from binding to Hip-1 (huntingtin-interacting protein-1), as normal huntingtin does, thereby allowing free Hip-1 to bind to Hippi (Hip-1 protein interactor). Since Hip-1/Hippi heterodimers can recruit procaspase-8 (cysteine aspartic acid–specific protease 8) (see below), the weakened interaction between mutated huntingtin and Hip-1 is a proapoptotic event (38). As apoptosis, including that associated with caspase-8, can occur via both a mitochondrial (i.e., cytochrome c–induced) and a nonmitochondrial (“extrinsic”) pathway, it is unclear at present whether the caspase-Hippi pathway is upstream or downstream of the known mitochondrial alterations in HD.
Parkinson disease
Nowhere is the role of mitochondrial respiratory chain dysfunction more persuasive, but also more contentious, than in the field of PD. PD is a neurodegenerative disorder characterized clinically by rigidity, tremor, and bradykinesia. The pathological hallmark of the disease is the loss of dopaminergic neurons in the substantia nigra pars compacta. As with ALS and AD, most PD cases are sporadic, but there are a few rare cases in which PD is inherited as a mendelian trait (familial PD [FPD]).
The mitochondrial connection to PD began almost 20 years ago, when it was discovered that 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) causes parkinsonism in humans and in laboratory animals. MPTP inhibits complex I of the respiratory chain, but only indirectly, as it is the MPTP metabolite N-methyl-4-phenylpyridinium ion (MPP+), produced by the mitochondrial outer membrane protein monoamine oxidase B (MAO-B), that causes the damage. Soon thereafter, investigators found complex I deficiency and oxidative damage in the substantia nigra of PD patients, as well as reduced immunoreactivity for complex I subunits in affected neurons. Because complexes I and III are the principal sources of free radicals in the cell, altered complex I function in the substantia nigra pars compacta could be responsible for the increased DNA damage and lipid peroxidation found in PD brains (39).
Mutations in FPD have been found in the genes encoding α-synuclein and parkin, but the role of these polypeptides in neurodegeneration is unknown. It is noteworthy, however, that both α-synuclein and parkin, which are components of Lewy bodies (abnormal aggregates found in PD neurons), can bind to each other in vitro (40), that α-synuclein can bind to CCO (41), and that mitochondrial respiratory chain inhibitors can stimulate aggregation of α-synuclein into Lewy bodies in vitro (42, 43).
As with AD, the role of mtDNA mutations in the respiratory chain defects of PD is quite controversial: while some groups showed that cybrids containing mtDNA derived from PD platelets had reduced complex I activity, others could not replicate this finding. Only three mutations in mtDNA were associated specifically with parkinsonism, and they did not present as “classical” PD. Patients with a heteroplasmic mutation in the 12S rRNA gene had parkinsonism as part of a syndrome characterized by deafness and neuropathy, while a boy with a heteroplasmic microdeletion in the cytochrome b gene had an overlap syndrome of parkinsonism and MELAS. A parkinsonian syndrome was also associated with one of the LHON mutations in complex I. On the other hand, it was recently reported that about 10% of patients diagnosed with clinical, biochemical, and/or morphological features of mitochondrial disease had features of parkinsonism, and that these patients responded positively to anti-Parkinson medications (44). Unfortunately, however, none of the 76 patients studied were diagnosed at the genetic level.
Progressive supranuclear palsy
Progressive supranuclear palsy (PSP) is a late-onset (age 50–70 years) neurological disorder with rapid progression, characterized by extrapyramidal symptoms, palsy of vertical gaze of supranuclear origin, and cognitive impairment. The pathological hallmark of PSP is the presence of neurofibrillary filaments containing hyperphosphorylated tau protein in the subcortical regions of the brain, with diffuse neuronal degeneration and gliosis. Genetic studies have linked an extended tau haplotype (H1) with the disease.
OXPHOS defects have been reported in muscle from PSP patients. Postmortem PSP brains had an increased content of malondialdehyde, a marker of lipid peroxidation, and reduced activity of α-ketoglutarate dehydrogenase, the rate-limiting enzyme of the Krebs cycle, but they also had normal respiratory chain activities (45, 46). While it is conceivable that a combination of mitochondrial dysfunction and oxidative stress could generate a vicious cycle leading to further oxidative damage and neuronal degeneration, at this point it is difficult to hypothesize a model consistent with both OXPHOS defects in muscle and normal complex I and IV activities in brain, or to explain why the Krebs cycle should be affected in particular.
Mechanisms of mitochondrion-mediated cell death
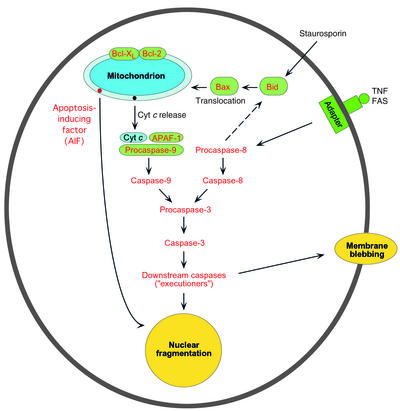
In neurodegenerative diseases, specific populations of neurons die, but by what mechanism? Apoptotic cell death has been invoked as the underlying basis for neuronal loss, but the data are both conflicting and confusing. Apoptosis in mammalian cells is a highly regulated process that operates to sculpt cell populations during the normal course of embryonic and fetal development, but it can also operate during adult life to eliminate cells that are under various stresses. In broad view, apoptosis can operate via two pathways, one mitochondrion-mediated and the other receptor-mediated but mitochondrion-independent (Figure 3). In the mitochondrion-dependent pathway, an external insult — elevated cytosolic calcium, to cite but one example — acts to cause release of cytochrome c, which is located in the IMS. Cytosolic cytochrome c can then bind APAF-1 (apoptotic protease-activating factor 1), which then binds to the inactive form of caspase-9. This “initiator” caspase complex, or “apoptosome,” can now activate a cascade of events, beginning with activation of downstream “effector” caspases, such as caspase-3, followed by activation of caspases further downstream, ultimately resulting in the hallmarks of apoptosis (condensation of nuclear and cytoplasmic contents, nDNA fragmentation, cell blebbing, and autophagy of membrane-bound bodies). Mitochondrion-mediated activation of caspase-9 can also occur via external (extracellular) receptor-mediated signals (e.g., TNF, growth factor deprivation, etc.) to target various ligands — e.g., Bad, Bax, Bik, Noxa — to the mitochondrion, thereby causing cytochrome c release and binding of APAF-1. Under other circumstances, a separate mitochondrion-independent pathway also operates (Figure 3).
Figure 3.
A schematic view of the main pathways of apoptosis: mitochondrion-mediated and mitochondrion-independent. See text for details.
Cell death in mitochondrial and in neurodegenerative disease
Among the numerous stresses known to participate in mitochondrion-mediated apoptosis, at least in vitro, bioenergetic failure and elevated ROS figure prominently (47). Thus, two key hypotheses immediately present themselves. First, one would expect a causal relationship between mitochondrial dysfunction and neurodegeneration via apoptosis, perhaps due, at least in part, to changes in ROS levels. Second, if neurodegeneration is related in any way to defects in respiratory chain function or in OXPHOS, then one would expect authentic mitochondrial diseases to show many, if not all, of the features of cell death that are postulated to occur in neurodegenerative diseases. So what do we know about apoptosis and ROS in mitochondrial disease and in neurodegeneration? Not unexpectedly, the literature is conflicting.
It is well known that muscle biopsies from patients with “classic” mitochondrial disorders, including those with RRFs, show little or no evidence of necrosis, fiber loss, elevated circulating creatine kinase, or inflammation. Nevertheless, markers of increased ROS have been found in muscle biopsies from patients with mitochondrial disease (48–50). It is clear that elevated ROS are injurious to mitochondria. For example, ablation of the mitochondrial SOD2 gene in mice caused rapid death via a fatal cardiomyopathy (51), and even loss of only one SOD2 allele was harmful (52). Encouragingly, pathology could be ameliorated, and lifespan extended, in SOD2–/– mice upon pharmacological intervention to reduce free radicals (53).
An analysis of muscle biopsies from patients with CCO deficiency due to various known pathogenic mutations in mtDNA (e.g., those causing MERRF, MELAS, KSS, and PEO) and in nucleus-encoded mitochondrially targeted proteins (e.g., those causing LS) showed features of apoptosis in CCO-negative RRFs — DNA fragmentation by terminal deoxynucleo-tidyl transferase–mediated dUTP nick end-labeling (TUNEL), condensed chromatin, and elevated immunostaining for caspase-1, caspase-3, and Fas ligand (54, 55). However, another group found no evidence for apoptosis (56). In a third study, some apoptotic markers (Bax, p53, Fas, and caspase-3) were not increased in CCO-negative RRFs, in spite of the fact that other apoptosis-related proteins (Bcl-2, Bcl-XL, DNase I) were increased, as was the amount of cytosolic cytochrome c (50). What little evidence there was for apoptosis in muscle seemed to be confined to a few RRFs (57).
If markers of elevated ROS and apoptosis can be found in these muscle biopsies, why do these muscle fibers not degenerate? This apparent paradox points out that a marker of apoptosis is merely that — a marker — and does not necessarily imply that apoptosis is actually occurring. In skeletal muscle, there are at least two reasons why apoptosis is not occurring in muscle. First, APAF-1 is either present at very low levels (58) or missing entirely (59) in skeletal muscle. In the absence of APAF-1, cytosolic cytochrome c has no partner with which to bind in order to activate caspase-9 and induce apoptosis (60) (although in some circumstances the absence of APAF-1 may trigger a “default pathway” of necrosis [ref. 61]). In other words, cytosolic cytochrome c in this situation is a marker of, but cannot induce, apoptosis. In support of this view, Nonaka’s group could detect APAF-1 and released cytochrome c only in RRFs from patients with MERRF, MELAS, or PEO, but not in the far more abundant relatively normal muscle fibers (57). Second, an inhibitor of apoptosis called ARC (apoptotic repressor with caspase recruitment domain), which is expressed almost exclusively in skeletal and heart muscle (62), not only inhibits apoptosis in these tissues but can also protect mitochondria from free radical damage (63).
What about neuronal death in mitochondrial disease? There is neuronal loss in KSS, MERRF, and MELAS, and the regions that are affected differ in each disease, but no studies have been published regarding apoptosis in these cells. Intriguingly, TUNEL assays in postmortem brains from these patients showed little if any positive staining in neurons, but staining was observed in non-neuronal cells, such as microglia (E. Bonilla, personal communication). Moreover, there are regions of the brain — the choroid plexus in KSS, for example — that are nearly homoplasmic for mutated mtDNA and are devoid of CCO immunoreactivity, and yet are both morphologically intact and TUNEL-negative (E. Bonilla, personal communication). Therefore, it is still unclear how neurons die in mitochondrial disorders.
Perspective
It is clear that impairment of mitochondrial energy metabolism is the key pathogenic factor in a number of neurodegenerative disorders (Table 1). This is most clearly seen in maternally inherited diseases that result from pathogenic mutations in mtDNA that interdict the normal functioning of the respiratory chain and OXPHOS. Examples of this group of diseases include LHON and maternally inherited LS. Similarly, mutations in nuclear genes that encode mitochondrially targeted proteins required for respiratory chain assembly and homeostasis can cause neurodegenerative disease. Examples in this class are autosomal-recessive LS, FRDA, hereditary spastic paraplegia, and Mohr-Tranebjaerg syndrome.
What is striking about both of these groups of neurodegenerative disorders is how diverse they are in their clinical manifestations, and how little they resemble the “classic” neurodegenerative disorders — AD, ALS, HD, PD, and PSP — that have no obvious connection to primary mitochondrial bioenergetic dysfunction. Put another way, it is remarkable that almost no pathogenic mutation in mtDNA causes the typical clinical manifestations associated with the most common neurodegenerative diseases. Of the more than 150 pathogenic mtDNA mutations that have been described to date (64), only one is associated with chorea and dementia; only three are associated with parkinsonism, but as part of a complex and atypical clinical picture; only three are associated with dystonia, but in the context of LHON or MELAS; only one is associated with spinocerebellar ataxia, but in the context of deafness, neuropathy, and dementia; and only one is associated with motor neuron disease, but in a syndrome with unusual secondary features. If respiratory chain dysfunction is a key factor in neurodegenerative diseases, why are there not more patients with “mitochondrial AD,” “mitochondrial PD,” or “mitochondrial ALS”? Conversely, why do so many mitochondrial diseases present as encephalopathies, myopathies, cardio-pathies, endocrinopathies, neuropathies, or combinations thereof, in which dementia and/or neurodegeneration is but one feature?
One response to these questions is that mitochondrial diseases that result from mtDNA mutations have features that are unique to the population genetics of mtDNA. In particular, the combination of heteroplasmy, mitotic segregation, and the threshold effect can conspire to mask features that might be more overt in disorders that obey the rules of mendelian genetics. In addition, it could be argued that many mitochondrial disorders strike early in life, thus precluding the features of dementia, chorea, and the like to evince themselves as they do in AD, ALS, or HD, which have later onset. While formally these are valid objections, they are not particularly convincing. There are many mitochondrial patients with extremely high levels of mutations in multiple tissues (and especially in the brain), and there are enough patients who live into their fourth and even fifth decades of life in whom the symptoms of AD, ALS, and PD should eventually be recapitulated.
This does not mean, however, that OXPHOS defects play no role in the late-onset neurodegenerative diseases. Rather, mitochondrial dysfunction probably plays an important, but secondary, role in these disorders. For example, it is clear that there is respiratory chain deficiency in hippocampal neurons in AD patients, as measured by biochemistry, histochemistry, and immunohistochemistry. It is likely, however, that these deficits are secondary to some underlying dysfunction, of which Aβ deposition is but one of many possible primary causes.
The failure to detect true cell death in muscle from patients with mitochondrial disease tells us little about neuronal cell death in these patients. It does, however, send a cautionary message regarding a too-eager willingness to equate cell death markers with the event itself. Conversely, the massive amounts of cell death in the neurodegenerative disorders mean that the very cells that are postulated to be dying due to mitochondrial dysfunction — e.g., motor neurons in ALS, striatal cells in HD, and, most pointedly, nigral cells in PD — are, in the final analysis, unavailable for examination, especially in the terminal stages of the disease and in autopsy tissue. Thus, many, if not all, studies that rely on patient material have, a fortiori, focused on the function, or lack of same, in a skewed population of surviving cells, such as glia, that are likely to be unrepresentative of the true pathogenetic course of events. Furthermore, studies of patient neuronal tissue or of model cells in vitro have their own limitations, not the least of which is the absence of the interactions among entire populations and subpopulations of cells that define the behavior of a vascularized tissue in vivo. These caveats point out the truly difficult nature of deducing pathogenesis in diseases as complex as neurodegenerative disorders. A mechanistic relationship between neurodegeneration and mitochondrial dysfunction still awaits a direct, clear, and unequivocal demonstration.
Acknowledgments
We thank Salvatore DiMauro, Michio Hirano, Michael T. Lin, and Serge Przedborski for insight and advice on the manuscript. This work was supported by grants from the NIH (NS-28828, NS-39854, and HD-32062, to E.A. Schon), from the Muscular Dystrophy Association (to E.A. Schon and G. Manfredi), and from the New York Academy of Medicine Speaker’s Fund (to G. Manfredi).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Huntington disease (HD); Parkinson disease (PD); familial Parkinson disease (FPD); Alzheimer disease (AD); familial Alzheimer disease (FAD); amyotrophic lateral sclerosis (ALS); sporadic amyotrophic lateral sclerosis (SALS); oxidative phosphorylation (OXPHOS); reactive oxygen species (ROS); mitochondrial DNA (mtDNA); ribosomal RNA (rRNA); transfer RNA (tRNA); nuclear DNA (nDNA); inner mitochondrial membrane (IMM); intermembrane space (IMS); mitochondrial encephalomyopathy with lactic acidosis and strokelike episodes (MELAS); myoclonus epilepsy with ragged-red fibers (MERRF); ragged-red fiber (RRF); Kearns-Sayre syndrome (KSS); progressive external ophthalmoplegia (PEO); cytochrome c oxidase (CCO); Leber hereditary optic neuropathy (LHON); Leigh syndrome (LS); Friedreich ataxia (FRDA); mitochondrial superoxide dismutase (Mn-SOD); Cu,Zn-superoxide dismutase (Cu,Zn-SOD); β-amyloid peptide (Aβ); progressive supranuclear palsy (PSP); terminal deoxynucleotidyl transferase–mediated dUTP nick end-labeling (TUNEL).
References
- 1.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Schon EA, Bonilla E, DiMauro S. Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomembr. 1997;29:131–149. doi: 10.1023/a:1022685929755. [DOI] [PubMed] [Google Scholar]
- 3.Cavadini P, O’Neill HA, Benada O, Isaya G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum. Mol. Genet. 2002;11:217–227. doi: 10.1093/hmg/11.3.217. [DOI] [PubMed] [Google Scholar]
- 4.Gakh O, et al. Physical evidence that yeast frataxin is an iron storage protein. Biochemistry. 2002;41:6798–6804. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- 5.Puccio H. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 6.Rustin P, et al. Effect of idebenone on cardiomyopathy in Friedreich’s ataxia: a preliminary study. Lancet. 1999;354:477–479. doi: 10.1016/S0140-6736(99)01341-0. [DOI] [PubMed] [Google Scholar]
- 7.Schulz JB, et al. Oxidative stress in patients with Friedreich ataxia. Neurology. 2000;55:1719–1721. doi: 10.1212/wnl.55.11.1719. [DOI] [PubMed] [Google Scholar]
- 8.Casari G, et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 9.Hansen JJ, et al. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am. J. Hum. Genet. 2002;70:1328–1332. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin H, et al. A novel X-linked gene, DDP, shows mutations in families with deafness (DFN-1), dystonia, mental deficiency and blindness. Nat. Genet. 1996;14:177–180. doi: 10.1038/ng1096-177. [DOI] [PubMed] [Google Scholar]
- 11.Roesch K, Curran SP, Tranebjaerg L, Koehler CM. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum. Mol. Genet. 2002;11:477–486. doi: 10.1093/hmg/11.5.477. [DOI] [PubMed] [Google Scholar]
- 12.Lutsenko S, Cooper MJ. Localization of the Wilson’s disease protein product to mitochondria. Proc. Natl. Acad. Sci. USA. 1998;95:6004–6009. doi: 10.1073/pnas.95.11.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orth M, Schapira AH. Mitochondria and degenerative disorders. Am. J. Med. Genet. 2001;106:27–36. doi: 10.1002/ajmg.1425. [DOI] [PubMed] [Google Scholar]
- 14.Smith MA, et al. Amyloid-beta, tau alterations and mitochondrial dysfunction in Alzheimer disease: the chickens or the eggs? Neurochem. Int. 2002;40:527–531. doi: 10.1016/s0197-0186(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 15.Busciglio J, et al. Altered metabolism of the amyloid β precursor protein is associated with mitochondrial dysfunction in Down’s syndrome. Neuron. 2002;33:677–688. doi: 10.1016/s0896-6273(02)00604-9. [DOI] [PubMed] [Google Scholar]
- 16.Casley CS, et al. Beta-amyloid inhibits integrated mitochondrial respiration and key enzyme activities. J. Neurochem. 2002;80:91–100. doi: 10.1046/j.0022-3042.2001.00681.x. [DOI] [PubMed] [Google Scholar]
- 17.Mutisya EM, Bowling AC, Beal MF. Cortical cytochrome oxidase activity is reduced in Alzheimer’s disease. J. Neurochem. 1994;63:2179–2184. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- 18.Bonilla E, et al. Mitochondrial involvement in Alzheimer’s disease. . Biochim. Biophys. Acta. 1999; 1410:171–182. doi: 10.1016/s0005-2728(98)00165-0. [DOI] [PubMed] [Google Scholar]
- 19.Ojaimi J, et al. Irregular distribution of cytochrome c oxidase protein subunits in aging and Alzheimer’s disease. Ann. Neurol. 1999;46:656–660. [PubMed] [Google Scholar]
- 20.Kong J, Xu Z. Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. J. Neurosci. 1998;18:3241–3250. doi: 10.1523/JNEUROSCI.18-09-03241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browne SE, et al. Metabolic dysfunction in familial, but not sporadic, amyotrophic lateral sclerosis. J. Neurochem. 1998;71:281–287. doi: 10.1046/j.1471-4159.1998.71010281.x. [DOI] [PubMed] [Google Scholar]
- 22.Mattiazzi M, et al. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J. Biol. Chem. 2002;27:29626–29633. doi: 10.1074/jbc.M203065200. [DOI] [PubMed] [Google Scholar]
- 23.Klivenyi P, et al. Neuroprotective effects of creatine in a transgenic animal model of amyotrophic lateral sclerosis. Nat. Med. 1999;5:347–350. doi: 10.1038/6568. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki S, Iwata M. Impairment of fast axonal transport in the proximal axons of anterior horn neurons in amyotrophic lateral sclerosis. Neurology. 1996;47:535–540. doi: 10.1212/wnl.47.2.535. [DOI] [PubMed] [Google Scholar]
- 25.Borthwick GM, et al. Mitochondrial enzyme activity in amyotrophic lateral sclerosis: implications for the role of mitochondria in neuronal cell death. Ann. Neurol. 1999;46:787–790. doi: 10.1002/1531-8249(199911)46:5<787::aid-ana17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 26.Wiedemann FR, et al. Mitochondrial DNA and respiratory chain function in spinal cords of ALS patients. J. Neurochem. 2002;80:616–625. doi: 10.1046/j.0022-3042.2001.00731.x. [DOI] [PubMed] [Google Scholar]
- 27.Okado-Matsumoto A, Fridovich I. Amyotrophic lateral sclerosis: a proposed mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:9010–9014. doi: 10.1073/pnas.132260399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunst CB, Mezey E, Brownstein MJ, Patterson D. Mutations in SOD1 associated with amyotrophic lateral sclerosis cause novel protein interactions. Nat. Genet. 1997;15:91–94. doi: 10.1038/ng0197-91. [DOI] [PubMed] [Google Scholar]
- 29.Menzies FM, Ince PG, Shaw PJ. Mitochondrial involvement in amyotrophic lateral sclerosis. Neurochem. Int. 2002;40:543–551. doi: 10.1016/s0197-0186(01)00125-5. [DOI] [PubMed] [Google Scholar]
- 30.Zhu S, et al. Minocycline inhibits cytochrome c release and delays progression of amyotrophic lateral sclerosis in mice. Nature. 2002;417:74–78. doi: 10.1038/417074a. [DOI] [PubMed] [Google Scholar]
- 31.Subramaniam JR, et al. Mutant SOD1 causes motor neuron disease independent of copper chaperone-mediated copper loading. Nat. Neurosci. 2002;5:301–307. doi: 10.1038/nn823. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR. Evidence for impairment of energy metabolism in vivo in Huntington’s disease using localized 1H NMR spectroscopy. Neurology. 1993;43:2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- 33.Koroshetz WJ, Jenkins BG, Rosen BR, Beal MF. Energy metabolism defects in Huntington’s disease and effects of coenzyme Q10. Ann. Neurol. 1997;41:160–165. doi: 10.1002/ana.410410206. [DOI] [PubMed] [Google Scholar]
- 34.Beal MF, et al. Age-dependent striatal excitotoxic lesions produced by the endogenous mitochondrial inhibitor malonate. J. Neurochem. 1993;61:1147–1150. doi: 10.1111/j.1471-4159.1993.tb03633.x. [DOI] [PubMed] [Google Scholar]
- 35.Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 36.Arnould T, et al. CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation. EMBO J. 2002;21:53–63. doi: 10.1093/emboj/21.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu ZX, Li SH, Nguyen HP, Li XJ. Huntingtin inclusions do not deplete polyglutamine-containing transcription factors in HD mice. Hum. Mol. Genet. 2002;11:905–914. doi: 10.1093/hmg/11.8.905. [DOI] [PubMed] [Google Scholar]
- 38.Gervais FG, et al. Recruitment and activation of caspase-8 by the Huntingtin-interacting protein Hip-1 and a novel partner Hippi. Nat. Cell Biol. 2002;4:95–105. doi: 10.1038/ncb735. [DOI] [PubMed] [Google Scholar]
- 39.Dexter DT, et al. Indices of oxidative stress and mitochondrial function in individuals with incidental Lewy body disease. Ann. Neurol. 1994;35:38–44. doi: 10.1002/ana.410350107. [DOI] [PubMed] [Google Scholar]
- 40.Choi P, et al. Co-association of parkin and alpha-synuclein. Neuroreport. 2001;12:2839–2843. doi: 10.1097/00001756-200109170-00017. [DOI] [PubMed] [Google Scholar]
- 41.Elkon H, et al. Mutant and wild-type alpha-synuclein interact with mitochondrial cytochrome c oxidase. J. Mol. Neurosci. 2002;18:229–238. doi: 10.1385/JMN:18:3:229. [DOI] [PubMed] [Google Scholar]
- 42.Lee HJ, Choi C, Lee SJ. Membrane-bound alpha-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J. Biol. Chem. 2002;277:671–678. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- 43.Lee HJ, et al. Formation and removal of alpha-synuclein aggregates in cells exposed to mitochondrial inhibitors. J. Biol. Chem. 2002;277:5411–5417. doi: 10.1074/jbc.M105326200. [DOI] [PubMed] [Google Scholar]
- 44.Finsterer J. Parkinson syndrome as a manifestation of mitochondriopathy. Acta Neurol. Scand. 2002;105:384–389. doi: 10.1034/j.1600-0404.2002.01221.x. [DOI] [PubMed] [Google Scholar]
- 45.Albers DS, et al. Frontal lobe dysfunction in progressive supranuclear palsy: evidence for oxidative stress and mitochondrial impairment. J. Neurochem. 2000;74:878–881. doi: 10.1046/j.1471-4159.2000.740878.x. [DOI] [PubMed] [Google Scholar]
- 46.Park LC, et al. Mitochondrial impairment in the cerebellum of the patients with progressive supranuclear palsy. J. Neurosci. Res. 2001;66:1028–1034. doi: 10.1002/jnr.10062. [DOI] [PubMed] [Google Scholar]
- 47.Kroemer G, Reed JC. Mitochondrial control of cell death. Nat. Med. 2000;6:513–519. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- 48.Mitsui T, et al. Oxidative damage to skeletal muscle DNA from patients with mitochondrial encephalomyopathies. J. Neurol. Sci. 1996;139:111–116. [PubMed] [Google Scholar]
- 49.Filosto M, et al. Antioxidant agents have a different expression pattern in muscle fibers of patients with mitochondrial diseases. Acta Neuropathol. (Berl.) 2002;103:215–220. doi: 10.1007/s004010100455. [DOI] [PubMed] [Google Scholar]
- 50.Umaki Y, et al. Apoptosis-related changes in skeletal muscles of patients with mitochondrial diseases. Acta Neuropathol. (Berl.) 2002;103:163–170. doi: 10.1007/s004010100446. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 52.Murakami K, et al. Mitochondrial susceptibility to oxidative stress exacerbates cerebral infarction that follows permanent focal cerebral ischemia in mutant mice with manganese superoxide dismutase deficiency. J. Neurosci. 1998;18:205–213. doi: 10.1523/JNEUROSCI.18-01-00205.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melov S, et al. Lifespan extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J. Neurosci. 2001;21:8348–8353. doi: 10.1523/JNEUROSCI.21-21-08348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mirabella M, et al. Apoptosis in mitochondrial encephalomyopathies with mitochondrial DNA mutations: a potential pathogenic mechanism. Brain. 2000;123:93–104. doi: 10.1093/brain/123.1.93. [DOI] [PubMed] [Google Scholar]
- 55.Di Giovanni S, et al. Apoptosis and ROS detoxification enzymes correlate with cytochrome c oxidase deficiency in mitochondrial encephalomyopathies. Mol. Cell. Neurosci. 2001;17:696–705. doi: 10.1006/mcne.2001.0970. [DOI] [PubMed] [Google Scholar]
- 56.Sciacco M, et al. Lack of apoptosis in mitochondrial encephalomyopathies. Neurology. 2001;56:1070–1074. doi: 10.1212/wnl.56.8.1070. [DOI] [PubMed] [Google Scholar]
- 57.Ikezoe K, et al. Apoptosis is suspended in muscle of mitochondrial encephalomyopathies. Acta Neuropathol. (Berl.) 2002;103:531–540. doi: 10.1007/s00401-001-0502-8. [DOI] [PubMed] [Google Scholar]
- 58.Zou H, et al. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 59.Burgess DH, et al. Human skeletal muscle cytosols are refractory to cytochrome c-dependent activation of type-II caspases and lack APAF-1. Cell Death Differ. 1999;6:256–261. doi: 10.1038/sj.cdd.4400489. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida H, et al. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- 61.Miyazaki K, et al. Caspase-independent cell death and mitochondrial disruptions observed in the Apaf1-deficient cells. J. Biochem. (Tokyo). 2001;129:963–969. doi: 10.1093/oxfordjournals.jbchem.a002944. [DOI] [PubMed] [Google Scholar]
- 62.Koseki T, Inohara N, Chen S, Nunez G. ARC, an inhibitor of apoptosis expressed in skeletal muscle and heart that interacts selectively with caspases. Proc. Natl. Acad. Sci. USA. 1998;95:5156–5160. doi: 10.1073/pnas.95.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neuss M, et al. The apoptotic regulatory protein ARC (apoptosis repressor with caspase recruitment domain) prevents oxidant stress-mediated cell death by preserving mitochondrial function. J. Biol. Chem. 2001;276:33915–33922. doi: 10.1074/jbc.M104080200. [DOI] [PubMed] [Google Scholar]
- 64.Servidei S. Mitochondrial encephalomyopathies: gene mutation. Neuromuscul. Disord. 2002;12:334–339. doi: 10.1016/s0960-8966(01)00334-0. [DOI] [PubMed] [Google Scholar]