Abstract
We have developed an efficient method to target lentivirus-mediated gene transduction to a desired cell type. It involves incorporation of antibody and fusogenic protein as two distinct molecules into the lentiviral surface. The fusogen is constructed by modifying viral envelope proteins, so that they lack the ability to bind to their cognate receptor but still retain the ability to trigger pH-dependent membrane fusion. Thus, the specificity of such a lentiviral vector is solely determined by the antibody, which is chosen to recognize a specific surface antigen of the desired cell type. This specific binding then induces endocytosis of the surface antigen, bringing the lentivirus into an endosome. There, the fusogen responds to the low pH environment and mediates membrane fusion, allowing the virus core to enter the cytosol. Using CD20 as a target antigen for human B cells, we have demonstrated that this targeting strategy is effective both in vitro and in intact animals. This methodology is flexible and can be extended to other forms of cell type-specific recognition to mediate targeting. The only requirement is that the antibody (or other binding protein) must be endocytosed after interaction with its cell surface-binding determinant.
Keywords: antibody, gene therapy, lentivirus, retrovirus, targeted gene delivery
Gene therapy is the introduction of a functional gene into a target cell to provide a therapeutic advantage (1). A particularly desirable gene therapy protocol would be to precisely deliver a gene of interest to specific cells or organs in vivo by means of administration of a designed gene delivery vehicle. Certain viruses are natural gene delivery systems, and much effort has been focused on engineering viral vectors as gene transfer vehicles (1, 2). Among these vectors, ones derived from oncoretroviruses and lentiviruses exhibit promising features because they have the ability to produce stable transduction, maintain long-term transgene expression and, for lentiviruses, to transduce nondividing cells. Targeting such viruses to particular cell types has proved to be challenging. We report here a general methodology that allows such targeting, even in vivo, and that is remarkably flexible.
Many attempts have been made to develop targetable transduction systems by using retroviral and lentiviral vectors (3, 4). Significant effort has been devoted to altering the envelope glycoprotein (Env), the protein that is responsible for binding the virus to cell surface receptors and for mediating entry. The plasticity of the surface domain of Env allows insertion of ligands, peptides and single-chain antibodies (5–14) that can direct the vectors to specific cell types. However, this manipulation adversely affects the fusion domain of Env, resulting in low viral titers. The unknown and delicate coupling mechanisms of binding and fusion make it extremely difficult to reconstitute fusion function once the surface domain of the same molecule has been altered (4). Another approach involves using a ligand protein or antibody as a bridge to attach the virus to specific cells (15–18). The challenge to this approach is that the Env, once complexed with the one end of the bridge molecule, fuses inefficiently. Because no practical strategies are available for targeted in vivo gene delivery, current gene therapy clinical trials are based on in vitro transduction of purified cells followed by infusion of the modified cells into the patient. This is an expensive procedure, with significant safety challenges.
Our strategy involves uncoupling the target cell recognition function from the fusion function by providing them in separate proteins. For recognition, we use antibodies, and, for fusion, we use a viral glycoprotein that has been mutated to inactivate its binding ability. We make lentiviral vectors that incorporate both molecules into their surface. Our working hypothesis was that the antibody should recognize a molecular constituent on the target cell membrane and attach the lentivirus to the cell surface (Fig. 5, which is published as supporting information on the PNAS web site). Antibody binding should then induce endocytosis, bringing the lentivirus into an endosome. There, the fusogenic molecule (FM) should respond to the low pH environment and trigger membrane fusion, allowing the virus core to enter the cytosol. After reverse transcription and migration of the product to the nucleus, the genome of the vector should integrate into the target cell genome, incorporating the vector's transgene into the cell's inheritance.
Results
Construction of pH-Dependent Fusogen.
Effective FMs for the proposed system should be able to incorporate into the lentivirus envelope and induce membrane fusion at low pH, independent of receptor binding. There are two classes of such FMs (19). The class I fusogens trigger membrane fusion using helical coiled-coil structures whereas the class II fusogens trigger fusion with β barrels. These two structures have different mechanics and kinetics (19), and both were evaluated to determine which would be better for the promotion of infection. One class I fusogen, HA from influenza A/fowl plague virus/Rostock/34 (FPV), was previously found to pseudotype murine leukemia virus (MLV) (20). Cannon and coworkers (21) created a binding defective version of FPV HA designated as HAmu (Fig. 1A). When incorporated into MLV displaying a functionally attenuated envelope glycoprotein, HAmu could enhance viral transduction efficiency (21). HAmu-mediated fusion is thought to be independent of receptor binding (3). The class II FM that we tested was the Sindbis virus glycoprotein from the alphavirus family (22) and is designated as SIN. SIN consists of two transmembrane proteins (23), one responsible for fusion (E1) and the other for cell binding (E2). SIN is known to pseudotype both oncoretroviruses and lentiviruses. By inserting the IgG-binding domain of protein A (ZZ domain) into the E2 protein and making several additional mutations to inactivate the receptor-binding sites, Chen and coworkers (16) made a binding-deficient and fusion-competent SIN. We adapted this form of SIN but replaced the ZZ domain with a 10-residue tag sequence, for which there exists a monoclonal antibody that allows monitoring of SIN expression; we designated it SINmu (Fig. 1C).
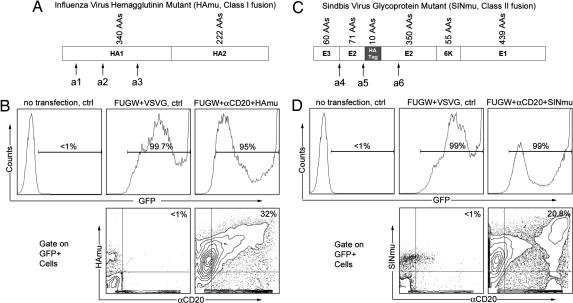
Fig. 1.
Coexpression of antibody and fusogenic protein on the surface of the virus packaging cell line. (A) The class I fusion protein HAmu derived from influenza A (FPV) HA. HA contains two glycoproteins after maturation: HA1 for binding to cell surface receptor, sialic acid; HA2 for triggering membrane fusion. Three point mutations within the receptor binding sites (a1, Y106F; a2, E199Q; a3, G237K) (21) were introduced to generate the binding-defective but fusion-competent HAmu. Single letter amino acid abbreviations are as follows: A, alanine; D, aspartic acid; E, glutamic acid; F, phenylalanine; G, glycine; K, lysine; M, methionine; P, proline; Q, glutamine; V, valine; Y, tyrosine. (B) FACS analysis of virus-producing cells. 293T cells were transiently transfected with plasmids encoding the following: the lentiviral vector FUGW; the membrane-bound antibody αCD20; the accessory proteins for antibodies, Igα and Igβ; the fusion protein HAmu; and viral gag, pol, and rev genes. Expression of αCD20 and HAmu was detected by using anti-human IgG antibody and anti-FPV HA antibody. (C) The class II fusion protein SINmu derived from SIN. SIN contains two membrane glycoproteins (E1 and E2) and a signal peptide (E3): E1 for mediating fusion, E2 for receptor binding, and E3 as a signal sequence for processing of E2 glycoprotein. A 10-residue tag sequence (MYPYDVPDYA) was inserted between amino acids 71 and 74 of the E2 glycoprotein. A series of alterations (a4: deletion of amino acids 61–64 of E3; a5: mutations of 68SLKQ71 into 68AAAA71; mutations of 157KE158 into 157AA158) (16) was introduced to yield the binding-defective and fusion-competent SINmu. (D) Directly analogous to B, except that SINmu was used for the fusion protein and was detected by an anti-tag antibody.
Construction of Membrane-Bound Antibody for Targeting.
The antibody that we have chosen for targeting in this study is the anti-human CD20 antibody (αCD20), a version of which is currently being used in the treatment of B cell lymphomas. We generated a construct that encodes a mouse/human chimeric anti-CD20 antibody with the human membrane-bound IgG constant region (pαCD20). Genes encoding human Igα and Igβ, the two associated proteins that are required for surface expression of antibodies, were cloned into a construct designated pIgαβ.
Preparation of Recombinant Lentiviral Vectors.
The production of lentiviruses enveloped with both anti-CD20 antibody and the candidate FM was achieved by cotransfection of 293T cells with the lentiviral vector FUGW, plasmids encoding viral gag, pol, and rev genes, pαCD20, pIgαβ and pFM (the plasmid encoding a FM, either HAmu or SINmu), by using a standard calcium phosphate precipitation method (24). FUGW is a self-inactivating and replication-incompetent lentiviral vector that carries the human ubiquitin-C promoter driving a GFP reporter gene (25). As a control, the Env derived from vesicular stomatitis virus (VSVG) was used as a joint recognition and fusion protein. FACS analysis of the transfected cells showed that virtually all expressed some level of GFP as an indicator of the presence of the viral vector (Figs. 1 B and D Upper). Some 30% of GFP-positive cells coexpressed HAmu and αCD20 on the cell surface (Fig. 1B Lower). A slightly smaller percentage (≈20%) of the 293T cells exhibited coexpression of GFP, SINmu, and αCD20 (Fig. 1D). The resultant viruses from these transfected production cells were designated FUGW/αCD20+HAmu and FUGW/αCD20+SINmu.
Coincorporation of Fusogen and Antibody into Lentiviral Vectors.
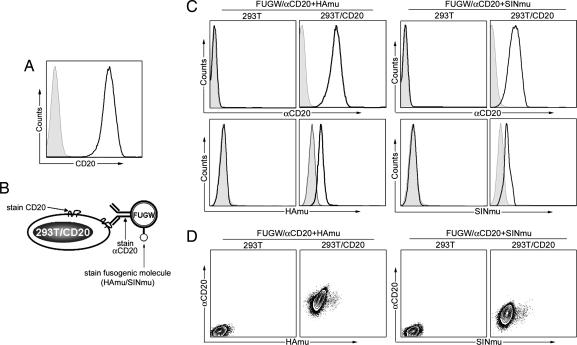
To examine whether αCD20 and the FM were incorporated in the same virion, we performed a virus–cell binding assay. As a target, we made a 293T cell line stably expressing the CD20 protein antigen (293T/CD20; Fig. 2A). The parental cell line 293T served as a negative control. The viral supernatants were incubated with the target cells at 4°C for half an hour. The resultant binding was assayed by means of a three-staining scheme (Fig. 2B). FACS analysis showed that lentivectors bearing αCD20 were in fact able to bind to CD20-expressing 293T cells (Fig. 2C Upper). The control of 293T cells with no CD20 expression displayed no detectable αCD20, showing that the virus binding to cells must be due to a specific interaction between the cell surface CD20 antigen and the viral surface αCD20 molecule. In another control, the virus bearing only FM exhibited no ability to bind either cell line, indicating that the HAmu and SINmu did lack the capacity for cell binding (L.Y., L.B., D.B., and P.W., unpublished work). FACS analysis also showed that the virus bound to the 293T/CD20 cell surface displayed the FMs (Fig. 2C Lower), suggesting that both αCD20 and FM were incorporated on the same virion, which was further confirmed by FACS plots of αCD20 versus FM (Fig. 2D). In addition to codisplay, these results suggest that the presence of the FM does not affect the αCD20 binding to CD20.
Fig. 2.
Virus–cell binding assay to study the codisplay of antibody and fusogenic protein on the lentiviral surface. (A) FACS analysis of target cell line 293T/CD20. CD20 expression was detected by using anti-CD20 antibody. Solid line, expression of CD20 in 293T/CD20; shaded area, CD20 expression in 293T cells (as a control). (B) Schematic representation of three-staining scheme used for analyzing virus–cell binding assay. Three stainings were used to detect the presence of CD20, αCD20, and the fusogenic molecule (HAmu or SINmu), respectively. (C Left) FACS plots of 293T/CD20 cells incubated with FUGW/αCD20+HAmu. The binding of virus to 293T/CD20 cells was probed with antibody against αCD20 (anti-IgG) and HAmu. Solid line, analysis on 293T/CD20; shaded area, analysis on 293T (as a control). (C Right) FACS plots of 293T/CD20 cells incubated with FUGW/aCD20+SINmu. The binding of virus to 293T/CD20 cells was detected by antibody against αCD20 and SINmu. Solid line, analysis on 293T/CD20; shaded area, analysis on 293T (as a control). (D) Codisplay of antibody and fusogenic protein was analyzed by a density plot correlating the presence of the two proteins.
Targeted Transduction of Lentiviral Vectors to Cell Line in Vitro.
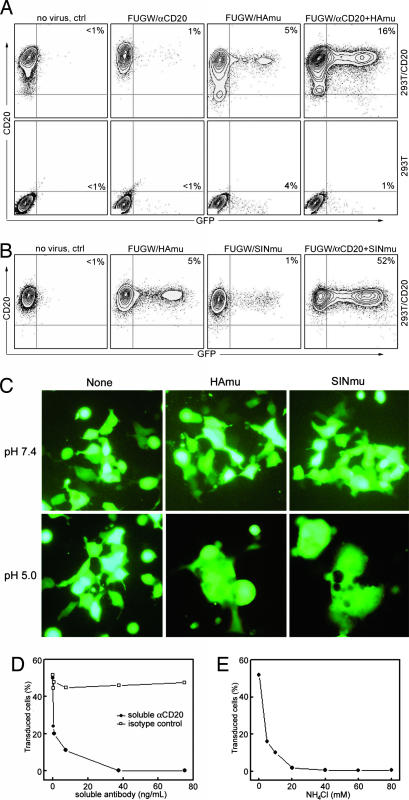
We next examined whether αCD20-bearing virus can transfer genes into cells expressing CD20 in a cell-specific manner. GFP expression was used to measure the transduction efficiency. The supernatants containing lentivectors bearing various surface proteins were incubated with CD20-expressing target cells, and 293T cells served as a control. Four days posttransduction, the efficiency of targeting was analyzed by FACS. Fig. 3A (rightmost image) shows that FUGW/αCD20+HAmu viral particles could specifically transduce 16% of 293T/CD20 cells. Images to the left show that transduction required the presence on the virions of HAmu, but there was some background transduction with virions lacking αCD20, likely because of residual weak binding of HAmu to its ligand, sialic acid. The titer for FUGW/αCD20+HAmu (fresh viral supernatant, no concentration) was estimated to be ≈1 × 105 transduction units (TU)/ml; the titer was determined by the percentage of GFP+ cells in the dilution ranges that showed a linear response. The 293T cells showed a small background infection level but no specific transduction by FUGW/αCD20+HAmu (Fig. 3A Lower). When SINmu was used as the fusion protein, substantial enhancement of specific transduction was observed (52%; Fig. 3B). The titer for FUGW/αCD20+SINmu was estimated to be ≈1 × 106 TU/ml. Also, we detected a much lower transduction in the absence of the binding protein (≈1%). Thus, the data in Fig. 3B show that SINmu is a better fusion protein to partner with αCD20 for targeting lentiviral vectors. When we monitored the transduction at various time points using FACS, we found that SINmu-containing virions exhibited faster transduction kinetics than those with HAmu (L.Y., L.B., D.B., and P.W., unpublished work). Both FUGW/αCD20+HAmu and FUGW/αCD20+SINmu could be concentrated by ultracentrifugation with a >90% recovery rate, which is important for in vivo application.
Fig. 3.
Targeting of lentivectors bearing both antibody and fusion protein to 293T/CD20 cells in vitro. (A) 293T/CD20 cells (2 × 105) were transduced with 500 μl of fresh unconcentrated FUGW/αCD20 (no HAmu), FUGW/HAmu (no αCD20), or FUGW/αCD20+HAmu. 293T cells (no expression of CD20) were included as controls. The resulting GFP expression was analyzed by FACS. The specific transduction titer for FUGW/αCD20+HAmu was estimated to be ≈1 × 105 TU/ml. (B) A similar transduction experiment was performed by using unconcentrated FUGW/SINmu (no αCD20) or FUGW/αCD20+SINmu. For comparison of targeting specificity, cells were also transduced with FUGW/HAmu. The specific transduction titer for FUGW/αCD20+SINmu was estimated to be ≈1 × 106 TU/ml. (C) Evidence of pH-dependent fusion of HAmu and SINmu by a cell–cell fusion assay. 293T cells (0.1 × 106) transiently transfected to express GFP and surface αCD20 and fusion protein (either HAmu or SINmu), and 293T/CD20 cells were mixed together, washed once with normal PBS (pH 7.4), and incubated in low pH PBS (pH 5.0) or normal pH PBS (as a control) for half an hour at 37°C. The cells were then washed and cultured in the regular medium for 1 day. Cells were visualized by epifluorescence microscope equipped with a GFP filter set. (D and E) Effect of addition of soluble αCD20 (D) or NH4Cl (E). αCD20 or NH4Cl was added into viral supernatants during transduction for 8 h. Then, the supernatants were replaced with fresh medium. The cells were analyzed for GFP expression after 2 days. Isotype-matched antibody was used as a control for D.
To assess whether αCD20 and the fusion protein (HAmu or SINmu) had to be incorporated into the same viral particle, and therefore functioned in cis to mediate transduction, we mixed FUGW/αCD20 with FUGW/HAmu or FUGW/SINmu, each displaying only one protein, and tested their transduction of 293T/CD20 cells. This procedure did not result in specific transduction, indicating that the specific transduction conferred by the engineered recombinant viruses requires the two proteins to be displayed on the same viral particle.
Antibody–Antigen Interaction Responsible for Targeted Transduction.
It seems that two distinct proteins can carry the binding and fusion events of engineered lentiviruses for targeted transduction. To further confirm that the specificity we observed was a consequence of interaction between αCD20 and CD20, we transduced 293T/CD20 cells in the presence of anti-CD20 blocking antibody. As expected, a dramatic decrease of infectivity was detected for both FUGW/αCD20+HAmu (L.Y., L.B., D.B., and P.W., unpublished work) and FUGW/αCD20+SINmu (Fig. 3D), suggesting the essential role of antibody–antigen binding for the targeted transduction.
Confirmation of pH Dependence of Fusogen.
To examine the requirement for a low pH comparment to allow the recombinant lentivectors to penetrate into cells, we incubated both FUGW/αCD20+HAmu and FUGW/αCD20+SINmu with 293T/CD20 cells in the absence or presence of ammonium chloride (NH4Cl), which can neutralize acidic endosomal compartments. Addition of NH4Cl to cells completely abolished transduction by either FUGW/αCD20+HAmu (L.Y., L.B., D.B., and P.W., unpublished work) or FUGW/αCD20+SINmu (Fig. 3E). These results are consistent with the low pH requirement of HA and SIN to trigger membrane fusion. More direct evidence for pH-dependent fusion was provided by a cell–cell fusion assay. 293T cells expressing GFP, surface αCD20, and FM were incubated with 293T/CD20 cells in a low-pH buffer for half an hour, followed by culturing in regular medium. Both HAmu and SINmu induced cell–cell fusion by forming multinucleated polykaryons (Fig. 3C). The interaction between αCD20 and CD20 dramatically enhances the probability of fusion, because a similar experiment with cells that lacked αCD20 and CD20 yielded a much lower level of fusion (L. Y., L.B., D.B., and P.W., unpublished work). The αCD20/CD20 interaction probably brings the cell membranes into close apposition, facilitating the action of the fusion protein.
Targeted Transduction of Lentiviral Vectors to Primary B Cells.
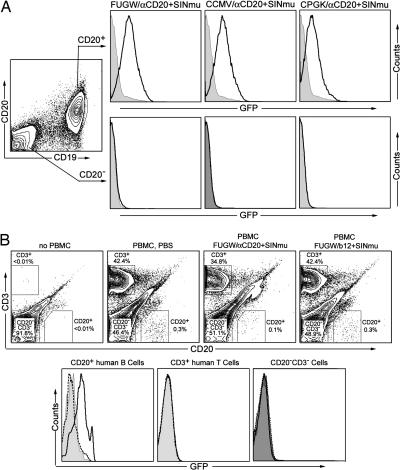
Having established the ability of the system to mediate CD20-specific transduction of artificially created cell lines, we next investigated the possibility of specific transduction of primary human B-lymphoid cells, cells that naturally carry the CD20 antigen. Fresh, unfractionated human peripheral blood mononuclear cells (PBMCs) were transduced with FUGW/αCD20+SINmu and then stimulated with LPS to expand the B cell population. Four days later, the cells were stained for CD19 (a B cell marker), CD20, and GFP expression (Fig. 4A). We found that >35% of cells were CD20+ B cells under our culture condition. When we gated on CD20+ B cells, the majority of them were GFP+. On the contrary, virtually no GFP+ cells were detected when we gated on CD20− non-B cells, confirming that the transduction was strictly dependent on CD20 expression. In another control experiment, fresh PBMCs were transduced with FUGW/αCD20+SINmu followed by stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin to expand T cells. FACS analysis of these T cells showed no expression of GFP (Fig. 6, which is published as supporting information on the PNAS web site), confirming transduction specificity.
Fig. 4.
Targeting CD20+ human primary B cells in vitro and in vivo using engineered lentivectors. (A) Fresh, unfractionated human PBMCs (2 × 106) were transduced by coculturing with concentrated FUGW/αCD20+SINmu, CCMV/αCD20+SINmu, or CPGK/αCD20+SINmu (10 × 106 TU). LPS (50 μg/ml) was added into the culture media for B cells to survive and grow. After 2 days, the B cell population was identified by costaining of CD19 and CD20. Solid line, analysis on transduced cells; shaded area, analysis on cells without transduction (as a control). (B) Fresh human PBMCs were transferred into irradiated RAG2−/−γc−/− mice (100 × 106 per mouse) via tail vein injection. Six hours later, concentrated virus (100 × 106 TU per mouse) was injected through the tail vein. Two days later, whole blood was collected from these mice via heart puncture, and the cells were stained for human CD3 and CD20 and then analyzed by FACS for GFP expression. Shaded area, no virus treatment; dashed line, treated with FUGW/b12+SINmu; solid line, treated with FUGW/αCD20+SINmu.
To demonstrate that the targeting method is not limited to the lentiviral vector FUGW, we evaluated two additional lentiviral vectors with different promoter configurations. Kohn and coworkers (26) have incorporated the Ig heavy chain enhancer (Eμ) with associated matrix attachment regions into lentivectors carrying either the human cytomegalovirus (CMV) promoter (CCMV) or the murine phosphoglycerate kinase promoter (CPGK). We adapted these two lentiviral vectors into our system and prepared recombinant lentiviruses CCMV/αCD20+SINmu and CPGK/αCD20+SINmu. Transduction of PBMC-derived B cells with these viral supernatants exhibited results similar to those observed previously with FUGW (Fig. 4A).
Targeted Transduction of Recombinant Lentiviral Vectors in Vivo.
The real test of this system is whether it will mediate specific transduction in vivo. For this purpose, we used a human PBMC xenograft in a mouse model. Fresh human PBMCs (100 × 106 per mouse) were transferred into irradiated immunodeficient RAG2−/−γc−/− mice through a tail vein injection. Engineered lentiviruses bearing αCD20 and SINmu were administered through the tail vein 6 h after human cell transfer. After 2 days, we collected the whole blood from these mice, and the cells were analyzed for surface antigens and GFP expression. Approximately 30–40% of the cells recovered from the mice were human T cells (CD3+), and ≈0.1–0.3% were CD20+ human B cells (Fig. 4B). Three populations were analyzed for GFP expression: CD20+, CD3+, and CD20−CD3−. None of the cells harvested from mice injected with virus bearing a control antibody and SINmu (FUGW/b12+SINmu) showed evidence of GFP expression in any of the three populations (Fig. 4B). In contrast, GFP expression was observed in at least 40% of the CD20+ cells isolated from mice injected with FUGW/αCD20+SINmu whereas no transduction was detected in the other two populations.
Discussion
This demonstration of targeting efficient gene delivery vehicles strictly to the desired cell types in vivo greatly enhances the therapeutic potential of lentivirus-mediated gene therapy and alleviates concerns of off-target effects. Possibly the most important implication of the work is that gene therapy could now be carried out as an inexpensive procedure, able to be considered even in the less-developed world.
In our approach, we break up the binding and fusion functions into two separate molecules that are inserted into the viral envelope. This methodology is particularly easy with lentiviruses (or other retroviruses) because these viruses readily incorporate into their envelope whatever proteins are found on the surface of producing cells (27). Other viruses have surfaces with many close-packed viral glycoproteins and exclude cellular proteins. A major advantage of this scheme over others where the viral protein is engineered with a foreign binding component is that the fusion protein maintains its full biological activity so that viral titer is not killed for increased specificity. The other key to the method is choosing a viral glycoprotein that mediates fusion in response to low pH and a cellular receptor that is efficiently endocytosed after antibody binding. The fusion molecule must exhibit fast enough kinetics that the viral contents can empty into the cytosol before the degradation of the viral particle. Our choice of CD20 as a target was arbitrary. We have already extended the method to other antibodies and cell surface receptor–ligand pairs.§ We envision that the flexibility (easy combination of antibody, or other binding protein, and fusiogenic molecule) and broadness (availability of monoclonal antibodies or ligands for many endocytosed cell-specific surface molecules) of this method will facilitate the application of targeted gene delivery for therapy and research.
Materials and Methods
Construct Preparation.
The cDNAs of the human κ light chain constant region and the membrane bound human IgG1 constant region were amplified and inserted downstream of human CMV and EF1αpromoters, respectively, in the pBudCE4.1 vector (Invitrogen). We cloned the light and heavy chain variable regions from the murine anti-CD20 antibody (clone 2H7) using PCR amplification and inserted them directly upstream of the corresponding constant regions. The resulting construct was designated pαCD20. We cloned cDNAs of human Igα and Igβ into the pBudCE4.1 vector (Invitrogen) to yield pIgαβ.
P. Cannon (University of Southern California and Childrens Hospital, Los Angeles) was kind enough to provide us with the construct encoding HAmu (21). We obtained the cDNA for wild-type SIN from J. Strauss's laboratory at the California Institute of Technology. PCR mutagenesis and assembly were used to generate the mutant SIN as described by Chen and colleagues (16), except a 10-residue tag sequence (MYPYDVPDYA) replaced the ZZ domain of protein A, which is located between amino acids 71 and 74 of the E2 glycoprotein of SIN. This version of SIN is designated SINmu.
Virus Production.
Lentivectors were generated by transfecting 293T cells by using a standard calcium phosphate precipitation technique (24). 293T cells (≈80% confluent) in 6-cm culture dishes were transfected with the appropriate lentiviral vector plasmid (5 μg), together with 2.5 μg each of pαCD20, pIgαβ, and the packaging vector plasmids (pMDLg/pRRE and pRSV-Rev) (28). The viral supernatants were harvested 48 and 72 h after transfection and filtered through a 0.45-μm pore size filter.
To prepare high-titer lentivectors, the viral supernatants were concentrated by using ultracentrifugation (Optima L-80 K preparative ultracentrifuge, Beckman Coulter) for 90 min at 50,000 × g. Particles were then resuspended in an appropriate volume of cold PBS.
Cell Line Construction.
The 293T/CD20 cell line was generated by stable transduction via vesicular stomatitis virus (VSVG)-pseudotyped lentivector. The cDNA of human CD20 was cloned downstream of the human ubiquitin-C promoter in the lentivector plasmid FUW to generate FUW-CD20. The lentiviral vector FUW-CD20 was then pseudotyped with VSVG and was used to transduce 293T. The resulting cells were subjected to cell sorting to obtain a uniform population of CD20+ cells designated 293T/CD20.
Virus–Cell Binding Assay.
Cells (293T/CD20 or 293T, 0.1 × 106) were incubated with 500 μl of viral supernatant at 4°C for half an hour and washed with 4 ml of cold PBS. The cells were then stained with the following three antibodies: an anti-human IgG antibody (BD PharMingen) to stain αCD20, an anti-human CD20 antibody (BD PharMingen) to stain CD20, and an anti-FPV HA polyclonal antibody (obtained from H.-D. Klenk, Institute of Virology, Philipps University, Marburg, Germany) to stain HAmu, or an anti-tag antibody (Roche Applied Science, Mannheim, Germany) to stain SINmu. After staining, cells were analyzed by FACS analysis.
Targeted Transduction of 293T/CD20 Cells in Vitro.
293T/CD20 cells (0.2 × 106 per well) or 293T cells (0.2 × 106 per well) were plated in a 24-well culture dish and spin-infected with viral supernatants (0.5 ml per well) at 2,500 rpm, 30°C for 90 min by using a Beckman Allegra 6R centrifuge. Then, the medium was removed and replaced with fresh medium and incubated for a further 3 days at 37°C with 5% CO2. The percentage of GFP+ cells was determined by FACS. The transduction titer was measured in dilution ranges that exhibited a linear response.
Effects of Soluble Antibody and NH4Cl on Viral Transduction.
293T/CD20 cells (0.2 × 106) and 0.5 ml of viral supernatants were incubated for 8 h in the absence or presence of graded amounts of αCD20 (BD PharMingen) or NH4Cl. The medium was replaced with fresh medium and incubated for another 2 days at 37°C with 5% CO2. FACS analysis was used to quantify transduction efficiency.
Cell–Cell Fusion Assay.
293T cells (0.1 × 106), transiently transfected to express GFP, surface αCD20, and fusion protein (either HAmu or SINmu), and 293T/CD20 cells (0.1 × 106) were mixed together, washed twice with normal PBS (pH 7.4), and incubated in 150 μl of low pH PBS (pH 5.0) or normal pH PBS (pH 7.4) (as a control) for half an hour at 37°C with 5% CO2. The cells were then washed extensively and cultured in the regular medium for 1 day. Cells were visualized by an epifluorescence microscope equipped with a GFP filter set.
Targeted Transduction of Primary Human B Cells in Vitro.
Fresh, unfractionated human PBMCs (2 × 106) (AllCells) were incubated with concentrated virus with total TUs of 10 × 106 (based on the titer on 293T/CD20 cells). LPS (50 μg/ml) was then added for B cells to survive and grow. After 2 days, cells were harvested and washed in PBS. B cell population was determined by FACS staining using anti-human CD20 and CD19 antibodies. Targeting transduction was quantified by gating on the different populations of cells and measuring their GFP expression.
Targeted Transduction of Primary Human B Cells in Vivo.
RAG2−/−γc−/− female mice (Taconic) of 6–8 weeks old were given 360 rad whole-body irradiation. On the following day, 100 × 106 fresh human PBMCs (AllCells) were transferred by tail vein injection into each mouse. After 6 h, concentrated virus (100 × 106 TU per mouse) or PBS (as control) was administered via the tail vein. Two days later, whole blood was collected from these mice via heart puncture, and the cells were stained for human CD3 and CD20 and then analyzed by FACS for CD3, CD20, and GFP expression. The mice were maintained on the mixed antibiotic sulfmethoxazole and trimethoprim oral suspension (Hi-Tech Pharmacal) in a sterile environment in the California Institute of Technology animal facility in accordance with institute regulations.
Supplementary Material
Acknowledgments
We thank Dr. James Strauss (California Institute of Technology, Pasadena, CA), Drs. Paula Cannon and Donald Kohn (Childrens Hospital, Los Angeles, CA), and H.-D. Klenk (Philipps University, Marburg, Germany) for providing reagents; Rochelle Diamond and Stephanie Adams for help with cell sorting; and Dr. Markus Covert and Eric Santiestevan for critical reading of this manuscript. This research was generously supported by the Skirball Foundation, a grant from the Bill and Melinda Gates Foundation through the Grand Challenges in Global Health Initiative, and a University of Southern California startup fund.
Abbreviations
- FM
fusogenic molecule
- Env
envelope glycoprotein
- SIN
Sindbis virus glycoprotein
- αCD20
anti-human CD20 antibody
- TU
transduction units
- PBMC
peripheral blood mononuclear cells
- FPV
influenza A/fowl plague virus/Rostock/34
Footnotes
Conflict of interest statement: No conflicts declared.
We have observed that this method can be exploited to target dendritic cells using a membrane-bound monoclonal antibody against the DEC-205 receptor. In addition, we found that incorporation of a membrane-bound form of stem cell factor could target c-kit-positive cells.
References
- 1.Verma I. M., Somia N. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 2.Somia N., Verma I. M. Nat. Rev. Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- 3.Lavillette D., Russell S. J., Cosset F. L. Curr. Opin. Biotech. 2001;12:461–466. doi: 10.1016/s0958-1669(00)00246-9. [DOI] [PubMed] [Google Scholar]
- 4.Sandrin V., Russell S. J., Cosset F. L. Curr. Top. Microbiol. Immunol. 2003;281:137–178. doi: 10.1007/978-3-642-19012-4_4. [DOI] [PubMed] [Google Scholar]
- 5.Somia N. V., Zoppe M., Verma I. M. Proc. Natl. Acad. Sci. USA. 1995;92:7570–7574. doi: 10.1073/pnas.92.16.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang A., Chu T. H. T., Nocken F., Cichutek K., Dornburg R. J. Virol. 1998;72:10148–10156. doi: 10.1128/jvi.72.12.10148-10156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen T. H., Pages J. C., Farge D., Briand P., Weber A. Hum. Gene Ther. 1998;9:2469–2479. doi: 10.1089/hum.1998.9.17-2469. [DOI] [PubMed] [Google Scholar]
- 8.Maurice M., Verhoeyen E., Salmon P., Trono D., Russell S. J., Cosset F. L. Blood. 2002;99:2342–2350. doi: 10.1182/blood.v99.7.2342. [DOI] [PubMed] [Google Scholar]
- 9.Fielding A. K., Chapel-Fernandes S., Chadwick M. P., Bullough F. J., Cosset F. L., Russell S. J. Hum. Gene Ther. 2000;11:817–826. doi: 10.1089/10430340050015437. [DOI] [PubMed] [Google Scholar]
- 10.Gollan T. J., Green M. R. J. Virol. 2002;76:3558–3563. doi: 10.1128/JVI.76.7.3558-3563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin F., Chowdhury S., Neil S., Phillipps N., Collins M. K. Mol. Ther. 2002;5:269–274. doi: 10.1006/mthe.2002.0550. [DOI] [PubMed] [Google Scholar]
- 12.Valsesiawittmann S., Drynda A., Deleage G., Aumailley M., Heard J. M., Danos O., Verdier G., Cosset F. L. J. Virol. 1994;68:4609–4619. doi: 10.1128/jvi.68.7.4609-4619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasahara N., Dozy A. M., Kan Y. W. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 14.Han X., Kasahara N., Kan Y. W. Proc. Natl. Acad. Sci. USA. 1995;92:9747–9751. doi: 10.1073/pnas.92.21.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morizono K., Bristol G., Xie Y. M., Kung S. K. P., Chen I. S. Y. J. Virol. 2001;75:8016–8020. doi: 10.1128/JVI.75.17.8016-8020.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morizono K., Xie Y. M., Ringpis G. E., Johnson M., Nassanian H., Lee B., Wu L., Chen I. S. Y. Nat. Med. 2005;11:346–352. doi: 10.1038/nm1192. [DOI] [PubMed] [Google Scholar]
- 17.Roux P., Jeanteur P., Piechaczyk M. Proc. Natl. Acad. Sci. USA. 1989;86:9079–9083. doi: 10.1073/pnas.86.23.9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boerger A. L., Snitkovsky S., Young J. A. Proc. Natl. Acad. Sci. USA. 1999;96:9867–9872. doi: 10.1073/pnas.96.17.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrov D. S. Nat. Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatziioannou T., Valsesia-Wittmann S., Russell S. J., Cosset F. L. J. Virol. 1998;72:5313–5317. doi: 10.1128/jvi.72.6.5313-5317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin A. H., Kasahara N., Wu W., Stripecke R., Empig C. L., Anderson W. F., Cannon P. M. Hum. Gene Ther. 2001;12:323–332. doi: 10.1089/10430340150503957. [DOI] [PubMed] [Google Scholar]
- 22.Wang K. S., Kuhn R. J., Strauss E. G., Ou S., Strauss J. H. J. Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay S., Kuhn R. J., Rossmann M. G. Nat. Rev. Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 24.Pear W. S., Nolan G. P., Scott M. L., Baltimore D. Proc. Natl. Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lois C., Hong E. J., Pease S., Brown E. J., Baltimore D. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 26.Lutzko C., Senadheera D., Skelton D., Petersen D., Kohn D. B. J. Virol. 2003;77:7341–7351. doi: 10.1128/JVI.77.13.7341-7351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coffin J. M., Hughes S. H., Varmus H. E. Retroviruses. Woodbury, NY: Cold Spring Harbor Lab. Press; 1997. [PubMed] [Google Scholar]
- 28.Klages N., Zufferey R., Trono D. Mol. Ther. 2000;2:170–176. doi: 10.1006/mthe.2000.0103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.