Abstract
Conditional N-Myc deletion limits the proliferation of granule neuron progenitors (GNPs), perturbs foliation, and leads to reduced cerebellar mass. We show that c-Myc mRNA levels increase in N-Myc-null GNPs and that simultaneous deletion of both c- and N-Myc exacerbates defective cerebellar development. Moreover, N-Myc loss has been shown to trigger the precocious expression of two cyclin-dependent kinase inhibitors, Kip1 and Ink4c, in the cerebellar primordium. We now further demonstrate that the engineered disruption of the Kip1 and Ink4c genes in N-Myc-null cerebella partially rescues GNP cell proliferation and cerebellar foliation. These results provide definitive genetic evidence that expression of N-Myc and concomitant down-regulation of Ink4c and Kip1 contribute to the proper development of the cerebellum.
Keywords: Myc, N-Myc deficiency, cerebellum, granule neuron progenitor, cell cycle inhibitor
Cerebellar development is initiated during embryogenesis, but, in contrast to the rest of the nervous system, the major proliferative phase occurs postnatally. In the mouse, granule neuron progenitors (GNPs) are born from the germinal neuroepithelium within the rhombic lip at embryonic day (E)10.5 and migrate outwardly within the cerebellar primordium between E12.5 and E15.5 to form the external germinal layer (EGL) (1). At birth [postnatal day 0 (P0)], the EGL is composed of only a single layer of GNPs overlaying the Purkinje neurons that secrete the mitogen sonic hedgehog (Shh). After birth, GNPs proliferate rapidly in response to Shh, and cells in the inner zone of the expanding EGL exit the cell cycle and migrate through the Purkinje cell layer to ultimately form the internal granule layer (IGL) of the adult cerebellum. This migratory process depletes the EGL of virtually all granule neurons by P21; however, their retrograde axons synapse with Purkinje cell dendrites within the outer molecular layer of the mature organ. Cerebellar foliation, due to the rapid expansion of the EGL and subsequent formation of the IGL, occurs during the first 2 weeks after birth, and, by 1 month of age, the cerebellum is completely formed (2–4). The basic adult foliation pattern is present by P7 and is distinguished by 10 folia (designated I to X), each separated from one another by fissures that form along the rostral–caudal axis.
N-Myc promotes the rapid cell division of GNPs (5, 6), whereas the related family member, c-Myc, is not detectably expressed within the proliferating outer zone of the EGL. Because it is a downstream target of Shh signaling, N-Myc overexpression can enforce the proliferation of GNPs independently of Shh signaling (5), and, conversely, its conditional loss early during embryonic cerebellar development results in a severe GNP deficiency and failure of proper organogenesis (7). The anatomic defects resulting from conditional N-Myc inactivation are associated with the ectopic expression of abnormally high levels of two cyclin-dependent kinase (CDK) inhibitors, p18Ink4c and p27Kip1, which can be detected by immunohistochemistry in the cerebellar primordium at E12.5. This expression contrasts with their pattern of expression during normal cerebellar development in which Ink4c is transiently expressed only within the postnatal EGL as GNPs exit the cell cycle (8) and where expression of p27Kip1 is restricted to postmitotic granule neurons. However, unlike p18Ink4c, p27Kip1 is maintained in these neurons throughout adult life (9). In mice lacking Ink4c or Kip1, the cerebella have a normal anatomic architecture, although these animals exhibit generalized organomegaly (10–13). GNPs isolated from Ink4c- or Kip1-null postnatal cerebella display an enhanced proliferative potential in vitro compared with those explanted from WT mice. These findings motivated us to test whether deletion of Ink4c and/or Kip1 might rescue aspects of cerebellar development disrupted by conditional N-Myc deletion.
Results and Discussion
Impaired Postnatal Cerebellar Development in Mice Conditionally Lacking N-Myc.
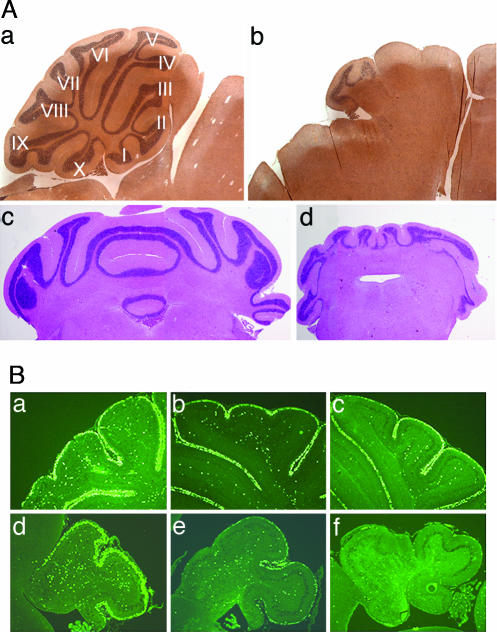
Sagittal and coronal sections prepared at P30 from the cerebella of mice containing “floxed” but functional N-Myc alleles (N-MycFL/FL) and lacking a Nestin-Cre transgene [Nes-Cre(−)] (herein designated WT mice) revealed a normal anatomical pattern consisting of 10 discrete folia (Fig. 1A a and c). In contrast, the cerebella of conditional N-Myc-null mice [N-MycFL/−, Nes-Cre(+)] were much smaller, displaying a significant reduction in cerebellar mass (see Table 1) and exhibiting a reduced number of folia, with the rostral cerebellum completely lacking an identifiable IGL (Fig. 1Ab). Coronal sections revealed empty spaces within the caudal IGL that would normally have contained a continuous zone of cells (Fig. 1A d vs. c) (14, 15).
Fig. 1.
Effects of conditional loss of N-Myc in the cerebellum. (A) N-Myc-null cerebella have a reduced number of folia. H&E staining of sagittal (a and b) and coronal (c and d) sections of adult cerebellum from WT (a and c) and N-Myc-null (b and d) mice. (B) Proliferation of the EGL is prematurely terminated in conditional N-Myc-null cerebella. In vivo BrdU labeling from a 2-h pulse were followed by immunostaining with anti-BrdU in P8 (a and d), P10 (b and e), and P12 (c and f) cerebella from WT (a–c) and N-Myc-null (d–f) mice. (Magnifications: adult brain, ×2; P8–P12 cerebellum, ×10; both adjusted digitally for presentation.)
Table 1.
Cerebellar and total-brain mass in adult mice (P30 or older) of various genotypes
| Genotype | Brain mass, mg* | Cerebellar mass, mg* | Ratio† | Relative increase in cerebellar mass‡ |
|---|---|---|---|---|
| N-Myc+/+, Kip1+/+ | 531 | 59 | 0.111 | 1.00 |
| N-Myc+/+, Kip1+/− | 602 | 81 | 0.134 | 1.21 |
| N-Myc+/+, Kip1−/− | 623 | 83 | 0.133 | 1.20 |
| N-Myc−/−, Kip1+/+ | 273 | 16 | 0.059 | 0.53 |
| N-Myc−/−, Kip1+/− | 337 | 21 | 0.062 | 0.56 |
| N-Myc−/−, Kip1−/− | 398 | 26 | 0.065 | 0.59 |
| N-Myc+/+, Ink4c+/+ | 425 | 57 | 0.134 | 1.00 |
| N-Myc+/+, Ink4c+/− | 456 | 61 | 0.133 | 1.00 |
| N-Myc+/+, Ink4−/− | 448 | 64 | 0.142 | 1.06 |
| N-Myc−/−, Ink4c+/+ | 272 | 16 | 0.059 | 0.44 |
| N-Myc−/−, Ink4c+/− | 270 | 18 | 0.067 | 0.50 |
| N-Myc−/−, Ink4c−/− | 265 | 20 | 0.075 | 0.56 |
| N-Myc+/+, Kip1−/−, Ink4c−/− | 624 | 75 | 0.120 | 1.0 |
| N-Myc−/−, Kip1−/−, Ink4c−/− | 284 | 24 | 0.085 | 0.71 |
*The average weights of the brains and cerebella taken from cohorts of mice of the indicated genotypes are indicated; all standard deviations were <10% of the means.
†Cerebellar mass/brain mass.
‡Comparisons of relative ratios with N-Myc+/+ phenotypes assigned a value of 1.00 for each cohort.
Because conditional N-Myc inactivation leads to the precocious up-regulation of CDK inhibitors in the cerebellar primordium (7), we reasoned that this might limit the pool of embryonic neuronal progenitors, ultimately shortening the postnatal window for genesis of the organ and resulting in formation of a smaller cerebellum. Indeed, when we counted the number of neural progenitors in the E12.5 rhombic lip and caudal part of the neuroepithelium, the progenitor pool was diminished by ≈40% when N-Myc was disrupted [N-MycFl/+, Nes-Cre(−) = 1,040 ± 24; N-MycFL/FL, Nes-Cre(−) = 1,001 ± 22; N-MycFL/−, Nes-Cre(+) = 671 ± 36]. Because of an absence of suitable markers, it is not known whether different cohorts of embryonic neuroblasts give rise to specific folia within the adult cerebellum, although studies have suggested that GNPs born at different times in the rhombic lip may have predetermined “addresses” that determine their final destination along the surface of the developing cerebellum (15). The development of few folia and absence of the rostral cerebellum in the conditional N-Myc-null setting suggest that the remaining embryonic progenitors preferentially populate only the caudal portion of the developing organ (16).
Quantification of the number of GNPs purified from the EGL of P10 cerebella by a two-step Percoll gradient revealed many fewer GNPs in the N-Myc-null (1 × 106) organ than in those of WT (4–5 × 106) animals. To determine whether the proliferation of residual GNPs in the postnatal cerebellum was also affected by N-Myc inactivation, WT and conditionally N-Myc-null mice at P8, -10, and -12 were injected i.p. with BrdU for 2 h to mark cells in S phase. Their brains were then excised and fixed, and sectioned cerebella were stained with an antibody to BrdU. In WT mice, incorporation of BrdU was detected in GNPs located in the outer layer of the EGL and in scattered astrocytes within the IGL from P8 to -12, although the levels of staining progressively decreased during this interval (Fig. 1B a–c). Despite the fact that N-Myc-null cerebella possessed a greatly reduced number of GNPs within the EGL, these showed similar levels of BrdU incorporation as WT GNPs at P8 (Fig. 1Bd). Immunoblotting of equal quantities of protein extracted from purified GNPs at P6 and -8 showed no differences in the levels of p18Ink4c and p27Kip1 expressed in cells of WT or N-Myc-null mice (data not shown). Similarly, flow cytometric analysis of the DNA content of GNPs purified from P8 cerebella of WT and N-Myc-null mice revealed no significant differences in their cell-cycle distribution, regardless of N-Myc genotype; as expected, the fraction of cells with a 2 N DNA content progressively increased during the P10–P12 interval, consistent with the withdrawal of GNPs from the division cycle (data not shown). However, at these later times, N-Myc-null cerebella exhibited a striking deficiency in proliferating neuronal precursors, with only scattered areas of BrdU-positive GNPs observed at P10 and even fewer at P12 (Fig. 1B e and f). Moreover, we found no evidence of increased βTUBIII, cleaved caspase (in vitro) or TUNEL staining (in vivo) in N-Myc-null GNPs, indicating that these cells are not undergoing apoptosis or premature differentiation (17). Therefore, N-Myc inactivation leads to a reduction in the number of neuronal progenitors in the primordial cerebellum and to the premature exhaustion of proliferative GNPs during postnatal development.
Up-Regulation of c-Myc in N-Myc-Null GNPs.
Although Myc activity is required for G1/S progression during the cell-division cycle (18–20), the proliferation of GNPs within the EGL of N-Myc-null mice at P8 was not noticeably compromised. Cre-mediated N-Myc excision, which is initiated at E9.5 and maximized by E10.5, did not eliminate all GNPs, so some progenitors may have been born earlier, or N-Myc excision could have been incomplete. Alternatively, another Myc gene might compensate for the loss of N-Myc during embryogenesis to allow the birth of some progenitors. Normally, N-Myc RNA expression predominates in the CNS and the peripheral nervous system, whereas c-Myc transcripts are undetectable (21). N-Myc is thought to directly and negatively cross-regulate c-Myc expression (21–23). In N-Myc homozygous mutant embryos, c-Myc is expressed in the neuroepithelium, a site where it is not normally detected (21). Enforced overexpression of c-Myc in neural progenitor cells promotes their proliferation (24).
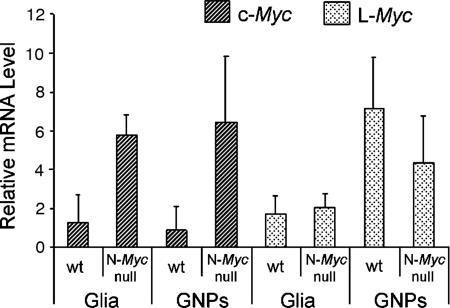
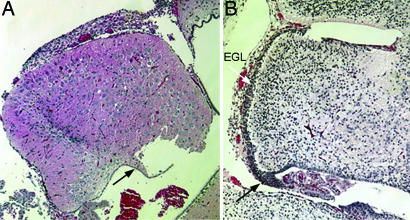
We therefore performed quantitative real-time PCR on total RNA extracted at P7 from GNPs purified on a Percoll gradient, as well as from a less-dense fraction containing glia, Purkinje cells, and large interneurons (Fig. 2). Although the levels of c-Myc were relatively low in cell fractions from WT cerebella, its expression was up-regulated 5- to 6-fold in cells obtained from N-Myc-deficient cerebella (Fig. 2, dark shaded bars). L-Myc was also expressed in GNPs, but its levels were not significantly changed in response to N-Myc disruption (Fig. 2, light shaded bars). Notably, L-Myc-deficient mice have no CNS defects or other obvious phenotypic abnormalities (25). These data are most consistent with the idea that increased c-Myc expression might partially compensate for N-Myc loss. To test this hypothesis directly, we generated mice in which both the N-Myc and c-Myc genes were flanked by lox sites and then conditionally inactivated the two genes simultaneously by using Nes-Cre. Although deletion of c-Myc alone had no effect on cerebellar mass relative to whole-brain mass (P.K. and R.N.E., unpublished data), the cerebella of doubly Myc-deficient mice at P21 were significantly smaller than those from conditional N-Myc-null animals and sustained a nearly complete failure of cerebellar development. Fig. 3 shows a comparison at equal magnification of the doubly Myc-deficient cerebellum at P23 (Fig. 3A) with that of the N-Myc-null organ at E17.5 (Fig. 3B), which, at this stage, exhibited only a small reduction in mass relative to the WT E17.5 cerebellum. The mature doubly Myc-deficient cerebellum was about the same size as that of the single N-Myc-deficient E17.5 primordial cerebellum and exhibited a strikingly similar, primitive morphology, including an aberrant residual rhombic lip devoid of GNPs. In contrast to the effect on GNPs, disruption of N-Myc, c-Myc, or both genes did not appear to affect the development of Purkinje cell neurons. Indeed, the large cells that continued to populate the P23 cerebella of the doubly Myc-deficient mice stained positively with antibodies to calbindin, a Purkinje cell marker not expressed by GNPs (data not shown). Therefore, although its expression does not restore normal cerebellar development, c-Myc up-regulation contributes specifically to the proliferative potential of N-Myc-null GNPs.
Fig. 2.
c-Myc is up-regulated in N-Myc-null GNPs. Quantitative real-time PCR analysis of c-Myc (dark bars) and L-Myc (light gray bars) amplified from total RNA extracted from Percoll gradient-fractionated cell populations from WT and N-Myc-null P7 cerebella.
Fig. 3.
Coinactivation of c-Myc exacerbates defective cerebellar development in N-Myc-null mice. (A) The adult (P23) cerebellum of doubly Myc-deficient mice lacks virtually all evidence of granule neuron maturation. Most of the cells populating the organ are Purkinje neurons. A residual vestige of the rhombic lip is marked by the black arrow. (B) The embryonic E17.5 cerebellum of the N-Myc-null mouse documented in ref. 7 is shown here at equal magnification. The black arrow indicates the rhombic lip, and the white arrow indicates the developing EGL.
Inactivation of Ink4c and Kip1 Partially Reverses Defects in Cerebellar Development Arising from N-Myc Deficiency.
Because previous results implied that N-Myc might control the number of GNPs by down-regulating the expression of p18Ink4c and p27Kip1 during embryonic stages of cerebellar development (7), we designed a breeding program that would yield conditional N-Myc-null mice also lacking Ink4c, Kip1, or both genes. At maturity, the N-Myc-null brain is ≈2-fold reduced in mass, but its cerebellar mass is disproportionately reduced by ≈3- to 4-fold (Table 1). Disruption of Kip1 in addition to N-Myc attenuated the decrease in overall brain mass, including that of the cerebellum, whereas lesser compensatory effects of Ink4c loss were limited to the cerebellum. These findings point toward a more specific role for p18Ink4c in the formation of the cerebellum, whereas p27Kip1 appears to act more globally throughout the brain.
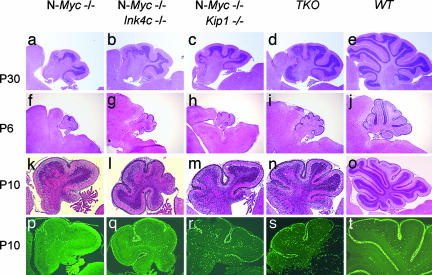
Staining of sections from 1-month-old Ink4c, Kip1, N-Myc triple-null cerebella revealed a partial but significant and reproducible rescue of the size and number of folia compared with matched N-Myc-null cerebella (Fig. 4d vs. a, and Table 1). The sizes of the cerebella of P6 mice were increased when Ink4c, Kip1, or both were inactivated (Fig. 4 f–i). Although the EGL was not intact along the surface of the N-Myc-null P10 cerebellum (Fig. 4k), a complete and thicker EGL was present on the cerebellar surface of N-Myc-null mice lacking Ink4c, Kip1, or both (Fig. 4 l–n). Although inactivation of Ink4c and Kip1 in the N-Myc-null setting improved cerebellar mass and foliation, defects were still apparent when compared with WT cerebella at similar stages of development (Fig. 4 e, j, and o). In vivo BrdU injections, followed by immunostaining, confirmed that loss of Ink4c and Kip1 prolonged proliferation within the EGL of P10 cerebella from N-Myc-null mice (Fig. 4 p–s).
Fig. 4.
Inactivation of Ink4c, Kip1, or both, partially rescues cerebellar development in N-Myc-deficient mice. Sagittal sections of cerebella taken at the indicated times of development (indicated to the left of the panels) were stained with H&E (a–o). BrdU incorporation into the cerebella of P10 mice detected by immunostaining is also illustrated (p–t). Genotypes of the different mouse strains are indicated above the panels. (Magnifications: P30, ×2; P6, ×2; P10, ×10; all adjusted digitally for presentation.)
Because Ink4c or Kip1 inactivation provides otherwise WT GNPs with an increased proliferative potential in culture (8, 9), and because previous studies suggested epistasis between Myc and these CDK inhibitors (7), we tested whether the same would hold true for GNPs lacking functional N-Myc. We assessed the proliferation in culture of GNPs purified by a two-step Percoll gradient from the P8 cerebella of mice with different genotypes. Equal numbers of cells were cultured for 2 days in the presence of Shh, and proliferation was measured by the percentage of cells incorporating BrdU after a pulse during the last 6 h in culture. The net incorporation of BrdU at this later time (approximately equivalent to the P10 state in vivo) provides a measurement of the relative fraction of cells that have remained in cycle.
A high percentage (45.2 ± 2.6%) of WT cells incorporated BrdU under these conditions, whereas the relative S-phase fraction of GNPs lacking N-Myc was markedly reduced (12.2 ± 2.2% BrdU-positive cells). In contrast, when Ink4c or Kip1 were lost, equal numbers of GNPs lacking N-Myc exhibited an increased proliferative potential (19.0 ± 7.4% and 19.2 ± 5.1%, respectively). Coinactivation of both CDK inhibitors further increased BrdU incorporation to 33.1% ± 2.1%. The observed differences between the levels of BrdU incorporated into GNPs of N-Myc-null and triple-null (TKO) mice, and between TKO and WT mice, are highly significant (both P < 0.0002, t test). Thus, the loss of Ink4c, Kip1, or both prolongs the time that Shh-stimulated N-Myc-null cells remain in cycle when cultured ex vivo.
In summary, a primary effect of N-Myc inactivation occurs during embryonic development when the number of neuroepithelial progenitors in the cerebellar anlage is reduced, and both p18Ink4c and p27Kip1 are ectopically up-regulated (7). N-Myc loss also limits the proliferative response of postnatal GNPs to Shh, further restricting the duration of the proliferative phase in the EGL, perturbing foliation, and leading to a dramatic reduction in cerebellar mass. However, conditional inactivation of N-Myc is compensated in part by the expression of c-Myc, which is normally absent in proliferating postnatal GNPs. In turn, disruption of Ink4c and Kip1 in an N-Myc-null background increased the number of folia and the overall mass of the organ. Although, this rescue of N-Myc deficiency was by no means complete, it is remarkable in the sense that the gene is a regulator of global chromatin remodeling (26) that affects the expression of many polymerase I, II, and III transcripts (26–28). Among N-Myc’s many effects is its ability to drive cell-cycle progression mediated not only by down-regulation of CDK inhibitors, but also through induction of D-type cyclins and CDKs (29) and by activation of many other genes governing cellular energy metabolism and growth (30). Interestingly, previously reported attempts to rescue proliferative function in Myc-depleted cells by introducing libraries of other genes or by Kip1 deletion have been unsuccessful (31, 32). Our results now provide direct genetic evidence that expression of N-Myc and concomitant down-regulation of Ink4c and Kip1 contribute to proper development of the cerebellum.
Materials and Methods
Mouse Husbandry.
N-Myc-null mice were generated by crosses between N-MycFL/FL and Nes-Cre transgenic animals as described (7). These mice were then bred with Ink4c-null (33), Kip1-null (10), or c-MycFL/FL (34) mice to generate compound-mutant animals, taking into account the fact that Kip1-null females are sterile and that Ink4c-Kip1 double-null mice develop pituitary gland tumors early in life (see Fig. 5, which is published as supporting information on the PNAS web site). Mouse genotyping was performed as reported for N-Myc-null (7), Ink4c-null (30), Kip1-null (10), and c-Myc-null (34) mice. N-MycFL/+ mice appear phenotypically identical to N-MycFL/FL or N-Myc+/+ strains.
Histology and in Vivo BrdU Immunostaining.
Labeling of GNPs in vivo was performed by i.p. injection of BrdU (40 μl of 5 mg/ml BrdU per g of mouse body weight) for 2 h. Brains were harvested and fixed overnight at 4°C in 4% paraformaldehyde. Brains were cut in half, and both sides were embedded in paraffin to obtain sagittal sections. For coronal sectioning, only the cerebellum was embedded. Sections of 5 μM were stained with H&E or with an antibody to BrdU as described (8).
Purification of GNPs and in Vitro Proliferation Assays.
Cerebellar granule cells were prepared by using a protocol (35) modified as described (8). Briefly, postnatal cerebella were triturated into single-cell suspensions that were loaded onto a two-step Percoll gradient (Sigma). Granule cells sedimenting in a dense fraction were further enriched by panning on tissue-culture plastic dishes, and their purity (>90%) was assessed by immunostaining with markers of neurons and glia. Cells (3 × 105 per well) were plated in eight-well Lab-Tek chamber slides precoated with 100 μg/ml poly d-lysine (Sigma) and Matrigel (Beckton Dickinson) in neural basal medium supplemented with 0.45% glucose, SPITE (Sigma), oleic acid albumin/linoleic acid (Sigma), B27 (Invitrogen), N-acetyl cysteine (Sigma) and 16 μg/ml Shh. Identification of purified granule cells in S phase was performed by incubation with BrdU (Amersham Pharmacia Biosciences) during the last 6 h of a 48-h tissue culture period before fixation. Fixed cells were processed as described (8), stained overnight at 4°C with a polyclonal antibody to BrdU (Bu-20 A, 1:250 dilution; Santa Cruz Biotechnology), and detected with secondary goat antibody to mouse IgG conjugated to Cy3 (1:200 dilution for 30 min; Jackson ImmunoResearch). Stained slides covered with mounting medium containing DAPI (Vector Laboratories) were enumerated under a microscope (500 cells per culture). The results reflect the averages of five independent experiments (each with duplicate cultures) performed with WT and N-Myc-null cells, eight experiments with N-Myc/Ink4c double-null cells, and three experiments each with N-Myc/Kip1 double- and triple-null cells.
Real-Time PCR.
Total RNA from the cerebella of WT and N-Myc-null mice at P7 was extracted by using Trizol (Invitrogen). Quantitative real-time PCR was performed by using a 7900HT Sequence Detection System (Applied Biosystems) and the TaqMan One Step PCR Master Mix Reagents kit (Applied Biosystems). Total RNA extracted from WT cerebella was used to generate standard curves for relative quantification, and 18S ribosomal RNA assay reagents (Applied Biosystems) were used as an internal standard. Oligonucleotide primers and a TaqMan probe for each gene were designed by using Primer Express version 2.0 software (Applied Biosystems). Primer/TaqMan probe sets for murine genes were c-Myc, 5′-GCCCCTAGTGCTGCATG (forward), 5′-CCACAGACACCACATCAATTTCTT (reverse), and 5′-CACCACCAGCAGCGACTCTGAAGAAG (TaqMan probe); L-Myc, 5′-CAGGCCTGCTCCGGGT (forward), 5′-TCCACGGTCACCACGTCA (reverse), and 5′-AGAGCCCCAGCGATTCTGAAGGTGAAG (TaqMan probe). Real-time PCR was performed with 50 ng of total RNA of each sample in triplicate reactions in a 50-μl volume containing 100 nM primers and 50 nM probe. Cycling conditions were 25°C for 10 min, 48°C for 30 min, and 95°C for 10 min, followed by a 40-cycle amplification of 95°C for 15 s and 60°C for 1 min. Data were analyzed by using SDS version 2.0 software (Applied Biosystems) and normalized to the internal 18S rRNA level (36).
Supplementary Material
Acknowledgments
We thank Rose Mathew for purification of cerebellar granule neurons and proliferation assays; Olivier Ayrault for providing immunoblots of proteins expressed in purified GNPs; Marie Assem for BrdU immunohistochemistry; Robert Jenson, Shelly Wilkerson, and Deborah Yons for excellent technical assistance with mouse colony management and genotyping; Adrianna Nance for preparing paraffin sections; and Mary E. Hatten for helpful advice. This work was supported, in part, by a Children’s Brain Tumor Foundation grant (to M.F.R.); National Institutes of Health Grants CA-96832, CA-72907 (both to M.F.R.), CA-20525 (to R.N.E.), and K01-CA-114400 (to P.S.K.); Cancer Core Grant CA-21765; and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital. C.J.S. is an Investigator of the Howard Hughes Medical Institute, and R.N.E. is an American Cancer Society Research Professor.
Abbreviations
- CDK
cyclin-dependent kinase
- En
embryonic day n
- EGL
external germinal layer
- GNP
granule neuron progenitor
- IGL
internal germinal layer
- Nes
Nestin
- Pn
postnatal day n
- Shh
sonic hedgehog.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Miale I. L., Sidman R. L. Exp. Neurol. 1961;4:277–296. doi: 10.1016/0014-4886(61)90055-3. [DOI] [PubMed] [Google Scholar]
- 2.Goldowitz D., Hamre K. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 3.Wang V. Y., Zoghbi H. Y. Nat. Rev. Neurosci. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- 4.Hatten M. E. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- 5.Kenney A. M., Cole M. D., Rowitch D. H. Development (Cambridge, U.K.) 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 6.Kenney A. M., Widlund H. R., Rowitch D. H. Development (Cambridge, U.K.) 2003;131:217–228. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- 7.Knoepfler P. S., Cheng P. F., Eisenman R. N. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uziel T., Zindy F., Xie S., Lee Y., Forget A., Magdaleno S., Rehg J. E., Calabrese C., Solecki D., Eberhart C. G., et al. Genes Dev. 2005;19:2656–2667. doi: 10.1101/gad.1368605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyazawa K., Himi T., Garcia V., Yamagishi H., Sato S., Ishizaki Y. J. Neurosci. 2000;20:5756–5763. doi: 10.1523/JNEUROSCI.20-15-05756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fero M. L., Rivkin M., Tasch M., Porter P., Carow C. E., Firpo E., Polyak K., Tsai L.-H., Broudy V., Perlmutter R. M., et al. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 11.Kiyokawa H., Kineman R. D., Manova-Todorova K. O., Soares V. C., Hoffman E. S., Ono M., Khanam D., Hayday A. C., Frohman L. A., Koff A. Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama K., Ishida N., Shirane M., Inomata A., Inoue T., Shishido N., Horii I., Loh D. Y. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 13.Franklin D. S., Godfrey V. L., Lee H., Kovalev G. I., Schoonhoven R., Chen-Kiang S., Su L., Xiong Y. Genes Dev. 1998;12:2899–2911. doi: 10.1101/gad.12.18.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S., Hillman D. E. Brain Res. 1988;468:201–212. doi: 10.1016/0165-3806(88)90132-0. [DOI] [PubMed] [Google Scholar]
- 15.Jensen P., Smeyne R., Goldowitz D. J. Neurosci. 2004;24:2202–2211. doi: 10.1523/JNEUROSCI.3427-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millen K., Wurst W., Herrup K., Joyner A. Development (Cambridge, U.K.) 1994;120:695–706. doi: 10.1242/dev.120.3.695. [DOI] [PubMed] [Google Scholar]
- 17.Knoepfler P. S., Zhang X.-Y., Cheng P. F., Gafken P. R., McMahon S. B., Eisenman R. N. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenman R. N. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Z. Q., Hurlin P. J. Trends Cell Biol. 2001;11:S10–S14. doi: 10.1016/s0962-8924(01)02121-3. [DOI] [PubMed] [Google Scholar]
- 20.Adhikary S., Eilers M. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 21.Stanton B. R., Perkins A. S., Tessarollo L., Sassoon D. A., Parada L. F. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 22.Dildrop R., Zimmerman K., DePinho R. A., Yancopoulos G. D., Tesfaye A., Alt F. W. Curr. Top. Microbiol. Immunol. 1988;141:100–109. doi: 10.1007/978-3-642-74006-0_14. [DOI] [PubMed] [Google Scholar]
- 23.Rosenbaum H., Webb E., Adams J. M., Cory S., Harris A. W. EMBO J. 1989;8:749–755. doi: 10.1002/j.1460-2075.1989.tb03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fults D., Pedone C., Dai C., Holland E. C. Neoplasia. 2002;4:32–39. doi: 10.1038/sj.neo.7900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatton K. S., Mahon K., Chin L., Chiu F. C., Lee H. W., Peng D., Morgenbesser S. D., Horner J., DePinho R. A. Mol. Cell. Biol. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Roman N., Grandori C., Eisenman R. N., White R. J. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 27.Arabi A. S., Wu S., Shiue C., Ridderstrale K., Larsson L.-G., Wright A. P. H. Nat. Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 28.Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., White R. J. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 29.Ciemerych M. A., Kenney A. M., Sicinska E., Kalaszczynska I., Bronson R. T., Rowitch D. H., Gardner H., Sicinski P. Genes Dev. 2002;16:3277–3289. doi: 10.1101/gad.1023602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel J. H., Loboda M. K., Showe L. C., McMahon S. B. Nat. Rev. Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 31.Berns K., Hijmans E. M., Koh E., Daley G. Q., Bernards R. Oncogene. 2000;19:3330–3334. doi: 10.1038/sj.onc.1203639. [DOI] [PubMed] [Google Scholar]
- 32.Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martine G. R., Bishop J. M. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 33.Latres E., Malumbres M., Sotillo R., Martin J., Ortega S., Martin-Caballero J., Flores J. M., Cordon-Cardo C., Barbacid M. EMBO J. 2000;19:3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Alboran I. M., O’Hagan R. C., Gartner F., Malynn B., Davidson L., Rickert R., Rajewsky K., DePinho R. A., Alt F. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 35.Hatten M. E., Gao W. Q., Morrison M. E., Mason C. A. In: Culturing Nerve Cells. Banker G., Goslin K., editors. Cambridge, MA: MIT Press; 1998. pp. 419–459. [Google Scholar]
- 36.Lee Y., Miller H. L., Jensen P., Hernan R., Connelly M., Wetmore C., Zindy F., Roussel M., Curran T., Gilbertson R., et al. Cancer Res. 2003;63:5428–5437. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.