Abstract
Spindle assembly checkpoint (SAC) ensures bipolar attachment of chromosomes to the mitotic spindle and is essential for faithful chromosome segregation, thereby preventing chromosome instability (CIN). Genetic evidence suggests a causal link between compromised SAC, CIN, and cancer. Bloom syndrome (BS) is a genetic disorder that predisposes affected individuals to cancer. BS cells exhibit elevated rates of sister chromatid exchange, chromosome breaks, and CIN. The BS gene product, BLM, is a member of the RecQ helicases that are required for maintenance of genome stability. The BLM helicase interacts with proteins involved in DNA replication, recombination, and repair and is required for the repair of stalled-replication forks and in the DNA damage response. Here we present biochemical evidence to suggest a role of BLM phosphorylation during mitosis in maintaining chromosome stability. BLM is associated with the SAC kinase MPS1 and is phosphorylated at S144 in a MPS1-dependent manner. Phosphorylated BLM interacts with polo-like kinase 1, a mitotic kinase that binds to phospho-serine/threonine through its polo-box domain (PBD). Furthermore, BS cells expressing BLM-S144A show normal levels of sister chromatid exchange but fail to maintain the mitotic arrest when SAC is activated and exhibit a broad distribution of chromosome numbers. We propose that MPS1-dependent BLM phosphorylation is important for ensuring accurate chromosome segregation, and its deregulation may contribute to cancer.
Keywords: Bloom syndrome, spindle assembly checkpoint
Bloom syndrome (BS) is a rare autosomal recessive disorder characterized by growth retardation, immunodeficiency, and cancer predisposition. BS cells exhibit elevated rates of sister chromatid exchange (SCE) and chromosome instability. The BS gene product, BLM, is a member of the RecQ helicases, which are required for the maintenance of genome integrity (1, 2). BLM is a caretaker type of tumor suppressor that prevents the accumulation of genetic alterations required for tumorigenesis. BLM interacts physically and functionally with proteins involved in DNA replication, repair, and damage signaling (3–9). Genetic and biochemical evidence suggest that BLM suppresses hyperrecombination by preventing DNA breakage during DNA replication (10, 11).
The spindle assembly checkpoint (SAC) is another important surveillance mechanism for maintaining chromosome stability. SAC monitors the proper attachment of chromosomes to mitotic spindle, thereby preventing cells from entering anaphase in the presence of unaligned chromosomes (12, 13). Compromised spindle assembly checkpoint is thought to be a major contributing factor for chromosome instability, including aneuploidy, a key feature of many cancer cells (14). Several recent studies provided important genetic evidence to suggest a causal link among SAC, chromosome instability, and cancer (15, 16). MPS1, the human homologue of the yeast Mps1p (monopolar spindle), is a key mitotic checkpoint kinase that is required for mitotic arrest in the presence of unaligned chromosomes (17–19). MPS1 level and kinase activity increase in M phase and peak upon SAC activation (19). MPS1 itself is extensively phosphorylated upon SAC activation. MPS1 also may participate in G2/M checkpoint regulation through the Chk2-signaling pathway (20). Despite its importance in SAC, the regulation and the identity of the physiological substrates of mammalian MPS1 remain elusive.
The polo-like kinase 1, PLK1, is a mammalian M phase kinase that is involved in several critical events during cell division (21). PLK1 localizes to various mitotic structures and is subjected to temporal and spatial regulations. The C-terminal noncatalytic region of PLK1 contains two tandem Polo boxes termed polo-box domain (PBD), which has been implicated in phospho-dependent substrate targeting (22).
BLM is phosphorylated during mitosis and is hyperphosphorylated in the presence of microtubule destabilization agents (23). In contrast to its better-studied role in DNA replication and repair, the role of mitotic BLM phosphorylation and the responsible kinases are not known. Here we report that MPS1, a key SAC kinase, physically associates with BLM and is required for BLM phosphorylation at S144 in vivo. Furthermore, BLM interacts with PLK1 in a phosphorylation and PBD-dependent manner, suggesting that MPS1-dependent phosphorylation of BLM may recruit PLK1, which, in turn, phosphorylates BLM at additional sites. Importantly, BS cells expressing BLM-S144A exit from mitotic arrest in the presence of SAC, which, in turn, correlates with a wide distribution of chromosome numbers. Together our data suggest that MPS1-dependent mitotic phosphorylation of BLM at S144 is required for the maintenance of chromosome stability.
Results
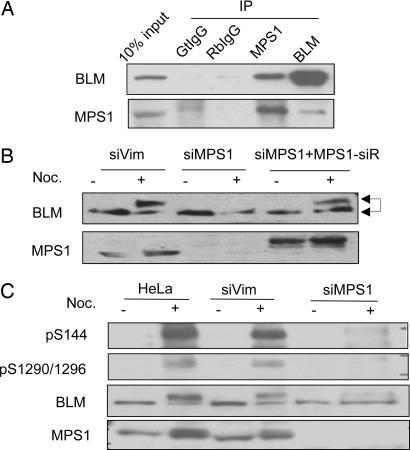
We identified BLM among several DNA repair proteins in the BRCA1-associated genome surveillance complex, BASC (4). Further experiments demonstrated that lesion-specific DNA-binding and repair proteins form signaling modules with DNA damage checkpoint kinases (ATM and ATR) and function upstream of the response (24, 25). To examine whether BLM also forms a complex with a checkpoint kinase, we immunoprecipitated BLM and identified copurified proteins by mass spectrometry. A spindle checkpoint kinase, MPS1, was identified in the BLM immunoprecipitate (data not shown). Western blotting confirmed that BLM and MPS1 can be reciprocally coprecipitated (Fig. 1A). Because BLM is phosphorylated during mitosis (23), we examined whether BLM phosphorylation depends on MPS1. When MPS1 was knocked down by siRNA transfection, phosphorylation of BLM in nocodazole-treated cells was diminished, as determined by SDS/PAGE mobility shifts (Fig. 1B). Because diminished BLM phosphorylation was restored by expression of a siRNA-resistant RFP-MPS1 (Fig. 1B), we conclude that MPS1 is required for mitotic BLM phosphorylation. To identify MPS1-dependent BLM phosphorylation sites, we cotransfected 293T cells with plasmids encoding full-length GFP-BLM and RFP-MPS1. The transfected cells were treated with nocodazole to enrich for mitotic cells. GFP-BLM was immunoprecipitated, and its phosphorylation was analyzed with mass spectrometry. We detected three phospho-peptides spanning amino acids 139–167, 286–317, and 1285–1305 by MALDI-TOF mass spectrometry. Furthermore, we unambiguously identified S304 as a phosphorylation site of BLM by liquid chromatography-coupled ion trap mass spectrometry (LC/MS/MS) (data not shown). We were unable to identify the phosphorylation sites on the other two phospho-peptides directly by LC/MS/MS. Because S304 is followed by a proline, we hypothesized that the SP motifs in the other two phospho-peptides may be phosphorylated. Three serines at S144, S1290, and S1296, in addition to S304, conform to the SP motif. We raised phospho-specific antibodies against peptides that were singly phosphorylated at S144, S304, S1290, and S1296 and doubly phosphorylated at both S1290 and S1296. These antibodies recognized phosphorylated residues and were specific to the targeted serines, because treating the lysate with phosphatase or mutating these serines to alanines abolished the signals by Western blotting (Fig. 6 A, B, and D, which is published as supporting information on the PNAS web site). To confirm that phosphorylation at these sites depends on MPS1, we depleted MPS1 in HeLa cells by siRNA transfection and examined BLM phosphorylation upon nocodazole treatment. Phosphorylations at S144 and S1290/S1296 were decreased markedly in siMPS1-transfected cells as compared with control siRNA-transfected cells (Fig. 1C). Ectopic expression of MPS1 increased the phosphorylation of S144 and S1290/S1296, but not S304 (Fig. 6C). Collectively, these results demonstrate that BLM can be phosphorylated at S144 and S1290/S1296 in a MPS1-dependent manner. In addition, various recombinant GST-BLM fragments (Fig. 2A) were tested for phosphorylation by recombinant GST-MPS1 in in vitro kinase assays (Fig. 2B). The GST-BLM fragment D1F1, which contains S144 (Fig. 2A), was phosphorylated; however, GST-BLM fragment D3F2, which contains S1290/S1296, was not phosphorylated (Fig. 2B). Together these data suggest that MPS1 can phosphorylate BLM directly at S144 but may be indirectly responsible or requires SAC activation for the phosphorylation of S1290 and S1296.
Fig. 1.
BLM is phosphorylated during mitosis and upon SAC activation in a MPS1-dependent manner. (A) BLM and MPS1 coimmunoprecipitate from HeLa nuclear extract. Immunoprecipitations with goat and rabbit preimmune sera serve as negative controls. (B) Hyperphosphorylation of BLM is diminished in MPS1-knockdown cells and is restored upon expression of siRNA-resistant RFP-MPS1. Knockdown of Vimentin (siVim) serves as a negative control. The arrows indicate hypo- and hyperphosphorylated BLM. (C) MPS1-dependent BLM phosphorylation at S144 and S1290/S1296. HeLa cells transfected with siMPS1 or siVim were treated with nocodazole and cell lysates were Western blotted with the indicated phospho-specific antibodies.
Fig. 2.
BLM is phosphorylated in vivo in a cell cycle-dependent manner and by MPS1 in vitro. (A) A schematic of BLM fragments used. (B) GST-BLM fragments expressed in Escherichia coli were incubated with recombinant WT- or kinase dead- (KD-) MPS1 in in vitro kinase assays, and the products were analyzed by autoradiography. Coomassie blue staining shows the amount of proteins used in each reaction. (∗, position of the GST-BLM-D3F2). (C) BLM phosphorylation during mitosis and upon SAC activation. HeLa cells blocked at prometaphase or G1/S boundary were released to drug-free media to allow cell cycle progression (see also Materials and Methods). Cells lysate prepared at indicated times after release from prometaphase (Noc. Release) or G1/S boundary (DB released) were immunoprecipitated with an antibody against total BLM and detected with phospho-specific antibodies. Cell cycle distribution was indicated under each lane. Asy, asynchronized cells; AT, attached cells after shake-off.
We examined BLM phosphorylation during the M to G1 and S to G2/M transitions by immunoprecipitating endogenous BLM at different stages of the cell cycle and Western blotting with the phospho-specific antibodies (Fig. 2C). Phosphorylation of all four sites was enhanced upon nocodazole treatment and diminished when cells entered G1 (Fig. 2C, lanes 3–7). When cells progressed from G1/S to G2/M (Fig. 2C, lanes 8–13), only S144 phosphorylation was noticeably enhanced 13 h after release (Fig. 2C, lane 12), at a time when cells enter mitosis. Phosphorylation of S1290 and S1296 was minimal in the absence of nocodazole-induced spindle checkpoint activation (Fig. 2C, lanes 8–13). In contrast, S304 phosphorylation appeared to persist throughout the cell cycle. Together these data suggest that S144 phosphorylation may have a distinct function in mitosis and that the kinase(s) that phosphorylate S144, S1290, and S1296 may be different from the kinase(s) that phosphorylate S304.
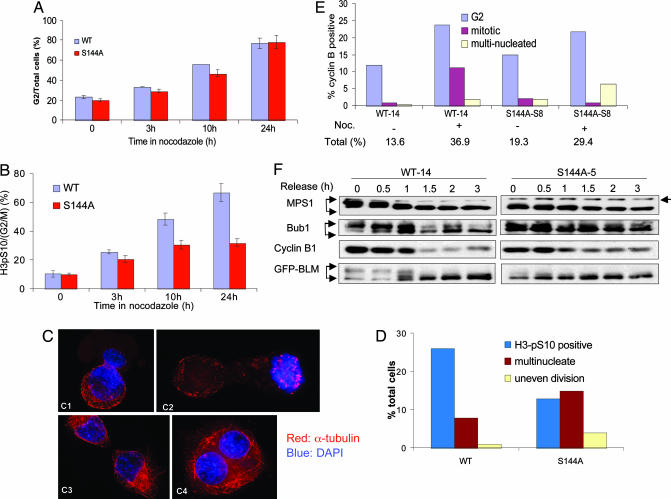
Members of the MPS1 family kinases regulate SAC from yeast to man (17, 19, 26, 27). We tested whether BLM phosphorylation is required for spindle assembly checkpoint activation. The SV40-transformed BS fibroblast cell line GM08505 was transfected with plasmids encoding GFP-BLM-WT, GFP-BLM-S144A, or GFP. Single colonies were isolated to establish stable cell lines. Several clones expressing similar levels of BLM-WT and BLM-S144A were analyzed for their mitotic functions (Fig. 7A, which is published as supporting information on the PNAS web site). These cells were treated with nocodazole for various times, and their mitotic indices were measured with FACScan. The G2/M population (as measured by the DNA content) accumulated at a similar rate among these cell lines (Fig. 3A). However, the mitotic population (as measured by phosphorylation on S10 of histone H3) of the BLM-S144A cells accumulated at a much slower rate than that of BLM-WT cells, indicating that mutant cells may be defective in establishing or maintaining mitotic arrest in the presence of a microtubule destabilizing agent (Fig. 3B). Examination of the cells by immunostaining revealed that after 10 h of nocodazole treatment, a significant percentage of the BLM-S144A cells were at postmetaphase stages of mitosis and displayed various abnormalities that included uneven segregation of chromosomes (Fig. 3 C1 and C2), aberrant cytokinesis (Fig. 3C3), multinucleation (Fig. 3C4), and the appearance of multiple micronuclei (data not shown). Under these conditions, there were approximately twice the number of BLM-S144A cells that exhibited mitotic defects as compared with the BLM-WT cells (Fig. 2D), suggesting that phosphorylation of BLM at S144 is required to maintain mitotic arrest in the presence of nocodazole.
Fig. 3.
BLM-S144A cells escape mitotic arrest in the presence of SAC. (A) Accumulation of G2/M population in BS cells expressing GFP-BLM-WT or GFP-BLM-S144A in response to nocodazole treatment as a function of time. (B) Percentage of mitotic cells among G2/M cells in A measured by phospho-S10 of histone H3 staining. (C) Mitotic defects in BS cells expressing GFP-BLM-144A after treated for 10 h with nocodazole. Images representative of unevenly divided daughter cells (C1 and C2), aberrant cytokinesis (C3), and multinucleation (C4) were shown. (D) Statistics of normal mitotic and various defective mitotic cells in BLM-WT and -S144A cells observed in C. (E) Percentage of BLM-WT and -S144A cells showing positive cyclin B1 staining with G2-, mitotic-, and multinucleated appearance before and after 10 h treatment with nocodazole. (F) Dephosphorylation of MPS1, Bub1, and BLM in BS cells expressing GFP-BLM-WT or GFP-BLM-S144A collected by mitotic shake-off after 10 h of nocodazole treatment. A band migrating above MPS1 indicates a cross-reacting protein recognized by the anti-MPS1 antibody and serves as a marker for MPS1 mobility (arrow at right).
The outcome of SAC activation is inhibition of cyclin B1 degradation. Cyclin B1, a key mitotic regulator, normally accumulates during G2 and M and is degraded at the onset of anaphase in an anaphase promoting complex/cyclosome-dependent manner to allow for mitotic exit (28). Western blotting showed that 10 h after nocodazole treatment, both BLM-WT and BLM-S144A cells maintained high level of cyclin B1 as well as Cdc2 kinase activity (Fig. 3E and data not shown), suggesting that SAC remained activated in both cell lines. However, immunofluorescence revealed that among cells that were positive for cyclin B1 staining after nocodazole treatment (Fig. 7B), ≈10% of BLM-WT vs. ≈1% of BLM-S144A cells were in M phase (Fig. 3E, red bars), whereas both cell lines had similar percentage of cells in G2 (Fig. 3E, blue bars). In addition, the percentage of multinucleated cells observed after nocodazole treatment was much higher in the BLM-S144A than in the BLM-WT cells (6.5% vs. 2%, respectively; Fig. 3E, yellow bars). These data indicate that the BLM-S144A cells have escaped mitotic arrest in the presence of high level of cyclin B1 and Cdc2 activity. We conclude that phosphorylation of S144 in BLM is required to maintain optimal mitotic arrest even when SAC appeared to be active.
The mitotic kinase Bub1 is another upstream component of SAC proteins (29). Bub1 phosphorylates CDC20 in the anaphase promoting complex/cyclosome to inhibit its ubiquitin ligase activity in vitro (30). Like MPS1, Bub1 also is hyperphosphorylated upon SAC activation (30, 31). We treated BLM-WT and BLM-S144A cells with nocodazole for 10 h and collected the mitotic cells by shake-off. The arrested cells then were released in drug-free media to allow for mitotic exit. As shown in Fig. 3F, both MPS1 and Bub1 in the BLM-S144A cells exhibited less prominent SDS/PAGE mobility shifts compared with BLM-WT cells at the time of release; upon release from the nocodazole block, which eliminates the SAC signal, Bub1 dephosphorylation and cyclin B1 degradation occurred ≈30 min earlier in the BLM-S144A cells than in the BLM-WT cells, suggesting that BLM-S144A cells may have exited mitosis earlier in the presence of unaligned chromosomes. Together, these results suggest that phosphorylation of BLM at S144 is required to prevent early mitotic exit.
We investigated downstream events that depend on BLM phosphorylation. Phospho-S144 and phospho-S1296 are preceded by a serine and followed by a proline (SpSP), a motif to which the conserved PBD is predicted to bind in a phosphorylation-dependent manner (22). We thus examined whether phosphorylation of S144 and S1296 regulates its binding to the PBD of the mitotic kinase PLK1, a key regulator of multiple stages of mitosis, including mitotic exit (21). A panel of peptides containing S144, S304, or S1290 and S1296, in either phosphorylated or unphosphorylated form, were immobilized in equal amounts on agarose beads and incubated with HeLa cell lysate in pull-down experiments. PLK1 was differentially recovered from beads immobilized with phospho-peptides containing S144 or diphosphorylated S1290 and S1296 (Fig. 4A). In contrast, a similar amount of PLK1 was recovered from both phospho- and nonphospho-S304 peptides. Notably, the sequence flanking S304 does not conform to the polo-box binding consensus (22). Furthermore, the GST-PBD of PLK1 was able to pull-down BLM from HeLa cell extract (Fig. 4B). This interaction was enhanced when the cells were treated with nocodazole and abolished when the extract was treated with phosphatase, or when the two key residues in PBD that mediate phosphate binding were mutated (Fig. 4B). Thus, BLM and PLK1 interact in a phosphorylation and polo-box-dependent manner in vitro. To test whether BLM phosphorylation regulates PLK1 binding in vivo, we cotransfected plasmids encoding Flag-PLK1 and GFP-BLM-WT, GFP-BLM-S144A, or GFP-BLM-4A (all four serines mutated to alanines) in 293T cells (Fig. 4C). Although BLM-WT coimmunoprecipitated with Flag-PLK1, the S144A mutation diminished binding to PLK1, and mutations of all four serines to alanines completely abolished binding to PLK1. More importantly, endogenous PLK1 and BLM can be immunoprecipitated reciprocally from nocodazole-treated HeLa cell lysate (Fig. 4D), demonstrating that BLM and PLK1 interacted in vivo. One way for PLK1 to achieve substrate specificity is by being recruited to its substrates via polo-box binding (22). We therefore tested whether BLM can serve as a substrate for PLK1. Flag-PLK1 expressed in 293T cells was able to phosphorylate a GST-BLM fragment (fragment D1F2 encompassing amino acids 226–690; Fig. 2A) in vitro (Fig. 4E). Together these data suggest that PLK1 may regulate BLM function through binding to prephosphorylated BLM (by MPS1) and results in further phosphorylation of BLM by PLK1.
Fig. 4.
MPS1-dependent phosphorylation of BLM results in binding of polo-like kinase 1 (PLK1) and subsequent phosphorylation of BLM. (A) BLM peptides containing either phospho- or nonphospho-S at amino acid residues indicated were immobilized on agarose beads and incubated with HeLa cell lysate. PLK1 bound on beads were eluted and detected by Western blotting. (B) Phosphorylation and polo-box-dependent interaction between BLM and PLK1 in vitro. GST, GST-PBD-WT, or GST-PBD-mutant (H538A/K540M) proteins on beads were incubated with lysates of HeLa cells treated with nocodazole, and the bound BLM protein was detected by Western blotting. Some of the lysates were treated with calf-intestine-phosphatase (CIP) to dephosphorylate BLM. (C) Interaction between PLK1 and phosphorylated BLM protein in 293T cells. 293T cells were cotransfected with Flag-PLK1 and WT-, S144A, or 4A-BLM and treated with nocodazole. BLM bound to the Flag beads were detected by Western blotting. (D) In vivo interaction of BLM and PLK1 in HeLa cells treated with nocodazole. Immunoprecipitation with goat (Gt) and rabbit (Rb) IgGs serve as negative controls. (E) In vitro phosphorylation of BLM by PLK1. GST-BLM fragments expressed in E. coli were incubated with WT- or KD-flag-PLK1 overexpressed in 293T cells. The GST constructs were the same as shown in Fig. 2A. Kinase reaction products were detected by autoradiography. (∗, position of the substrates).
The hallmarks of BS cells are an increased rate of SCE and chromosome instability, which includes chromosome breakage and aneuploidy (32). The increased rate of loss of heterozygosity resulting from somatic recombination has been proposed as the underlying mechanism for increased tumor susceptibility for Blm−/− mice (10). We examined whether BLM phosphorylation regulates SCE and chromosome stability. The high rate of SCE was largely corrected in BS cells expressing either GFP-BLM-WT or GFP-BLM-S144A (Fig. 5A; see also Fig. 8, which is published as supporting information on the PNAS web site). Metaphase spreads did not reveal significant difference in chromosome breakage between BLM-WT and -S144A cells. However, we consistently observed a broader distribution of chromosome numbers in BLM-S144A than in BLM-WT cells (Fig. 5B), suggesting that missegregation of chromosomes in the BLM-S144A cells as a result of their inability to remain arrested in the presence of spindle checkpoint may lead to chromosome instability.
Fig. 5.
BS cells expressing GFP-S144A-BLM exhibit chromosome instability. (A) Frequencies of SCE in BS cells stably expressing GFP, GFP-BLM-WT, or GFP-BLM-S144A. N denotes the number of chromosomes counted. (B) Distributions of chromosome numbers in BS cells expressing BLM-WT or BLM-S144A. N denotes the number of cells counted.
Discussion
BS is a cancer predisposition syndrome that increases the likelihood of patients to develop wide spectrum of cancers. Our results show that mitotic BLM phosphorylation is not required for correction of SCE but is required for stability of chromosome number. These findings have several implications. Recent genetic data have demonstrated that mutations of genes involved SAC that resulted in aneuploidy are associated with cancer predisposition (15). A frequent feature of human tumor cells and cell lines is their inability to sustain mitotic checkpoint signaling despite their ability to produce the signal (14). It is conceivable that, during normal growth conditions, cells with a weakened checkpoint may gain or lose a few chromosomes during each cell division and are able to survive in the absence of functional p53. However, the mechanism by which cells adapt to mitotic arrest and eventually exit mitosis is unclear (13). Our data demonstrates that MPS1-dependent BLM phosphorylation is required for preventing mitotic exit, providing biochemical evidence to support the notion that compromised SAC, which leads to chromosome missegregation, is an important step in tumorigenesis. A high level of SCE is observed in most, but not all BS cells, and this defect is often considered to be the underlying mechanism for cancer predisposition in the BS patients (10). Notably, individuals carrying one BLM mutant allele have increased risk of colorectal cancer (33), and heterozygous Blm+/− mice show enhanced risk of tumor formation but display a normal level of SCE (34). Our data demonstrates that the inability to phosphorylate BLM during mitosis by a SAC kinase gives rise to chromosome instability but does not affect the frequency of SCE. These observations suggest that additional mechanism(s) is responsible for the increased cancer risk in the absence of high SCE. We propose that BLM plays a role in chromosome segregation in addition to its more extensively studied function during DNA replication and repair; this task is accomplished through a biochemical pathway in which BLM is phosphorylated in a SAC kinase MPS1-dependent manner, allowing the mitotic kinase PLK1 to bind BLM via its PBD and to further phosphorylate BLM in a concerted effort to prevent early mitotic exit.
Methods
Cell Cultures and Transfections.
HeLa and 293T cells were grown in DMEM supplemented with 5% and 10% FBS, respectively. Transfection of siRNAs were carried out with Oligofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's suggestions. DNA transfections were carried out with Lipofectamine (Invitrogen).
The SV40-transformed BS cell line GM08505C was purchased from Coriell Cell Repositories and were grown in DMEM supplemented with 15% FBS. To establish stable BS cell lines with various BLM constructs, BS cells were transfected with vectors encoding BLM-WT, BLM-S144A in pEGFP (Clontech, Mountain View, CA), or vector alone and were selected in growth medium containing G418 (800 μg/ml). Individual colonies with a similar expression level of BLM were isolated and maintained in growth medium containing 400 μg/ml of G418.
Recombinant DNA.
Full-length human MPS1 was isolated from a HeLa cDNA pool and cloned into pDsRed1 (Clontech) (RFP-MPS1). pGEX4T-MPS1 was a gift from S. Shieh (Institute of Biomedical Sciences, Academia Sinica; Taipei, Taiwan). siRNA oligonucleotides of MPS1 (5′-CAAGAGCCAGAUGAUGCACGUG-3′) and BLM (5′-AAGGAUGACUCAGAAUGGUUA-3′) were synthesized by Dharmacon (Chicago, IL). A siRNA-resistant MPS1 (RFP-MPS1siR) was created by site-directed mutagenesis of bases 428 (T to C), 431 (A to C), and 434 (T to C).
Full-length pEGFP-BLM was a gift from Nathan Ellis (Memorial Sloan–Kettering Cancer Center, New York, NY). GFP-BLM-S144A and GFP-BLM-4A were generated by site-directed mutagenesis. GST-BLM fragments were cloned into pGEX-4T-1.
PLK1 was amplified by PCR from a Myc-tagged PLK1 construct (pSC2-myc-PLK1) (a gift from Hongtao Yu, University of Texas Southwestern, Dallas, TX) and cloned into the pSG5 (Stratagene, La Jolla, CA). The PBD of PLK1 (amino acids 326–603) was amplified by PCR and cloned into pGEX-4T-1 (Amersham Pharmacia, Piscataway, NJ). A PBD mutant that abolishes its phospho-peptide binding (35) was generated by mutating H538 to A and K540 to M by site-directed mutagenesis.
Cell Synchronization and Cell Cycle Analysis.
To enrich for prometaphase cells, HeLa and BLM stable cell lines were cultured in the presence of nocodazole (100 ng/ml) for 16 and 10 h, respectively. Mitotic cells were collected by shake-off, washed three times with warm PBS, and released into drug-free media. To synchronize cells to the G1/S boundary, HeLa cells first were blocked in nocodazole (100 ng/ml) for 12 h, collected by shake-off, washed with PBS for three times, and released into the fresh medium containing mimosine (4 mM) for 10 h. After washing for three times with PBS, cells were released into drug-free media to allow for cell cycle progression.
To measure the mitotic index, ethanol fixed cells were permeablized with 0.25% Triton X-100 in PBS for 15 min on ice. Cells were incubated with Histone H3-pS10 antibody for 1 h and incubated with Alexa-488 conjugated anti-Rabbit IgG antibody (Molecular Probes, Carlsbad, CA) for 30 min. Finally, cells were stained with propidium iodide and RNaseA and subjected to FACS analysis.
Immunofluorescence.
Cells were grown on poly-d-lysine-coated coverslips and fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. They were permeablized with 0.3% Triton X-100 in PBS for 5 min on ice and incubated with primary antibodies for 20 min at 37°C, followed by incubation with Alexa-488 or Cy3-conjugated secondary antibodies for 20 min at 37°C. Finally, cells were counterstained with DAPI and mounted to glass slides with Vectashield. Microscopic images were captured on an Olympus IX70 microscope with a Deltavision (Applied Precision, Issaquah, WA) deconvolution system.
Immunoprecipitations and Antibodies.
HeLa nuclear extract and immunoprecipitation procedures were described in ref. 4.
BLM antibodies were raised against GST-fusion proteins spanning amino acids 1–158 or 108–255 of BLM and affinity purified. Phospho-specific antibodies against phospho-S144, -S304, -S1290, and -S1296 were generated by using synthetic phospho-peptides and affinity-purified (Bethyl Laboratories, Montgomery, TX).
Other antibodies used in this study are mouse monoclonal antibody against α-tubulin (Sigma, St. Louis, MO), rabbit polyclonal antibodies against MPS1, Bub1, PLK1, and histone H3pS10 (Bethyl Labs), cyclin B1 (Santa Cruz Biotechnology, Santa Cruz, CA), and goat polyclonal against MPS1 (Bethyl Laboratories).
Mitotic Spread.
Exponentially growing cells were treated with colcemid (Invitrogen) at a final concentration of 100 ng/ml for 2 h. Mitotic cells were collected and resuspended in 75 mM KCl and incubated at 37°C for 25 min. Cells then were fixed in methanol:acetic acid (3:1) three times for total of 30 min. Fixed cells were dropped on glass slides and mounted with Vectashield containing propidium iodide (Vector Laboratories, Burlingame, CA).
SCE.
Cells were seeded at 16 h before being labeled with BrdU (3 μg/ml) for 40 h. Colcemid were added for 30 min at a final concentration of 0.03 μg/ml. Trypsinized cell pellet was resuspended in 75 mM KCl and incubated at 37°C for 15 min. Cells were fixed in methanol:acetic acid (3:1) three times and dropped on a glass slide. After air drying, cells were stained with acridine orange (0.1 mg/ml) for 5 min, were rinsed briefly with water, and were mounted in 2.8% sodium phosphate dibasic, pH 11.
Mass Spectrometry Analysis.
Mass spectrometry analysis was carried out as described in ref. 36.
Supplementary Material
Acknowledgments
We thank N. Ellis, S. Shieh, and H. Yu for providing reagents; X. He for providing access to and technical assistance with deconvolution microscopy; and P. Zhang, S. Sazer, X. He, and A. Malovannaya for critical reading of the manuscript. This work is supported in part by National Institutes of Health Grant CA84199 (to J.Q.).
Abbreviations
- BS
Bloom syndrome
- PBD
polo-box domain
- SAC
spindle assembly checkpoint
- SCE
sister chromatid exchange
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., Ciocci S., Proytcheva M., German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 2.Neff N. F., Ellis N. A., Ye T. Z., Noonan J., Huang K., Sanz M., Proytcheva M. Mol. Biol. Cell. 1999;10:665–676. doi: 10.1091/mbc.10.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sengupta S., Linke S. P., Pedeux R., Yang Q., Farnsworth J., Garfield S. H., Valerie K., Shay J. W., Ellis N. A., Wasylyk B., et al. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 5.Bischof O., Kim S. H., Irving J., Beresten S., Ellis N. A., Campisi J. J. Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichierri P., Franchitto A., Rosselli F. EMBO J. 2004;23:3154–3163. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies S. L., North P. S., Dart A., Lakin N. D., Hickson I. D. Mol. Cell. Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu P., Beresten S. F., van Brabant A. J., Ye T. Z., Pandolfi P. P., Johnson F. B., Guarente L., Ellis N. A. Hum. Mol. Genet. 2001;10:1287–1298. doi: 10.1093/hmg/10.12.1287. [DOI] [PubMed] [Google Scholar]
- 9.Wu L., Davies S. L., North P. S., Goulaouic H., Riou J. F., Turley H., Gatter K. C., Hickson I. D. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 10.Luo G., Santoro I. M., McDaniel L. D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R. A., Bradley A. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 11.Hickson I. D. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 12.Kadura S., Sazer S. Cell Motil. Cytoskeleton. 2005;61:145–160. doi: 10.1002/cm.20072. [DOI] [PubMed] [Google Scholar]
- 13.Rieder C. L., Maiato H. Dev. Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Kops G. J., Weaver B. A., Cleveland D. W. Nat. Rev. Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 15.Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D., Kidd A., Mehes K., Nash R., Robin N., et al. Nat. Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopalan H., Jallepalli P. V., Rago C., Velculescu V. E., Kinzler K. W., Vogelstein B., Lengauer C. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 17.Fisk H. A., Mattison C. P., Winey M. Proc. Natl. Acad. Sci. USA. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S. T., Chan G. K., Hittle J. C., Fujii G., Lees E., Yen T. J. Mol. Biol. Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stucke V. M., Sillje H. H., Arnaud L., Nigg E. A. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei J. H., Chou Y. F., Ou Y. H., Yeh Y. H., Tyan S. W., Sun T. P., Shen C. Y., Shieh S. Y. J. Biol. Chem. 2005;280:7748–7757. doi: 10.1074/jbc.M410152200. [DOI] [PubMed] [Google Scholar]
- 21.Barr F. A., Sillje H. H., Nigg E. A. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 22.Elia A. E., Cantley L. C., Yaffe M. B. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 23.Dutertre S., Ababou M., Onclercq R., Delic J., Chatton B., Jaulin C., Amor-Gueret M. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Qin J. Proc. Natl. Acad. Sci. USA. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yazdi P. T., Wang Y., Zhao S., Patel N., Lee E. Y., Qin J. Genes Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X., Jones M. H., Winey M., Sazer S. J. Cell Sci. 1998;111:1635–1647. doi: 10.1242/jcs.111.12.1635. [DOI] [PubMed] [Google Scholar]
- 27.Abrieu A., Magnaghi-Jaulin L., Kahana J. A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D. W., Labbe J. C. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 28.Yu H., King R. W., Peters J. M., Kirschner M. W. Curr. Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 29.Ouyang B., Lan Z., Meadows J., Pan H., Fukasawa K., Li W., Dai W. Cell Growth Differ. 1998;9:877–885. [PubMed] [Google Scholar]
- 30.Tang Z., Shu H., Oncel D., Chen S., Yu H. Mol. Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 31.Chen R. H. EMBO J. 2004;23:3113–3121. doi: 10.1038/sj.emboj.7600308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn E. M., Therman E. Cancer Genet. Cytogenet. 1986;22:1–18. doi: 10.1016/0165-4608(86)90132-9. [DOI] [PubMed] [Google Scholar]
- 33.Gruber S. B., Ellis N. A., Scott K. K., Almog R., Kolachana P., Bonner J. D., Kirchhoff T., Tomsho L. P., Nafa K., Pierce H., et al. Science. 2002;297:2013. doi: 10.1126/science.1074399. [DOI] [PubMed] [Google Scholar]
- 34.Goss K. H., Risinger M. A., Kordich J. J., Sanz M. M., Straughen J. E., Slovek L. E., Capobianco A. J., German J., Boivin G. P., Groden J. Science. 2002;297:2051–2053. doi: 10.1126/science.1074340. [DOI] [PubMed] [Google Scholar]
- 35.Elia A. E., Rellos P., Haire L. F., Chao J. W., Ivins F. J., Hoepker K., Mohammad D., Cantley L. C., Smerdon S. J., Yaffe M. B. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X., Herring C. J., Romano P. R., Szczepanowska J., Brzeska H., Hinnebusch A. G., Qin J. Anal. Chem. 1998;70:2050–2059. doi: 10.1021/ac971207m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.