Abstract
Yersinia pestis causes bubonic plague, characterized by an enlarged, painful lymph node, termed a bubo, that develops after bacterial dissemination from a fleabite site. In susceptible animals, the bacteria rapidly escape containment in the lymph node, spread systemically through the blood, and produce fatal sepsis. The fulminant progression of disease has been largely ascribed to the ability of Y. pestis to avoid phagocytosis and exposure to antimicrobial effectors of innate immunity. In vivo microarray analysis of Y. pestis gene expression, however, revealed an adaptive response to nitric oxide (NO)-derived reactive nitrogen species and to iron limitation in the extracellular environment of the bubo. Polymorphonuclear neutrophils recruited to the infected lymph node expressed abundant inducible NO synthase, and several Y. pestis homologs of genes involved in the protective response to reactive nitrogen species were up-regulated in the bubo. Mutation of one of these genes, which encodes the Hmp flavohemoglobin that detoxifies NO, attenuated virulence. Thus, the ability of Y. pestis to destroy immune cells and remain extracellular in the bubo appears to limit exposure to some but not all innate immune effectors. High NO levels induced during plague may also influence the developing adaptive immune response and contribute to septic shock.
Keywords: inducible nitric oxide synthase, reactive nitrogen species
Yersinia pestis, the agent of plague, primarily affects rodents but is also one of the most invasive and virulent bacterial pathogens of humans (1). Bubonic plague, the most common form of the disease in rodents and humans, is usually acquired from the bite of an infected flea. Disease progression and histopathology of plague in the rat closely resemble those of human bubonic plague (2). From an intradermal inoculation site, Y. pestis rapidly disseminates through afferent lymphatic vessels to the regional draining lymph nodes. After entering the subcapsular sinus of the node, the bacteria rapidly multiply and spread throughout the entire parenchyma of the node, resulting in lymphadenitis characterized by a painful, swollen lymph node termed a bubo. Histologically, primary buboes of rats and humans contain extensive masses of extracellular Y. pestis, necrotic lymphoid tissue, hemorrhage and fibrin, and are surrounded by an edematous perinodal capsule (2). Without early antibiotic treatment, bubonic plague usually progresses rapidly to septicemic plague, a form of the disease characterized by bacteremia, systemic spread, and life-threatening Gram-negative sepsis. Hematogenous spread to the lungs can also result in pneumonic plague.
Current models of plague pathogenesis emphasize the importance of the bacterial Type III secretion system (TTSS) in evading innate immunity, the first line of defense against infection (1, 3, 4). The Y. pestis TTSS consists of at least 42 genes arranged in several operons on a 70.5-kb plasmid that is essential for virulence (5). These genes encode virulence proteins, termed Yops, and a delivery system to inject the Yops into eukaryotic cells (3). The translocated Yops disrupt a variety of eukaryotic cell signaling pathways to inhibit phagocytosis, attenuate the phagocytic oxidative burst that generates antimicrobial reactive oxygen species (ROS), down-regulate the normal proinflammatory response, and induce apoptosis of dendritic cells, NK cells, macrophages, and polymorphonuclear neutrophils (PMNs) (3, 4, 6–8). Although substantial, most of the experimental evidence for these models comes from in vitro studies with the less virulent enteric pathogens Yersinia pseudotuberculosis and Yersinia enterocolitica or from studies using attenuated strains of Y. pestis injected directly into the blood stream.
Bubonic plague does not invariably progress to septicemic plague, even in susceptible animals, indicating that the lymph node is the deciding arena of immune response success or failure. To better understand host–pathogen interactions in this key arena, we investigated Y. pestis adaptation to the innate immune response and other host factors in the bubo by in vivo gene expression profiling and virulence testing of select mutants using a previously characterized rat model of bubonic plague (2).
Results
Transcriptional Profile of Y. pestis in the Bubo.
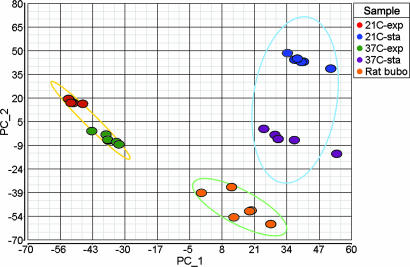
Within a few days after injecting Y. pestis into the dermis of the upper pelvic region, rats develop classic signs of bubonic plague, including a grossly enlarged inguinal lymph node and limping and reluctance to move the associated hind leg. The primary bubo in rats with terminal plague contains 107 to 109 Y. pestis (2), and sufficient bacterial RNA was recovered from the contents of a single inguinal bubo for microarray analysis (Fig. 5, which is published as supporting information on the PNAS web site). To identify bacterial genes that are differentially expressed during disease, we compared the transcriptional profiles of Y. pestis isolated from buboes and from laboratory cultures. The gene expression patterns were reproducible and discrete for each of the different in vivo and in vitro conditions, with the bubonic profile most similar to that of stationary-phase bacteria cultured at 37°C (Fig. 1).
Fig. 1.
Principal component analysis of microarray data from six biological replicate samples for each of the in vitro and in vivo conditions. The first two principal components show the separation of the bubo-specific expression profile from the expression profiles in exponential and stationary-phase cultures grown at 21°C and 37°C. Ellipses encompass exponential, stationary, and rat samples.
Among the Y. pestis genes most highly expressed in the bubo were those for known virulence factors (1), such as the antiphagocytic F1 protein capsule, a plasminogen activator required for systemic invasion from the lymph node, the iron acquisition system encoded on the Y. pestis pathogenicity island, and the TTSS (Table 1, which is published as supporting information on the PNAS web site). The histopathology and transcriptome of Y. pestis in the bubo are consistent with the predicted antiphagocytic activities of the TTSS. Y. pestis is extracellular in the bubo; thus, the bacteria successfully escape internalization and killing by macrophages and PMNs and instead cause necrosis and apoptosis of immune cells in the lymph node (2). Although highly expressed in the bubo and up-regulated compared with in vitro growth at 21°C, expression of the F1 capsule and the TTSS genes was not up-regulated compared with in vitro growth at 37°C. F1 transcription appears to be regulated primarily by temperature, and the TTSS appears to be regulated by temperature and low calcium in vitro and by host cell contact in vivo (1, 3). The high TTSS expression levels we observed in vitro may be due to the fact that the culture medium was not supplemented with calcium. In addition, the TTSS of bacteria within the dense extracellular masses in the bubo may not be maximally activated because they are not in contact with host cells (2).
Evidence for Iron Starvation in the Bubo.
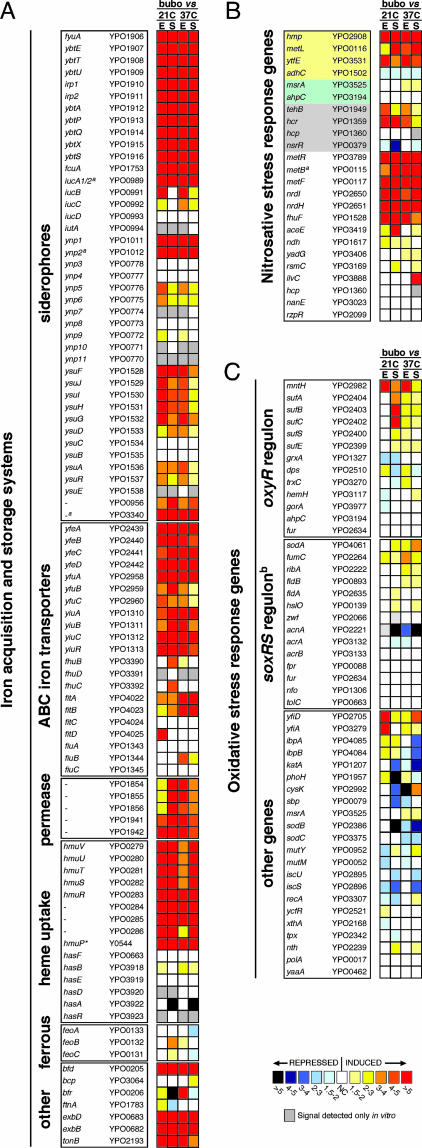
Approximately 13% of Y. pestis genes were differentially expressed in the bubo: 119 were up-regulated and 203 were down-regulated at least 2-fold compared with the four in vitro growth conditions (Tables 2 and 3, which are published as supporting information on the PNAS web site). The in vivo transcriptome indicated that Y. pestis faces iron limitation in the bubo. Expression of 10 of the 15 proven or potential iron or heme transport systems, including three siderophore-based systems, four ATP-binding cassette iron transporters, two iron permeases, and a heme transport system was up-regulated >4-fold (Fig. 2A). Only the ferrous ion uptake system and the ATP-binding cassette-, hemophore-, and siderophore-dependent transport systems Fhu, Has, and Ynp, which are probably nonfunctional in Y. pestis (9), were not up-regulated in the bubo compared with in vitro conditions.
Fig. 2.
Differential expression in the bubo of genes implicated in the response to iron limitation (A) (9), nitrosative stress (B) (13–20), and oxidative stress (C) (22, 39). The mean fold increase or decrease in individual gene expression in the bubo compared with exponential (E) and stationary (S) phase cultures at 21°C and 37°C is indicated by the color scale bar. For the nitrosative stress gene list (B), genes specifically required for resistance to NO and GSNO or to peroxynitrite are highlighted in yellow and green, respectively. Genes highlighted in gray, along with hmp and ytfE, are predicted to be up-regulated in response to RNS by repression of the NO-sensitive negative regulator encoded by nsrR (17, 18); the other genes were shown to be up-regulated at least 2-fold in response to NO or GSNO stress in three different in vitro microarray studies (13–15). The YPO and Y numbers refer to the Y. pestis CO92 and KIM genome annotations, respectively (10, 11). ∗, not annotated in Y. pestis CO92; a, pseudogene; b, the Y. pestis response to superoxide may not occur via SoxRS regulation because no soxRS homologs were identified in the genome (10, 11).
Evidence for Nitric Oxide (NO)-Derived Nitrosative Stress in the Bubo.
A marked number of the genes that have been implicated in the bacterial protective response to the reactive nitrogen species (RNS) NO and nitrosoglutathione (GSNO) were up-regulated in vivo (Fig. 2B). Y. pestis does not encode the NO or GSNO reductases (nor and nrf genes) that many other bacteria use to detoxify RNS (10, 11); but expression of hmp, which encodes a flavoglobin that is induced by NO and confers resistance to RNS in vitro and in macrophages (12–15), increased 10- to 20-fold in the bubo. In addition, metL, which is required for synthesis of the NO antagonist homocysteine and for Salmonella resistance to GSNO (16), and ytfE (dnrN), which is required for Escherichia coli resistance to NO (14), were both up-regulated 3- to 12-fold. Up-regulation of hmp, ytfe (dnrN), and other genes that may be involved in protection against RNS, such as hcp, hcr, and tehB, has recently been attributed to repression of the NO-sensitive negative regulator encoded by yjeB (nsrR) (17, 18). Expression of the Y. pestis nsrR homolog (YPO0379) was down-regulated 1.7- to 4-fold in the bubo compared with stationary-phase, in vitro cultures, and hcr, tehB, hmp, and ytfE (dnrN) were up-regulated (Fig. 2B). Y. pestis homologs of other genes, which have been consistently found to be induced in E. coli by in vitro exposure to RNS (13–15), such as the nrdHIEF ribonucleotide reductase operon, were also up-regulated in the bubo (Fig. 2B).
Localized and Systemic Induction of Inducible NO Synthase (iNOS) During Bubonic Plague.
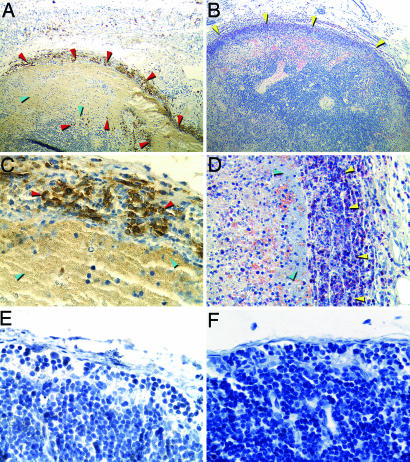
NO-derived antimicrobial activity is generated by macrophages, PMNs, and other host cells by the iNOS in response to infection and is an important component of the innate immune response to intracellular pathogens (19, 20). Both PMNs and macrophages are recruited to the rat bubo (2), and we detected large numbers of iNOS-expressing cells, which appeared to be primarily PMNs, in close association with extracellular masses of Y. pestis in infected lymph nodes (Fig. 3). iNOS induction and RNS production by rat PMNs have previously been shown to occur in vivo after exposure to LPS and IFN-γ (21), a cytokine present at high levels in the serum of rats infected by Y. pestis (2). Serum NO levels also were elevated in rats with bubonic plague (mean, 260 mM/ml NOx; range, 64–605; n = 5) compared with uninfected rats (mean, 74 mM/ml; range, 57–87; n = 5).
Fig. 3.
iNOS expression by PMNs in the bubo. Shown are sections of a bubo (A–D) or uninfected lymph node (E and F) stained to detect iNOS (A, C, and E) or PMNs (B, D, and F; yellow arrowheads). PMNs producing iNOS (dark brown) are indicated by red arrowheads. Masses of extracellular bacteria adjacent to PMNs are indicated by blue arrowheads. (Magnification: A and B, ×100; D, ×400; C, E, and F, ×600.)
Limited Oxidative Stress Response in the Bubo.
In contrast with the case for RNS, there was less evidence for exposure to ROS in the bubo. Bacteria respond to exogenous oxidative stress generated by activated macrophages and PMNs by two major regulatory pathways. The adaptive response to H2O2 or OCl− follows oxidative activation of the OxyR transcriptional regulator, resulting in the induction of bacterial catalase and peroxidase-detoxifying enzymes as well as the glutathione and thioredoxin systems (22). None of these Y. pestis genes were up-regulated in the bubo (Fig. 2C). The response to superoxide (O2−) is controlled by the SoxRS two-component regulatory system in most Gram-negative bacteria, leading to the induction of superoxide dismutases (Sod), O2−-resistant isoforms of metabolic enzymes, and other protective enzymes and membrane proteins (22). Y. pestis does not appear to have an intact SoxRS response regulator (10, 11); but, of the O2−-responsive genes in other bacteria, only sodA and other genes coinduced by RNS had higher expression levels in the bubo. Furthermore, genes specifically implicated in the protective response to peroxynitrite (ONOO−), an antimicrobial product of O2− and NO, were not up-regulated (Fig. 2B), suggesting that Y. pestis was not exposed to O2− in the bubo.
Role of the Y. pestis Hmp Flavoglobin in Virulence.
We next compared the relative effect on virulence of loss of protection against RNS and ROS. Y. pestis Δhmp and ΔsodA mutants were 5- to 15-fold more susceptible to RNS and ROS, respectively, and were equally susceptible to killing by macrophages (Fig. 4A and B). Complementation of the Δhmp and ΔsodA mutants restored wild-type resistance to RNS and ROS. Notably, deletion of hmp had a greater effect on virulence. The Y. pestis Δhmp mutant had a 10-fold higher LD50 (73, compared with 6 for wild type) and a longer incubation period in rats (Fig. 4C). In contrast, although SodA is required for full virulence of Y. enterocolitica (23), loss of sodA in Y. pestis did not increase the LD50 or the survival rate of infected rats and resulted only in a slightly longer time to disease (Fig. 4D).
Fig. 4.
Decreased resistance of Y. pestis to RNS attenuates virulence more than decreased resistance to ROS. (A and B) Susceptibility of Y. pestis wild type (black bars), Δhmp (gray bars), and ΔsodA (white bars) to RNS and ROS in vitro (A) and to intracellular killing by macrophages (B). The mean and standard error of three experiments are shown. (C and D) Incidence of plague in rats injected intradermally with ≈150 Y. pestis wild type (black circles), Δhmp (gray circles), or ΔsodA (white circles).
Discussion
Generation of RNS is considered to be more important for defense against intracellular pathogens that reside in macrophages, whereas ROS are primarily associated with killing of bacteria that are ingested by PMNs (20). Y. pestis is a facultative intracellular pathogen, but uptake and replication in phagocytes appears to be limited to initial stages of infection at the dermal infection site (24). Resistance to macrophage-generated ROS (25) and RNS (24, 26) is likely to be important during this early intracellular phase (Fig. 4B). By the time the bacteria disseminate to the lymph node, however, they are predominantly extracellular, presumably because of the antiphagocytic properties of the TTSS and F1 capsule, which are in fact highly expressed in the bubo (Table 1).
The extracellular niche and antiinflammatory activities of Y. pestis have been predicted to limit its exposure to phagocyte-derived ROS and RNS in the lymph node, thus enabling rapid multiplication and systemic spread (3, 4, 6). Our results indicate that, although ROS are not a significant factor in the bubo, exposure to RNS is sustained throughout bubonic and septicemic plague. Of the bacterial genes with proven roles in protection against RNS, only adhC, the glutathione-dependent formaldehyde dehydrogenase that reduces GSNO, was not up-regulated in the bubo compared with in vitro growth conditions. However, adhC does not appear to be induced by NO or nitrosative stress (12–15). Expression of the Y. pestis ripA gene, which is required for reduction of NO levels and replication within activated macrophages in vitro (26), was not up-regulated in the bubo. Although best known as an intracellular effector, NO as well as bacteriostatic iron-chelating agents, such as lactoferrin and lipocalin 2 (NGAL), may be released into the extracellular environment from degranulating or degrading PMNs (21, 27–29). Extracellular NO-related antimicrobial stress could also derive from macrophages (30) and from the high serum NO levels present at the terminal stage of disease. Endogenous NO produced as a minor by-product of bacterial anaerobic respiration using nitrate as a terminal electron acceptor may also contribute to the RNS response (18). However, the Y. pestis respiratory nitrate reductases were not up-regulated in the bubo.
The transcriptional profile of Y. pestis recovered from rat buboes shows similarities and differences with that of Y. pestis recovered from bronchoalveolar washings of mice with primary pneumonic plague (31). In keeping with the differences in lymph node and lung environments, there was limited overlap between the up- and down-regulated genes. Only 34 of the 119 Y. pestis genes that were up-regulated, and 19 of the 203 genes that were down-regulated in the rat bubo had similar differential regulation in the mouse lung. The Y. pestis TTSS and F1 capsule genes were highly expressed in both in vivo environments, whereas the plasminogen activator was down-regulated in the lung but not in the lymph node. Notably, up-regulation of genes required for iron acquisition and for resistance to nitrosative stress, but not oxidative stress, was common to both infection models. However, only 2 of the 10 iron acquisition systems that were up-regulated in the bubo were up-regulated in the lung. Among the nitrosative stress response genes, hmp, the met genes, and tehB were up-regulated in the lung, but ytfE (dnrN), hcr, and the nrd genes were not. Furthermore, hmp expression was increased only 2.5-fold in the lung (31) but 10 to 20-fold in the bubo (Table 2) compared with stationary-phase growth at 37°C in vitro, suggesting that iron deprivation and NO-induced stress are more severe in the rat bubo than in the mouse lung.
Much of the antibacterial activity of NO is due to nitrosylation of cysteine thiols and transition metal centers of proteins required for respiratory metabolism, DNA synthesis, and transcriptional regulation (20). These effects could reinforce the shift to anaerobic metabolism (Fig. 6, which is published as supporting information on the PNAS web site) and the robust iron-acquisition response we observed in the bubo. For example, NO inhibits the ferric ion uptake regulator, Fur, leading to induction of iron-acquisition pathways (13, 32). Two of the Y. pestis iron-acquisition systems have been shown to be required for bubonic plague pathogenesis, however, indicating that iron limitation does occur in vivo (9). NO has also been shown to activate OxyR in E. coli (33), but we did not detect evidence for this (Fig. 2C), in agreement with recent studies showing that the transcriptional responses to RNS and ROS are largely distinct (13).
Conclusions
Our results implicate NO-derived RNS as an important innate immune effector mechanism that Y. pestis encounters and responds to in the bubo, where it is primarily extracellular. Inhibition of phagocytosis by the TTSS and other known virulence factors appears to limit exposure to ROS, but not RNS, during bubonic plague. Thus, although best known as an intracellular effector, release of NO into infected tissue may also be an important innate immune mechanism against extracellular pathogens. In addition to its antibacterial role in innate immunity, NO has known concentration-dependent immunosuppressive and other regulatory effects on the developing adaptive immune response (19). Systemic iNOS activation during plague also likely contributes to septic shock, the ultimate cause of plague mortality, as it does in other Gram-negative septicemias (34). Further characterization of the effects of NO on the bacteria and the host during plague will lead to a better understanding of Y. pestis pathogenesis and a potential new target for anti-plague therapy.
Materials and Methods
Y. pestis Strains and in Vitro Growth Conditions.
The fully virulent Y. pestis strain 195/P was used in this study (2). RNA was extracted from exponential and stationary-phase Y. pestis grown at 21°C or 37°C in LB/0.1 M Mops, pH 7.4. From frozen stocks, bacteria were first serially cultured in brain–heart infusion (BHI) and LB broth for 18 h at 28°C with aeration. Four milliliters of the LB culture was added to 50 ml of LB/Mops and incubated with aeration at 21°C (flea temperature) or 37°C (rat temperature). When the cultures reached exponential (A600 = 0.5) phase and 3 h after entering stationary phase (A600 = 1.3), one volume of culture was added to two volumes of RNAprotect Bacteria Reagent according to the manufacturer’s protocol (Qiagen). Bacteria were harvested from six independent replicate cultures for each of the four in vitro conditions.
Individual hmp− and sodA− mutant strains, in which a 1,118- or 573-bp internal segment, respectively, of the single-gene operons was replaced with the aph kanamycin-resistance cassette from pUC4K, were produced by allelic exchange (35). The aph cassette was introduced in the same orientation as the deleted gene, and the rho-independent transcription terminator of each gene was maintained in the deleted alleles to avoid polar effects. Mutations were verified by PCR. Complemented versions of the Y. pestis hmp and sodA mutants were produced by transformation with recombinant pACYC177 plasmids containing a wild-type copy of the respective gene.
Rat Model of Bubonic Plague and Isolation of Y. pestis from the Bubo.
For in vivo RNA samples, 12 female Brown Norway rats, each 9 weeks old (Charles River Laboratories), were injected intradermally in the lower back with ≈500 Y. pestis as previously described (2). Signs of terminal plague (roughcast fur, watery eyes, hunched posture, reluctance to move, and limping of the hind leg adjacent to the bubo) occurred between 48–72 h after infection. At this point, each rat was anesthetized, and the inguinal bubo was quickly dissected and disrupted on a cell strainer (70 μm mesh), simultaneously releasing the bubo contents into 9 ml of RNAprotect. The filter was washed three times with 1 ml of RNAprotect. Cell suspensions were centrifuged for 10 min. at 1,500 × g at 20°C and resuspended in 600 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0) containing 2 mg/ml lysozyme. After 10 min at room temperature, RNA was extracted by using an RNeasy kit (Qiagen). RNA samples were DNase-treated twice with the DNA-free kit (Ambion), then purified and concentrated to 14 μl with the RNeasy cleanup kit (Qiagen). RNA quality and quantity was assayed on a Bioanalyzer 2100 using RNA LabChips (Agilent Technologies) and by spectrophotometry. Absence of DNA was verified by real-time TaqMan PCR using primers and probe specific for the Y. pestis proS gene (Table 4, which is published as supporting information on the PNAS web site).
Microarray Analysis.
A custom antisense oligonucleotide array representing 149,856 perfect match and mismatch probes, targeting 4,683 ORFs of the Y. pestis CO92 and KIM strains (10, 11), and manufactured by Affymetrix was used as previously described (36). Biotin–ddUTP-labeled cDNA was prepared from purified RNA samples from six biological replicates of each in vitro culture condition and from the buboes of six rats. The cDNA was then fragmented, hybridized to Affymetrix chips (one chip per sample; six biological replicates for each sample type), and scanned per manufacturer protocols. Data were analyzed with GeneChip Operating Software v1.1 (Affymetrix), GeneSpring v6.0 (Agilent Technologies), and Partek Pro (Partek, Inc.). Data were normalized with the mean signal from the Y. pestis probe sets multiplied by a scale factor to obtain the target signal. Default detection thresholds of P < 0.06 and P ≥ 0.06 were used to call individual genes as present or absent, respectively. The mean signal intensities of all genes called present in three or more of the six replicate microarray experiments were used to calculate differential gene expression. A 2-fold difference (P ≤ 0.05 by two-tailed t test with Benjamini and Hochberg false discovery rate correction) was considered significant.
Real-Time RT-PCR.
In vivo microarray results for hmp and 11 randomly selected Y. pestis genes that covered the range of expression level in the bubo were verified by TaqMan RT-PCR analysis using an ABI 7700 TaqMan instrument and RNA isolated from the buboes of six additional rats (Fig. 7, which is published as supporting information on the PNAS web site) (37). The quantity of each mRNA was determined relative to that of the reference gene crr (YPO2995, encoding a phosphotransferase system component), whose expression level was not affected by in vivo or in vitro growth conditions.
Virulence Determination and Histopathology.
Groups of 10 female Brown Norway rats, each 8 to 10 weeks old, were infected by intradermal injection of 50 μl of PBS containing different numbers of Y. pestis. Rats were killed upon signs of terminal plague, and the quantity of Y. pestis in heart blood and spleen were determined by counting colony-forming units (2). LD50 values were calculated by using the Reed–Muench equation, and survival rate differences were determined by logrank test. Tissue sections of the inguinal lymph node proximal to the inoculation site were stained by Naphthol AS-D chloroacetate esterase (Sigma) to detect PMNs or by immunohistochemistry using anti-iNOS antibody 15323 (Abcam) and a peroxidase-conjugated secondary antibody. All experiments were performed at Biosafety Level 3 and were approved by the Rocky Mountain Laboratories Biosafety and Animal Care and Use Committees in accordance with National Institutes of Health guidelines.
Susceptibility to ROS and RNS.
Minimum inhibitory concentrations of GSNO, diethylenetriamine/NO adduct, H2O2, and paraquat (Sigma) were determined in LB at 37°C. Susceptibility of Y. pestis grown at 37°C to killing by J774A.1 macrophages was determined as previously described (38). The ratio of viable intracellular bacteria at 6 h compared with 2 h after infection was determined by colony-forming unit counts from three replicate wells in three independent experiments.
NO Assay.
Cumulative NO production was evaluated by measuring the total nitrite/nitrate (NOx) in the sera of uninfected and diseased rats by the Griess reaction (Total Nitric Oxide Assay kit; Endogen).
Supplementary Material
Acknowledgments
We thank F. DeLeo, F. Gherardini, and D. Erickson for the critical reading of the manuscript. This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (National Institutes of Health) and by a Ellison Medical Foundation New Scholars Award in Global Infectious Diseases (to B.J.H.).
Abbreviations
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- iNOS
inducible nitric oxide synthase
- TTSS
Type III secretion system
- PMN
polymorphonuclear neutrophil
- GSNO
nitrosoglutathione.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The MIAME (Minimum Information About a Microarray Experiment) data have been submitted to the Gene Expression Omnibus repository (accession no. GSE3793).
References
- 1.Perry R. D., Fetherston J. D. Clin. Microbiol. Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sebbane F., Gardner D., Long D., Gowen B. B., Hinnebusch B. J. Am. J. Pathol. 2005;166:1427–1439. doi: 10.1016/S0002-9440(10)62360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornelis G. R., Boland A., Boyd A. P., Geuijen C., Iriarte M., Neyt C., Sory M. P., Stainier I. Microbiol. Mol. Biol. Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarro L., Alto N. M., Dixon J. E. Curr. Opin. Microbiol. 2005;8:21–27. doi: 10.1016/j.mib.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Une T., Brubaker R. R. Infect. Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brubaker R. R. Infect. Immun. 2003;71:3673–3681. doi: 10.1128/IAI.71.7.3673-3681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerschen E. J., Cohen D. A., Kaplan A. M., Straley S. C. Infect. Immun. 2004;72:4589–4602. doi: 10.1128/IAI.72.8.4589-4602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marketon M. M., DePaolo R. W., DeBord K. L., Jabri B., Schneewind O. Science. 2005;309:1739–1741. doi: 10.1126/science.1114580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry R. D. In: Iron Transport in Bacteria. Crosa J. H., Mey A. R., Payne S. M., editors. Washington, DC: Am. Soc. Microbiol.; 2004. pp. 219–240. [Google Scholar]
- 10.Parkhill J., Wren B. W., Thomson N. R., Titball R. W., Holden M. T. G., Prentice M. B., Sebhaihia M., James K. D., Churcher C., Mungall K. L., et al. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 11.Deng W., Burland V., Plunkett G., Boutin A., Mayhew G. F., Liss P., Perna N. T., Rose D. J., Mau B., Zhou S., et al. J. Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole R. K. Biochem. Soc. Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay P., Zheng M., Bedzyk L. A., LaRossa R. A., Storz G. Proc. Natl. Acad. Sci. USA. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Justino M. C., Vicente J. B., Teixeira M., Saraiva L. M. J. Biol. Chem. 2005;280:2636–2643. doi: 10.1074/jbc.M411070200. [DOI] [PubMed] [Google Scholar]
- 15.Flatley J., Barrett J., Pullan S. T., Hughes M. N., Green J., Poole R. K. J. Biol. Chem. 2005;280:10065–10072. doi: 10.1074/jbc.M410393200. [DOI] [PubMed] [Google Scholar]
- 16.De Groote M. A., Testerman T., Xu Y., Stauffer G., Fang F. C. Science. 1996;272:414–417. doi: 10.1126/science.272.5260.414. [DOI] [PubMed] [Google Scholar]
- 17.Bodenmiller D. M., Spiro S. J. Bacteriol. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodionov Q. A., Dubchak I. L., Arkin A. P., Alm E. J., Gelfand M. S. PLoS Comp. Biol. 2005;1:415–431. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogdan C. Nat. Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 20.De Groote M. A., Fang F. C. In: Nitric Oxide and Infection. Fang F. C., editor. New York: Kluwer Academic/Plenum; 1999. pp. 231–261. [Google Scholar]
- 21.Fierro I. M., Nascimento-DaSilva V., Arruda M. A., Freitas M. S., Plotkowski M. C., Cunha F. Q., Barja-Fidalgo C. J. Leukocyte Biol. 1999;65:508–514. doi: 10.1002/jlb.65.4.508. [DOI] [PubMed] [Google Scholar]
- 22.Storz G., Zheng M. In: Bacterial Stress Responses. Storz G., Hengge-Aronis R., editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 47–59. [Google Scholar]
- 23.Roggenkamp A., Bittner T., Leitritz L., Sing A., Heesemann J. Infect. Immun. 1997;65:4705–4710. doi: 10.1128/iai.65.11.4705-4710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pujol C., Bliska J. B. Clin. Immunol. 2005;114:216–226. doi: 10.1016/j.clim.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Oyston P. C. F., Dorrell N., Williams K., Li S.-R., Green M., Titball R. W., Wren B. W. Infect. Immun. 2000;68:3419–3425. doi: 10.1128/iai.68.6.3419-3425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pujol C., Grabenstein J. P., Perry R. D., Bliska J. B. Proc. Natl. Acad. Sci. USA. 2005;102:12909–12914. doi: 10.1073/pnas.0502849102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., Zychlinsky A. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 28.Faurschou M., Borregaard N. Microbes Infect. 2003;5:1317–1327. doi: 10.1016/j.micinf.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., Strong R. K. Mol. Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 30.Webb J. L., Harvey M. W., Holden D. W., Evans T. J. Infect. Immun. 2001;69:6391–6400. doi: 10.1128/IAI.69.10.6391-6400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lathem W. W., Crosby S. D., Miller V. L., Goldman W. E. Proc. Natl. Acad. Sci. USA. 2005;102:17786–17791. doi: 10.1073/pnas.0506840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Autreaux B., Touati D., Bersch B., Latour J. M., Michaud-Soret I. Proc. Natl. Acad. Sci. USA. 2002;99:16619–16624. doi: 10.1073/pnas.252591299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hausladen A., Privalle C. T., Keng T., DeAngelo J., Stamler J. S. Cell. 1996;86:719–729. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 34.Titheradge M. A. Biochim. Biophys. Acta. 1999;1411:437–455. doi: 10.1016/s0005-2728(99)00031-6. [DOI] [PubMed] [Google Scholar]
- 35.Sebbane F., Jarrett C. O., Linkenhoker J. R., Hinnebusch B. J. Infect. Immun. 2004;72:7334–7337. doi: 10.1128/IAI.72.12.7334-7337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham M. R., Virtaneva K., Porcella S. F., Barry W. T., Gowen B. B., Johnson C. R., Wright F. A., Musser J. M. Am. J. Pathol. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaussee M. S., Watson R. O., Smoot J. C., Musser J. M. Infect. Immun. 2001;69:822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pujol C., Bliska J. B. Infect. Immun. 2003;71:5892–5899. doi: 10.1128/IAI.71.10.5892-5899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. J. Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.