Abstract
Cyclin D1 promotes nuclear DNA synthesis through phosphorylation and inactivation of the pRb tumor suppressor. Herein, cyclin D1 deficiency increased mitochondrial size and activity that was rescued by cyclin D1 in a Cdk-dependent manner. Nuclear respiratory factor 1 (NRF-1), which induces nuclear-encoded mitochondrial genes, was repressed in expression and activity by cyclin D1. Cyclin D1-dependent kinase phosphorylates NRF-1 at S47. Cyclin D1 abundance thus coordinates nuclear DNA synthesis and mitochondrial function.
Keywords: mitochondria, phosphorylation
Mitochondria function as central components of mammalian cellular survival and through production of ATP and reoxidized NAD and govern cell death through mitochondrial membrane-dependent cellular death signals. The mammalian cell contains 103 to 104 copies of mtDNA, 2 rRNA, and 22 tRNAs required for mitochondrial protein synthesis. Nuclear-encoded genes regulate mitochondrial biogenesis and functions, respiratory genes, the citric acid cycle and upstream glycotic steps, and amino acid metabolism (1). Mitochondrial transcription factor A (mtTFA; also known as Tfam) and nuclear respiratory factors (NRFs) (2, 3) regulate nuclear genes governing mitochondrial function and promoters within the mitochondrial D loop region, thereby promoting replication and transcription of mtDNA. mtTFA is essential for yeast mtDNA maintenance (4), and mtTFA−/− mice have reduced mitochondrial respiratory chain function with depleted mtDNA and oxidative phosphorylation (2).
NRF-1 increases mitochondrial respiratory capacity, induces expression of a subset of genes governing mitochondrial activity in a cell-type specific manner (reviewed in ref. 5), and enhances mitochondrial responses to the peroxisome proliferator activated receptor γ coactivator PGC-1 (6). NRF-1-independent mitochondrial gene expression pathways are regulated by peroxisome proliferator activated receptors, Sp 1, and other factors. Although mitochondrial gene function is codependent upon the nuclear genome and mitochondrial activity alters nuclear gene expression (“retrograde communication”; ref. 7), the mechanism coordinating nuclear DNA synthesis, growth responses, and mitochondrial synthesis is unknown.
The cyclin D1 gene encodes a labile growth factor- and oncogene-inducible component of the holoenzyme that phosphorylates and inactivates the retinoblastoma protein (pRb), promoting nuclear DNA synthesis (8). Cyclin D1−/− mouse embryo fibroblasts (MEFs) have a normal cell size and reduced DNA synthesis rates, which are rescued by cyclin D1 (ref. 9 and data not shown). Cyclin D1−/− mice are resistant to oncogene-induced tumors of the skin, gut, and mammary gland (10, 11). We examined the possibility that cyclin D1 may function to regulate mitochondrial activity and, thereby, couple biosynthetic priorities between the nucleus and mitochondria.
Results
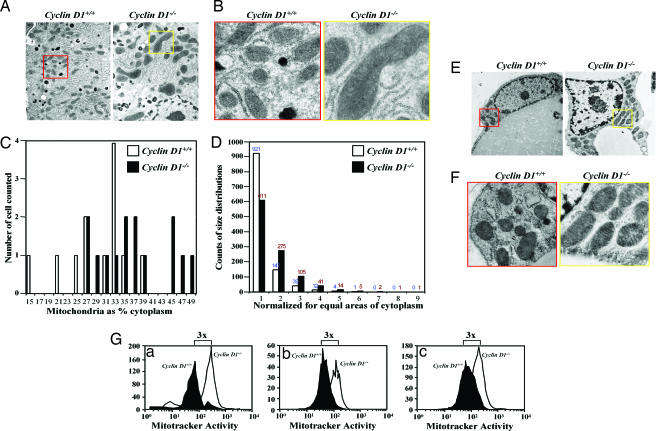
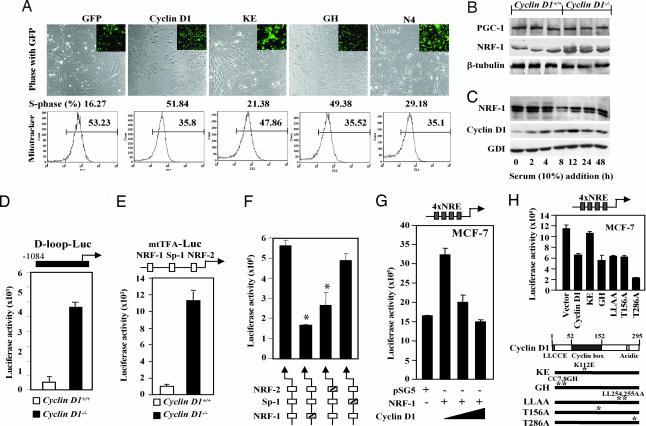
To explore the role of cyclin D1 in mitochondrial gene function, we examined the mitochondrial size in cyclin D1−/− and littermate wild-type control animals. Mitochondrial size was increased 2- to 3-fold as a proportion of cellular cytoplasm in hepatocytes (Fig. 1A–D) and mammary gland adipocyte (Fig. 1 E and F) in cyclin D1−/− mice. MitoTracker Deep Red 633, used to stain the functioning mitochondria in living cells through a fluorescent signal that is proportional to the activity of mitochondria, showed increased mitochondrial activity in cyclin D1-deficient MEFs, hepatocytes, and bone marrow macrophages compared with cyclin D1-expressing cells from littermate control animal (Fig. 1G). The cyclin D1 wild-type and mutant cDNAs were transduced by retroviral infection into cyclin D1−/− MEFs in the MCSV-IRES-GFP vector to allow GFP sorting (Fig. 2A). Mitochondrial function, assessed by using MitoTracker (6), was conducted 1 week after viral transduction. Cyclin D1 inhibited mitochondrial function 52% (Fig. 2A) and induced DNA synthesis (16% to 52%). The pRb-binding-deficient (GH) and C-terminal deletion (N4) of cyclin D1 repressed mitochondrial function, whereas the Cdk-binding-defective KE mutant failed to inhibit mitochondrial activity or induce DNA synthesis, indicating that the kinase-associated function of cyclin D1 inhibits mitochondrial function.
Fig. 1.
Cyclin D1-deficiency enhances mitochondrial size and function. (A and B) Transmission electron microscopic (TEM) images of hepatocytes from liver tissue of cyclin D1+/+ (Left; red box, enlarged area) and cyclin D1−/− (Right; yellow box, enlarged area) show increased mitochondria size in cyclin D1−/−. Catalase-positive peroxisomes (dark spherical structures) are evident in A. (Magnification: B, ×5,000.) (C and D) Mitochondrial size is increased as a proportion of cellular cytoplasm by stereoscopy in cyclin D1−/− and in cyclin D1+/+ hepatocytes. (E) TEM of mammary gland adipocyte with enlarged view (F) revealing increased mitochondria size. (G) Mitochondrial activity in hepatocytes (Ga) and bone marrow macrophages (Gb) and MEFs (Gc) derived from either cyclin D1+/+ or cyclin D1−/− assessed by using MitoTracker (Deep Red, 50 nM).
Fig. 2.
NRF-1 activity is enhanced in cyclin D1-deficient cells. (A) Vectors encoding cyclin D1 and mutants in the vector MSCV-IRES-GFP were used to transduce cyclin D1−/− MEFs for 1 week. Cells were stained with Mitotracker Red CMX Ros for 30 min. The GFP-positive cells were sorted, and MitoTracker fluorescence and cell cycle were analyzed by flow cytometry. The cyclin D1 K112 residue is required for inhibition of mitochondrial activity and induction of DNA synthesis. (B and C) MCF-7 cells were starved in DMEM supplemented with 0.2% FBS for 48 h. Cells were harvested at time points as indicated. Total cell lysates were prepared and subjected to Western blotting analysis to detect cyclin D1 and NRF-1 expression. β-tubulin and GDI were included as loading controls. (D–F) Cyclin D1+/+ and cyclin D1−/− 3T3 cells were transfected with promoters driving luciferase reporter genes for wild-type mtTFA, D loop (D), CMV (E), or mtTFA promoter reporter plasmids encoding mutant response elements for NRF-1, Sp-1, or NRF-2 (F). The luciferase activity is mean ± SEM (n ≥ 6). (G) MCF-7 cells were transfected in 24-well plates with 1 μg of a luciferase reporter gene containing four copies of NRF-1 response elements, 0.5 μg of pSG5 vector or pSG5-NRF-1 expression plasmid, and cyclin D1 expression vector. Luciferase activity is normalized to a cotransfected pRL-TK Luc control. (H) Mutant cyclin D1 expression plasmids were compared for NRF-1 repression function. (mean ± SEM; n > 3 separate experiments each performed in quadruplicate).
In keeping with increased mitochondrial size in cyclin D1−/− hepatocytes, abundance of NRF-1 was increased 3-fold in cyclin D1−/− compared with wild-type liver (Fig. 2B). The NRF-1 coactivators, P/CAF and PGC-1 were unchanged (data not shown and Fig. 2B). Addition of 10% serum to starved cells induced cyclin D1 expression and DNA synthesis and reduced NRF-1 (Fig. 2C). The NRF-1 target gene reporters, the D loop reporter and the mtTFA promoter, were 10-fold more active respectively in cyclin D1−/− cells compared with cyclin D1+/+ cells (Fig. 2 D and E), suggesting that cyclin D1 inhibits D loop transcriptional activity and mtTFA activity. Mutation of the mtTFA promoter NRF-1 or NRF-2, but not the Sp1 site, reduced promoter activity, indicating the enhanced mtTFA promoter activity in cyclin D1−/− cells involves increased NRF-1/NRF-2 activity (Fig. 2F). NRF-1 enhanced 4xNRE-LUC activity 2- to 3-fold in MCF-7 cells, which was inhibited by cyclin D1 expression (Fig. 2G) without affecting expression of NRF-1 from the expression vector (data not shown). Cyclin D1 represses activity of transcription factors, including peroxisome proliferator-activated receptor γ, in a Cdk-independent manner through HDAC recruitment (12). In contrast, cyclin D1 repression of 4xNRE-LUC activity was abrogated by point mutation of the Cdk-binding domain (cyclin D1 KE) (Fig. 2H) but not mutation of the pRb-binding domain (GH), the SRC-1 co-activator binding site (LLAA), or CAK association (T156A) site. The GH mutant induced DNA synthesis, suggesting the reduced pRb-binding affinity of this mutant is insufficient to abrogate the DNA synthesis-promoting function. A T286A mutant, which remains nuclear throughout the cell cycle in cultured cells (13), repressed NRF-1 activity more than cyclin D1 WT. Expression of the cyclin D1 mutants was similar to wild type in cultured cells (12). NRF-1 transactivation function, assessed with a heterologous DNA-binding domain, was repressed by cyclin D1 but not the KE mutant, implicating cyclin D1-dependent kinase function in NRF-1 repression (data not shown).
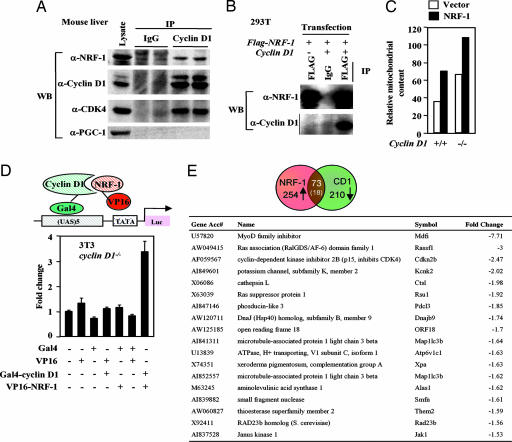
Cyclin D1 associated with NRF-1 in vivo, coprecipitating Cdk4 and NRF-1, but not PGC-1 (12) in immunoprecipitation (IP)-Western blotting of murine liver extracts (Fig. 3A). FLAG-tagged NRF-1, cotransfected into cells with cyclin D1, coprecipitated cyclin D1 (Fig. 3B). NRF-1 WT was inserted into the MSCV-IRES-GFP vectors and used to transduce cyclin D1−/− and cyclin D1+/+ MEFs. NRF-1 increased MitoTracker fluorescence 50% more in cyclin D1-deficient cells (Fig. 3C).
Fig. 3.
Cyclin D1 interacts with NRF-1. (A) Lysates from mouse liver were subjected to cyclin D1 antibody IP. IP products were resolved on SDS/PAGE with Western blot to NRF-1 or PGC-1. Cyclin D1 associates with NRF-1 but not PGC-1 in vivo. (B) HEK293T cells transfected with FLAG-tagged NRF-1 and cyclin D1. Cell lysates were subjected to either IgG or anti-FLAG antibody IP. IP products were resolved on SDS/PAGE followed by Western blot to either FLAG for NRF-1 or cyclin D1. (C) Vectors encoding NRF-1 in the vector MSCV-IRES-GFP were used to transduce cyclin D1−/− and cyclin D1+/[supi]+ MEFs. Cells were stained with MitoTracker Red CMX Ros for 30 min. The GFP-positive cells were sorted, and MitoTracker fluorescence was analyzed by flow cytometry. (D) The Gal4-cyclin D1 and VP16-NRF-1 fusion constructs were transfected with the pG5luc reporter into cyclin D1−/− 3T3 cells. Firefly luciferase activity was quantitated by using the Dual-Luciferase Reporter Assay System (Promega). Interaction between these two proteins (lane 7) increases luciferase activity over the negative controls. (E) Comparison of cyclin D1-regulated genes determined by microarray analysis (available upon request) with genomewide location analysis of candidate NRF-1 target genes comparing genes found on the HU13K and MGU74 chips.
In vivo association of NRF-1 with cyclin D1 was demonstrated by the mammalian two-hybrid system by using GAL4-cyclin D1 and VP16-NRF-1 fusion constructs in cyclin D1−/− 3T3 cells (Fig. 3D). Microarray analysis of cyclin D1−/− MEFs, transduced with a retroviral expression vector for cyclin D1 after 48 h compared with empty vector, identified 210 genes repressed by cyclin D1, 73 of which were similar and 18 identical to the putative NRF-1 site containing gene promoters (Fig. 3E). As NRF-1 induces genes in a cell-type specific manner, we identified NRF-1-responsive genes in MEFs by transducing cells with a retrovirus expressing NRF-1 and conducting microarray. Mitotracker activity was induced 3-fold (data not shown). Of the 131 genes induced by NRF-1 (Fig. 6, which is published as supporting information on the PNAS web site), 22 genes were similar to cyclin D1-repressed genes in MEFs.
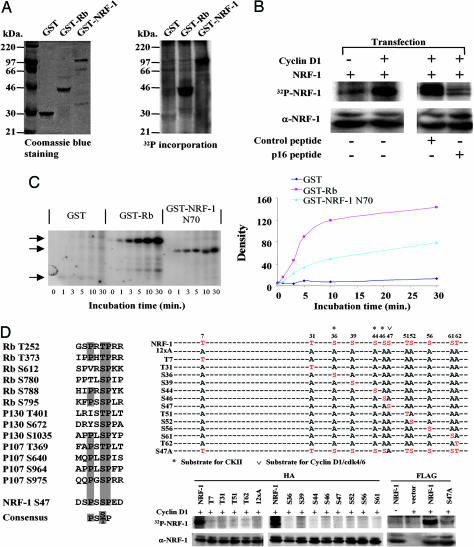
Because the Cdk4 binding-deficient mutant of cyclin D1 failed to repress NRF-1-mediated transcriptional activity and NRF-1 is phosphorylated (14), we compared GST-pRb and GST-NRF-1 as substrates of cyclin D1/Cdk4 kinase. NRF-1 was efficiently phosphorylated by cyclin D1-IP kinase (Fig. 4A). Cyclin D1 cotransfection enhanced 32P-orthophosphate-labeled NRF-1 (Fig. 4B Left). The p16INK4a peptide inhibits cyclin D1-dependent kinase activity and pRb phosphorylation (15, 16). The p16INK4a peptide (P20), D92A, substitution lowers the IC50 (16) and linked to the Antennapedia carrier sequence, inhibiting pRb phosphorylation (ref. 16 and data not shown) and NRF-1 phosphorylation (Fig. 4B). The p16INK4a peptide, P21, (A95/96) with increased IC50 for the inhibition of pRB phosphorylation by cyclin-dependent kinases in vitro (16), failed to inhibit NRF-1 phosphorylation (Fig. 4B). A comparison was made between NRF-1 and pRb as substrates in cyclin D1-dependent kinase assays. The pRb peptide, which contains two sites of phosphorylation, incorporated γ-32P at approximately twice the rate of equal moles of NRF-1 (Fig. 4C). Alignment of the major Cdk4 phosphorylation sites of pRb, p130, and p107 with NRF-1 identified a motif including S47 (Fig. 4D). Point mutation of NRF-1 S47 reduced phosphorylation by cyclin D1-dependent kinase activity 85% (Fig. 4D, right two lanes). To determine whether any of the other serine residues within the carboxyl-terminal fragment of NRF-1, which was sufficient for phosphorylation, could provide rescue function, analysis was conducted of alanine insertion mutants of all 12 candidate phosphorylation sites (12xA), with reintroduction of single wild-type residues into the NRF-1 12xA mutant. The proteins were expressed equally by hemagglutinin-Western blot analysis but served as poor cyclin D1-dependent kinase activity substrates compared with the hemagglutinin-tagged wild-type NRF-1 (Fig. 4D) indicating S47 is necessary but not sufficient for phosphorylation of NRF-1.
Fig. 4.
NRF-1 serves as substrate of cyclin D1-dependent kinase. (A) GST-NRF-1 was incubated with immunoprecipitated cyclin D1/Cdk4 kinase complex in the presence of [γ-32P]ATP. (A Left) Coomassie blue staining of input GST protein. (A Right) γ-32P incorporation into GST-NRF-1. GST and GST-RB were negative and positive control for kinase activity. (B Left) HEK293T cells were transfected with FLAG-NRF-1 with cyclin D1 or control empty vector. (B Right) HEK293T cells transfected with FLAG-NRF-1 and cyclin D1 were treated with p16INK4a peptide (20 μM) corresponding to amino acids 84–103 of the human p16INK4a protein (DAAREGFLATLVVLHRAGAR) with a C-terminal 16-aa Penetratin (RQIKIWFQNRRMKWKK) or control peptide (Biosynthesis, Lewisville, TX) (16). NRF-1 phosphorylation was abrogated by p16INK4a peptide. Cells were pulse-labeled with [γ-32P]orthophosphate. NRF-1 protein was precipitated with anti-FLAG M2 antibody and subjected to autoradiography. (C Left) Equal amounts of either the GST-NRF-1 N70 fusion protein or GST protein were incubated with 200 ng of purified cyclin D1/Cdk4 complex and [γ32P]ATP. The arrows indicate the autoradiogram of the phosphorylated fusion protein. (C Right) GST-Rb serves as a positive control. The phosphorylated bands were quantified by densitometry scanning. (D) Alignment of Cdk4 phosphorylation sites for pRb, p107, p130, and NRF-1. HEK293T cells were transfected with expression vectors encoding hemagglutinin or FLAG-tagged NRF-1 and mutants in which a single potential phosphorylation site was restored in all sites mutated version (12xA) or a single point mutant of S47, together with cyclin D1. NRF-1 and mutant protein were precipitated with anti-hemagglutinin antibody and subjected to autoradiography.
Discussion
The mechanisms integrating cell-cycle progression and/or exit with mitochondrial biogenesis were previously unknown (17, 18). Herein, cyclin D1 repressed mitochondrial function and size in vivo. This previously undescribed function of cyclin D1 represents a bona fide nonnuclear function for cyclin D1. The kinase function of cyclin D1 was required for repression of mitochondrial activity and phosphorylation of NRF-1. Cyclin D1 bound NRF-1 in cells assessed either by immunoprecipitation Western blot analysis or by mammalian 2-hybrid. To provide additional corroborative evidence that cyclin D1 inhibited NRF-1-regulated functions, we determined the molecular genetic signature regulated by cyclin D1 and compared this group of genes with putative NRF-1-regulated genes based on genomewide location analysis of candidate NRF-1 target genes (19). The genes regulated by cyclin D1 were determined by introducing a cyclin D1-expressing retrovirus into cyclin D1−/− MEFs and conducting microarray analysis. A comparison of cyclin D1-repressed genes and genes potentially regulated by NRF-1, based on analysis of NRF-1-binding sites in their promoters, showed substantial overlap (Fig. 3E). We next compared the molecular genetic signature of genes actually regulated by cyclin D1 and those genes actually regulated by NRF-1. The genes regulated by NRF-1 were determined by transducing MEFs with an NRF-1-expressing retrovirus and conducting microarray analysis. Of the 254 candidate NRF-1-inducible genes and 210 genes repressed by cyclin D1, 73 similar genes and 18 identical genes were shared between the two gene sets.
We examined the physiological regulation of cyclin D1 and NRF-1 expression. NRF-1 was increased 3-fold in cyclin D1 null cells. Cyclin D1 levels and NRF-1 expression are inversely correlated during cell cycle progression with serum-induced cyclin D1 expression peaking at 12 h and NRF-1 expression decreasing in 8–12 h. The reduction in NRF-1 abundance at 8 h shortly after cyclin D1 levels increase (Fig. 2C) is consistent with our model in which cyclin D1 inhibits NRF-1 function (Fig. 5). Together these studies identified the genes regulated by NRF-1 and showed significant overlap with cyclin D1-regulated genes. The prior studies of NRF-1 regulation by serum in other cell types showed no change in levels or activity of cytochrome oxidase and several NRF-1 targets within 12 h of serum stimulation in some studies (20). Chromatin IP assays in Cam et al. (19) show no change in NRF-1 binding to NRF-1 sites during serum treatment. At this time, no NRF-1 regulatory serum-inducible kinase has been isolated, and multiple phosphorylation sites exist in NRF-1 likely regulated by distinct kinases. The finding herein that NRF-1 is inactivated by cyclin D1 and phosphorylated by cyclin D1-dependent kinase does not preclude the possibility that NRF-1 may be regulated by other serum-regulated kinases because many proteins are under regulation by distinct kinases regulating proliferative and antiproliferative signals. It remains to be determined whether other cell cycle-regulatory kinases phosphorylate and repress NRF-1.
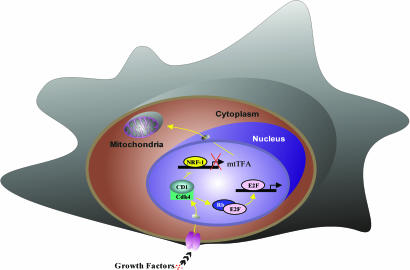
Fig. 5.
Schematic representation of the mechanism by which cyclin D1 inhibits mitochondrial function. Cyclin D1-dependent kinase phosphorylates and inhibits NRF-1 and, thereby, mtTFA and mitochondrial activity. Additional NRF-1-independent mechanisms regulating mitochondrial activity remain to be defined.
Cyclin D1 repressed NRF-1-induced genes by microarray analysis. In addition, cyclin D1 repressed mitochondrial genes that were not considered direct NRF-1 targets as defined by either the presence of putative NRF-1 sites in their promoters or by microarray analysis in MEFs. Cyclin D1 expression repressed mtTFA abundance and promoter activity requiring primarily the NRF-1- and NRF-2-binding sites. mtTFA binds to sequences in the divergent heavy- and light-chain promoters within the mitochondrial D loop region to stimulate transcription from mitochondrial DNA templates. Consistent with the finding that cyclin D1 repressed NRF-1 and mtTFA, cyclin D1 inhibited D loop transcriptional activity. mtTFA promotes differentiation in myoblasts (21–23), and cyclin D1 inhibits differentiation of myocytes (24) and adipocytes (12). Cyclin D1 antagonizes NRF-1 and mitochondrial function while inducing DNA synthesis, thereby potentially coupling biosynthetic priorities within the cell.
In this study, we proposed that cyclin D1 may integrate cell-cycle progression and/or exit with mitochondrial biogenesis (Fig. 5). Increased abundance of cyclin D1 correlates with reduced mitochondrial activity. The outcome of reduced mitochondrial activity would be anticipated to shift glucose metabolism toward cytosolic glycolysis. Whether such a triage of substrate utilization or metabolic prioritization occurs in the presence of increased cyclin D1 abundance remains to be determined. A shift toward cytosolic glycolysis is known to occur during tumor progression and is a component of the metabolic change described by O. Warberg in 1930. Known components of the mitochondria-to-nucleus retrograde pathway include CREB, mRpL12, and aconitase (which produces α-ketoglutarate) (25–27). In Drosophila, in which proliferation rates are partially uncoupled from growth, cyclin D1 induces cell size and Hif-1 prolyl hydroxylase (HPH) activity (25). But in mammalian cells, HPH is not required for proliferation by cyclin D1/Cdk4, mitochondria inhibit HPH hydroxylase, and cyclin D1 does not affect cell size (28, 29). Mitochondria produce ATP (30), regulate single-carbon metabolism, fatty acid metabolism, and oxidative glycolysis, (5) and are a potential target for cancer therapies (31). Because cyclin D1 is inhibited by differentiation and induced by oncogenes, this previously undescribed function of cyclin D1 may provide a mechanism by which select oncogenes and growth factors contribute to tumor maintenance.
Materials and Methods
Mice.
Mice homozygously deleted of the cyclin D1 gene (cyclin D1−/−) (32) on a mixed C57BL/6 × 129/SvJ background were maintained as described in ref. 9.
Cell Culture.
Day 5 bone marrow macrophage from cyclin D1+/+ and cyclin D1−/− mice were prepared as described in ref. 34 and cultured in supplemented α-modified MEM (α+MEM) (Life Technologies, Gaithersburg, MD) containing 15% FBS (Life Technologies) and 120 ng/ml human recombinant CSF-1 (gift of Chiron, Emeryville, CA). MCF-7 and HEK293T cells were maintained in DMEM containing penicillin and streptomycin (100 mg of each per liter) and supplemented with 10% FBS.
Primary MEFs were isolated by following the protocol described in ref. 33 from day 14 post coitus mouse embryos. Briefly, embryos were separated and minced and then incubated in a solution with 0.05% trypsin and 1 mM EDTA at 37°C. The supernatant was collected by centrifugation at 1,000 × g for 3 min.
Plasmids, Transfections, and Reporter Assays.
The expression vectors pCMV-cyclin D1, CMV-cyclin D1-KE, CMV-cyclin D1 GH, pCMV-cyclin E, pCMV-cyclin A, and RSV- and CMV-Renilla luciferase reporter were described in ref. 34. The human cyclin D1 mutants were derived by PCR-directed amplification by using sequence-specific primers and cloned into pRC/CMV. The reporter plasmids NRF4-LUC containing four tandem NRF-1 sites (37), mtTFA-RC4wt/PGL3 (38), reporters with NRF-1, NRF-2, or Sp1 mutation (6), and the expression vector pSG5 NRF-1 (37) were described previously. Wild-type and mutant cyclin D1 and NRF-1 cDNA fragment were inserted into the EcoRI site of pMSCV-IRES-GFP vector to make pMSCV-IRES-GFP-cyclin D1 and pMSCV-IRES-GFP-NRF-1.
Mammalian two-hybrid was performed by following manufacturer’s instructions (Promega, Madison, WI). Cyclin D1 cDNA was cloned into pBind vector, and NRF-1 cDNA was cloned into pACT vector to generate fusion proteins with the DNA binding domain of GAL4 (Gal4-cyclin D1) and the activation domain of VP16 (VP16-NRF-1), respectively. The pG5luc vector contains five GAL4 binding sites upstream of a minimal TATA box that, in turn, is upstream of the firefly luciferase gene.
DNA transfection and luciferase assays were performed as described in ref. 38.
Cytochemistry and Electron Microscopy.
Liver and mammary gland from cyclin D1 WT and cyclin D1−/− mice were removed under anesthesia. Slices (≈2 mm thick) were prepared by hand cutting with a razor and were immediately placed into cold fixative consisting of a mixture of 4% paraformadehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 3 h with continuous shaking. Nonfrozen sections (≈20 μm) of the aldehyde fixed tissue slices were prepared by using a Lancer vibratome sectioning instrument (Polysciences, Warrington, PA). Sections were tested for catalase activity by using an incubating medium containing 3,3′-diaminobenzidine tetrahydrochloride (Sigma, St. Louis, MO) as substrate at pH 9.7 to visualize peroxisomes and microperoxisomes. The incubated sections then were fixed in 1% osmium tetraoxide (Polysciences), alcohol dehydrated, and embedded in Epon. Ultrathin sections (≈500 Å) were prepared by using an LKB ultramicrotome (LKB), stained with lead citrate, and examined with a Philips 300 electron microscope (39). All pictures were taken at the same magnification, i.e., at ×5,000. To determine mitochondrial size and number, stereology was performed according to the method of Weibel (12). A grid of ≈1 cm square was placed over each print photo (magnification of ×13,500), and mitochondria within the grid were counted.
FACS Analysis.
Bone marrow macrophages, hepatocytes, and MEFs derived from cyclin D1+/+ and cyclin D1−/− mice were incubated in medium containing 50 nM of MitoTracker Deep Red 633 (Molecular Probes) for 30 min at 37°C. Cells were then trypsinized and resuspended in PBS. The MitoTracker fluorescence of these positive cells was analyzed on a flow cytometry.
Western Blot.
The antibodies used in Western blot analysis were cyclin D1 antibody Ab3 (NeoMarkers, Fremont, CA), cyclin D1 antibody 72–13G, PGC-1 (H-300) (Santa Cruz Biotechnology, Santa Cruz, CA), and NRF-1 (gift of R. C. Scarpulla, Northwestern University, Feinberg School of Medicine, Chicago, IL). Proteins were visualized by the enhanced chemiluminescence system (Amersham Pharmacia Biotech, Piscataway, NJ). The abundance of immunoreactive protein was quantified by using a densitometer (ImageQuant 1.11; Molecular Dynamics Computing Densitometer, Sunnyvale, CA).
Retroviral Infection and FACS Sorting.
Briefly, pMSCV-IRES-GFP was cotransfected with an ecotropic, replication-defective helper virus pSV-ψ—E-MLV into 293T cells by using calcium phosphate precipitation. The retroviral supernatants were harvested 48 h after transfection and filtered through a 0.45-μm filter. cyclin D1−/− MEFs were incubated with fresh retroviral supernatants and 8 μg/ml polybrene at 37°C overnight. After 2 days of culture in DMEM with 10% FBS, GFP-positive cells were sorted by FACStar (Becton Dickinson, Franklin Lakes, NJ).
High-Density Array Expression Analysis.
Total RNA was isolated from retrovirus vector-infected MEFs (infected with either MSCV- IRES-GFP vector or MSCV-IRES-FLAG-NRF-1) by using TRIzol and used to probe GeneChip Mouse Expression Set 430 array (Affymetrix, Santa Clara, CA). RNA quality was determined by gel electrophoresis. Probe synthesis and hybridization were performed according to the manufacturer’s manual (see eukaryotic target preparation section at www.affymetrix.com/support). Three arrays were used for each condition. Analysis of the arrays was performed as described in ref. 40.
In Vivo Labeling.
293T cells were plated 24 h before transfection. Twenty-four hours after transfection, cells were washed, incubated with phosphate-free DMEM for 2 h, and then 32P-orthophosphate was added to the cell culture medium at 1 mCi/ml (1 Ci = 37 GBq). Lysates were prepared and precipitated with M2 antibody for at least 6 h. Beads were washed with six changes of lysis buffer, and protein-bound beads were denatured in SDS buffer. Protein was resolved by SDS/PAGE. Gels were dried and subjected to autoradiography.
Supplementary Material
Acknowledgments
We thank Dr. C. Sherr for thoughtful comments and advice and Dawn Scardino for assistance with manuscript preparation. Support for these studies was from Susan Komen Breast Cancer Foundation Grant BCTR0504227 (to C.W.), National Institutes of Health Grants R01CA70896, R01CA75503, R01CA86072, R01CA93596, and R01CA107382, Kimmel Cancer Center Support Grant P30 CA56036 (to R.G.P.), and the Breast Cancer Alliance through an Exceptional Project Grant (to A.Q.).
Abbreviations
- IP
immunoprecipitation
- MEF
mouse embryo fibroblast
- mtTFA
mitochondrial transcription factor A
- NRF
nuclear respiratory factor.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Shadel G. S., Clayton D. A. Annu. Rev. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 2.Larsson N. G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G. S., Clayton D. A. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 3.Evans M. J., Scarpulla R. C. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 4.Diffley J. F., Stillman B. Proc. Natl. Acad. Sci. USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scarpulla R. C. Biochim. Biophys. Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 6.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 7.Poyton R. O., McEwen J. E. Annu. Rev. Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 8.Sherr C. J. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 9.Albanese C., D’Amico M., Reutens A. T., Fu M., Watanabe G., Lee R. J., Kitsis R. N., Henglein B., Avantaggiati M., Somasundaram K., et al. J. Biol. Chem. 1999;274:34186–34195. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- 10.Yu Q., Geng Y., Sicinski P. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 11.Hulit J., Wang C., Li Z., Albanese C., Rao M., Di Vizio D., Shah S., Byers S. W., Mahmood R., Augenlicht L. H., et al. Mol. Cell. Biol. 2004;24:7598–7611. doi: 10.1128/MCB.24.17.7598-7611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C., Pattabiraman N., Fu M., Zhou J. N., Sakamaki T., Albanese C., Li Z., Wu K., Hulit J., Neumeister P., Novikoff P. M., et al. Mol. Cell. Biol. 2003;23:6159–6173. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alt J. R., Cleveland J. L., Hannink M., Diehl J. A. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gugneja S., Scarpulla R. C. J. Biol. Chem. 1997;272:18732–18739. doi: 10.1074/jbc.272.30.18732. [DOI] [PubMed] [Google Scholar]
- 15.Fahraeus R., Paramio J. M., Ball K. L., Lain S., Lane D. P. Curr. Biol. 1996;6:84–91. doi: 10.1016/s0960-9822(02)00425-6. [DOI] [PubMed] [Google Scholar]
- 16.Fahraeus R., Lain S., Ball K. L., Lane D. P. Oncogene. 1998;16:587–596. doi: 10.1038/sj.onc.1201580. [DOI] [PubMed] [Google Scholar]
- 17.Mancini M., Anderson B. O., Caldwell E., Sedghinasab M., Paty P. B., Hockenbery D. M. J. Cell Biol. 1997;138:449–469. doi: 10.1083/jcb.138.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Wangenheim K. H., Peterson H. P. J. Theor. Biol. 1998;193:663–678. doi: 10.1006/jtbi.1998.0731. [DOI] [PubMed] [Google Scholar]
- 19.Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R. C., Young R., Kluger Y., Dynlacht B. D. Mol. Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 20.Herzig R. P., Scacco S., Scarpulla R. C. J. Biol. Chem. 2000;275:13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- 21.Herzberg N. H., Middelkoop E., Adorf M., Dekker H. L., Van Galen M. J., Van den Berg M., Bolhuis P. A., Van den Bogert C. Eur. J. Cell Biol. 1993;61:400–408. [PubMed] [Google Scholar]
- 22.Korohoda W., Pietrzkowski Z., Reiss K. Folia Histochem. Cytobiol. 1993;31:9–13. [PubMed] [Google Scholar]
- 23.Laeng H., Schneider E., Bolli R., Zimmermann A., Schaffner T., Schindler R. Exp. Cell Res. 1988;179:222–232. doi: 10.1016/0014-4827(88)90361-8. [DOI] [PubMed] [Google Scholar]
- 24.Skapek S., Rhee J., Kim P., Novitch B., Lassar A. Mol. Cell. Biol. 1996;16:7043–7053. doi: 10.1128/mcb.16.12.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frei C., Galloni M., Hafen E., Edgar B. A. EMBO J. 2005;24:623–634. doi: 10.1038/sj.emboj.7600523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X. J., Wang X., Kaufman B. A., Butow R. A. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- 27.Arnould T., Vankoningsloo S., Renard P., Houbion A., Ninane N., Demazy C., Remacle J., Raes M. EMBO J. 2002;21:53–63. doi: 10.1093/emboj/21.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroedl C., McClintock D. S., Budinger G. R., Chandel N. S. Am. J. Physiol. 2002;283:L922–L931. doi: 10.1152/ajplung.00014.2002. [DOI] [PubMed] [Google Scholar]
- 29.Chandel N. S., McClintock D. S., Feliciano C. E., Wood T. M., Melendez J. A., Rodriguez A. M., Schumacker P. T. J. Biol. Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 30.Green D. R., Reed J. C. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 31.Costantini P., Jacotot E., Decaudin D., Kroemer G. J. Natl. Cancer Inst. 2000;92:1042–1053. doi: 10.1093/jnci/92.13.1042. [DOI] [PubMed] [Google Scholar]
- 32.Sicinski P., Donaher J. L., Parker S. B., Li T., Fazeli A., Gardner H., Haslam S. Z., Bronson R. T., Elledge S. J., Weinberg R. A. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 33.Neumeister P., Pixley F. J., Xiong Y., Xie H., Wu K., Ashton A., Cammer M., Chan A., Symons M., Stanley E. R., Pestell R. G. Mol. Biol. Cell. 2003;14:2005–2015. doi: 10.1091/mbc.02-07-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reutens A. T., Fu M., Wang C., Albanese C., McPhaul M. J., Sun Z., Balk S. P., Janne O. A., Palvimo J. J., Pestell R. G. Mol. Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 35.Gugneja S., Virbasius C. M., Scarpulla R. C. Mol. Cell. Biol. 1996;16:5708–5716. doi: 10.1128/mcb.16.10.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Virbasius J. V., Scarpulla R. C. Proc. Natl. Acad. Sci. USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson U., Scarpulla R. C. Mol. Cell. Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C., Fu M., D’Amico M., Albanese C., Zhou J. N., Brownlee M., Lisanti M. P., Chatterjee V. K., Lazar M. A., Pestell R. G. Mol. Cell. Biol. 2001;21:3057–3070. doi: 10.1128/MCB.21.9.3057-3070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novikoff P. M., Cammer M., Tao L., Oda H., Stockert R. J., Wolkoff A. W., Satir P. J. Cell Sci. 1996;109:21–32. doi: 10.1242/jcs.109.1.21. [DOI] [PubMed] [Google Scholar]
- 40.Bouras T., Fu M., Sauve A. A., Wang F., Quong A. A., Perkins N. D., Hay R. T., Gu W., Pestell R. G. J. Biol. Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.