Abstract
B cell chronic lymphocytic leukemia (B-CLL) is a clonal overgrowth of CD5+ B lymphocytes. In this disease, the B cell antigen receptor (BCR) is intimately linked to disease severity, because patients with BCRs, comprised of unmutated VH genes, follow a much more aggressive course. This and related observations suggest that B-CLL derives from a B cell subset comprised of restricted BCR structural diversity and that antigen-selection and drive are major factors promoting the disease. Nevertheless, the initiating event(s) that lead to the development of B-CLL are still unclear, in part because of the lack of an animal model that spontaneously evolves the molecular abnormalities that occur in the human disease. Because overexpression of the TCL1 gene in murine B cells leads to a CD5+ B cell lymphoproliferative disorder with many of the features of human B-CLL, we studied leukemias emerging in these mice to examine the extent to which their BCRs resemble those in B-CLL. Our data indicate that the immunoglobulin heavy and light chain rearrangements in TCL1 mice display minimal levels of somatic mutations and exhibit several molecular features found in the human disease. Like human B-CLL, TCL1 leukemic rearrangements from different mice can be very similar structurally and closely resemble autoantibodies and antibodies reactive with microbial antigens. Antigen-binding analyses confirm that selected TCL1 clones react with glycerophospholipid, lipoprotein, and polysaccharides that can be autoantigens and be expressed by microbes. This (auto)antigen-driven mouse model reliably captures the BCR characteristics of aggressive, treatment-resistant human B-CLL.
Keywords: autoantibodies, mouse model, oncogene deregulation, B1 cells, autoantigens
B cell chronic lymphocytic leukemia (B-CLL), the most common adult leukemia in the Western world, is problematic for patients and clinicians because it has a heterogeneous clinical course and lacks a curative therapy (1). Some patients live for decades, dying with the disease and not because of it, whereas others have a much more aggressive course and shorter survival despite repeated efforts at providing beneficial relief. Accordingly, the management of patients is often relegated to a “wait and watch” mode, because initiating therapy early in the illness, before it becomes clear which clinical course an individual patient will follow, has not led to therapeutic advantage (2).
Significant advances in our understanding of this disease have come from studying the leukemic B cell’s antigen receptor (BCR; refs. 3–5). For example, these observations enabled the categorization of B-CLL patients into subgroups based on the presence or absence of Ig VH gene mutations (6, 7), termed “unmutated B-CLL” (U-CLL) and “mutated B-CLL,” that presage and influence disease heterogeneity and severity. Patients with U-CLL follow the more aggressive clinical course with shorter survival (8, 9). In addition, BCRs with remarkable structural similarity can be found between patients (10–17), and these patients can have poor outcomes, regardless of mutation status (11, 12). This striking similarity in BCR structure among different patients led to the conclusion that antigen selection and drive are major factors promoting B-CLL (3, 5). Nevertheless, the specific factors initiating the disease remain undefined (18).
Several challenges have thwarted defining the transforming event giving rise to B-CLL: B-CLL cells are resistant to immortalization in vitro and xenogeneic transfer in vivo, and an animal model that reproducibly replicates abnormalities occurring in the human disease, particularly the aggressive, treatment-resistant form, is lacking. Although prior attempts at producing mouse models [e.g., NZB mice, and TRAFDN/Bcl-2 double transgenic (Tg) animals; refs. 19 and 20] successfully reproduced some of the phenotypic features of the human disease, none demonstrate the detailed BCR features that are a hallmark of B-CLL. Recently, Bichi et al. (21) reported that overexpression of TCL1, a gene implicated in a number of human lymphoid malignancies (22, 23) that correlates with proliferative potential in B-CLL (24), leads to a murine disease with many of the characteristics of the human leukemia. These animals spontaneously develop B cell hyperplasia of the CD5+ lineage, initially in the peritoneal cavity and then in lymph nodes, spleen, bone marrow, and blood, that becomes oligoclonal and eventually monoclonal with age (8–16 months). Affected animals ultimately succumb because of the consequences of massive B cell clonal expansion. However, detailed information regarding the structure and mutation status of the BCR in the model has been unavailable.
The present study was designed to determine the extent to which VHDJH and VLJL rearrangements from a series of lethal TCL1-derived leukemias resemble those found in patients with B-CLL. Our findings indicate that the molecular features of the BCRs of TCL1 Tg mice resemble those from human B-CLL patients with the more aggressive form of the disease (U-CLL). Specifically, (i) somatic hypermutation has not significantly altered the structure of Ig VL and VH, (ii) biases exist in IgV gene use, D and J segment association, and CDR3 characteristics, (iii) sets of leukemic clones display remarkably similar BCRs, (iv) the structures of the leukemic Igs resemble those of antibodies to autoantigens and microbes, and (v) these Igs react with autoantigens and antigens found in bacterial cell membranes.
Results
(i) Characteristics of BCRs Expressed by Leukemic B Cells of TCL1 Tg Mice.
Ig V gene mutation status.
Patients with B-CLL segregate into two groups based on the presence or absence of significant numbers of Ig VH mutations (6, 7), and those patients with U-CLL have a radically worse clinical course (8, 9). In this study, 20 of 21 TCL1 mice showed a dominant VHDJH rearrangement, which we designated the major clone. Animal TCL1-021 died of a nonlymphoid malignancy (histiocytic sarcoma), which occasionally occurs in TCL1 mice (25), and the spleen of this animal exhibited 11 distinct B cell clones of approximately equal frequency that were not analyzed further.
Analysis of Ig VH mutation status requires knowledge of the number of germ-line segments and their DNA sequences. For humans, such analyses are robust because the number of functional germ-line VH genes is relatively few (≈43 and 51 for IMGT and VBase databases, respectively), and the corresponding loci have been sequenced (26–29). However, for mice, these analyses are less precise because the germ-line IgV repertoire is much larger (≈300 VH genes; refs. 30–32) and not completely defined (32). Therefore, in these murine studies we considered a level of somatic difference of <2.0% to be “unmutated,” an approach that was taken in human B-CLL studies, allowing for unknown polymorphisms or germ-line genes (6). Although our initial DNA sequence analyses indicated that 17 of 20 clones from the TCL1 leukemias expressed VH genes that were either identical or differed only minimally (<2%) from their most similar germ-line genes, three samples (TCL1-012, -015, and -017) exhibited sequences that were very different from those in the public databases. Therefore, we searched genomic DNA from non-Tg syngeneic mice for previously undescribed polymorphic variants of known genes or previously undefined germ-line genes that might have greater homology with the three apparently mutated clones. In each instance, we identified VH gene sequences that were not recorded in the public databases, indicating that the DNA sequences of TCL1-012, -015, and -017 differed by <2% from these previously undescribed germ-line genes (Table 1, VH genes). In the final analysis, all major leukemic clones expressed VH genes that are identical or only slightly divergent from those in the germ line (0.0–1.7%; mean = 0.5%).
Table 1.
IgVH and IgVL use and mutation status of TCL1 leukemic clones
| VH genes | VL genes | ||||||
|---|---|---|---|---|---|---|---|
| Animal no. | VH family | VH gene | % difference from germ line | Animal no. | Vκ family | Vκ gene | % difference from germ line |
| TCL1-001 | 12 | NC1-A7 | 0.0 | TCL1-001 | Vκ4 | V4-91 | 0.0 |
| TCL1-002 | 12 | NCl-A7 | 0.0 | TCL1-002 | Vκ4 | V4-91 | 0.0 |
| TCL1-003 | 4 | V4S1 | 0.3 | TCL1-003 | Vκ6 | V6-32 | 0.0 |
| TCL1-004 | 4 | V4S1 | 0.3 | TCL1-004 | Vκ6 | V6-32 | 0.0 |
| TCL1-005 | 11 | V11S1 | 0.7 | TCL1-005 | Vκ6 | V6-17 | 0.0 |
| TCL1-006 | 11 | V11S1 | 0.7 | TCL1-006 | Vκ3 | V3-5 | 0.0 |
| TCL1-007 | 1 | V165.1 | 0.0 | TCL1-007 | Vκ12 | V12-89 | 0.0 |
| TCL1-008 | 1 | V1S61 | 0.3 | TCL1-008 | Vκ1 | V1-135 | 0.0 |
| TCL1-009 | 1 | V165.1 | 0.0 | TCL1-009 | Vκ3 | V3-12 | 0.0 |
| TCL1-010 | 1 | V165.1 | 0.0 | TCL1-010 | Vκ12 | V12-89 | 0.0 |
| TCL1-011 | 1 | V165.1 | 0.0 | TCL1-011 | Not available | ||
| TCL1-012 | 1 | XE1+ | 0.7 | TCL1-012 | Vκ17 | V17-121 | 0.0 |
| TCL1-013 | 1 | V1S16 | 1.0 | TCL1-013 | Vκ8 | V8-27 | 0.0 |
| TCL1-014 | 1 | VMU-3.2 | 0.3 | TCL1-014 | Vκ10 | V10-96 | 0.0 |
| TCL1-015 | 1 | XE3+ | 1.7 | TCL1-015 | Vκ4 | V4-80 | 0.0 |
| TCL1-016 | 1 | J558.51 | 0.3 | TCL1-016 | Vκ8 | V8-28 | 0.0 |
| TCL1-017 | 3 | XE2+ | 1.0 | TCL1-017 | Vκ5 | V5-39 | 0.7 |
| TCL1-018 | 5 | VH7183.3b | 0.3 | TCL1-018 | Vκ10 | V10-96 | 0.0 |
| TCL1-019 | 6 | V6S1 | 1.4 | TCL1-019 | Not available | ||
| TCL1-020 | 7 | V7S1 | 0.0 | TCL1-020 | Not available | ||
| TCL1-021 | 11 different clones | TCL1-021 | 13 different clones | ||||
The expressed VL genes from these cases were even more similar to their germ-line counterparts (Table 1, VL genes). Only 1 of the 17 TCL1 clones for which we defined an L chain sequence (no. 017) exhibited any somatic changes (0.7% difference; mean for all animals = 0.1%). Because the murine Vκ locus has been sequenced (31, 33, 34), these data may be more informative than those for VH regarding B cell exposure to the somatic hypermutation process. Thus, both VH and VL genes of the TCL1-derived leukemias are identical and minimally divergent from the germ line, similar to U-CLL.
Ig V gene segment use.
In human B-CLL, Ig VH expression differs from that of normal circulating B cells (3–5). The major clones from 50% (10 of 20) of the animals expressed a VH1 family gene, which is similar to the representation of VH1 genes in the known murine germ-line repertoire (≈67% as per IMGT and NCBI databases). However, four of the VH1+ clones (nos. 007, 009, 010, and 011) expressed V165.1, a frequency four times that of normal murine B cells (≈10%; ref. 35). In the remaining 10 mice, three pairs expressed the same VH gene: TCL1-001 and -002 expressed NC1-A7, the sole member of the VH12 family; TCL1-005 and -006 expressed V11S1, the only member of the VH11 family; and TCL1-003 and -004 expressed V4S1, one of only two members in the VH4 family (Table 1, VH genes).
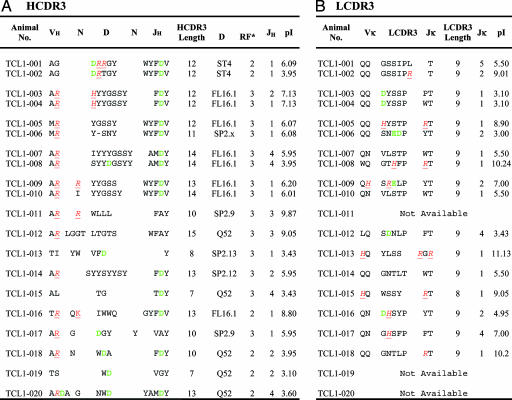
The 20 TCL1 clones used four D segment families (Table 2, HCDR3): DFL (40%), DSP (25%), DQ52 (25%), and DST (10%). In addition, 50% of the clones expressed JH1. Because in normal murine B cells the DSP, DQ52, and DST families and the JH1 segment are expressed at different frequencies (DSP, ≈50%; DQ52, 4–10%; DST, <2%; and JH1, 16%; ref. 36), a D and JH segment bias appears to exist among TCL1 leukemias.
Table 2.
H and L CDR3 rearrangements in TCL1 leukemic clones
Red letters indicate basic, positively charged amino acids; green letters indicate acidic, negatively charged amino acids. RF, D segment reading frame. RF no. 1 starts from the first nucleotide of germ-line gene, no. 2 starts from the second, and no. 3 starts from the third nucleotide.
Although not analyzed to as great an extent as Ig VH, the VL repertoire in human B-CLL also appears to differ from that of the normal adult B cells (37). All of the major TCL1-derived leukemic clones used Vκ genes and four genes, V10-96, V4-91, V6-32, and V12-89, were found twice in the 17 clones for which L chain sequences were available (Table 1). Thus, like human B-CLL, the use of specific IgV gene segments in TCL1 mice diverges from that of the normal B cell repertoire.
CDR3 features.
H and L CDR3s of human B-CLL cells can be distinctive in length, amino acid composition and charge, and D and JH segment pairing (7, 38, 39). The details of these differences vary between aggressive U-CLL and indolent mutated B-CLL.
Long HCDR3s are often found in VH unmutated, poor outcome B-CLL (7, 38), because of the use of longer human germ-line JH segments (e.g., JH6) and the insertion of nontemplated nucleotides at the VH–D and D–JH junctions. In normal adult murine B cells, mean HCDR3 length is ≈12 aa (ref. 40). HCDR3s of 4 of the 20 TCL1-derived major clones (Table 2, HCDR3) comprised 14 or more aa (nos. 007, 008, 010 and 012) and 4 others comprised 13 aa (nos. 009, 014, 016, and 020). For TCL1-007 and -008, this increase in HCDR3 length is due to the use of JH4, the longest JH segment in the murine repertoire. However, for the other leukemic clones, except no. 012, use of a long JH segment (JH1; Table 1, VH genes) and the addition of N nucleotides (Fig. 1, which is published as supporting information on the PNAS web site) were responsible for the longer HCDR3s. TCL1-012 is interesting because despite the use of DQ52, the shortest murine D segment, and JH3, one of the shorter JH segments, its HCDR3 is 15 aa, owing to extensive N insertions (Table 2, HCDR3). Indeed, 9 of the 20 TCL1-derived clones exhibit insertions of nontemplated nucleotides at the VH–D and, to a much lesser extent, at the D–JH junctions.
Nine leukemic clones had HCDR3s containing three or more charged amino acids, e.g., Arg (R), Lys (K), and Asp (D; Table 2, HCDR3). Germ-line segments coded some of these charged residues (e.g., Asp in JH1), whereas others were created by somatic events (e.g., Asp and Arg in TCL1-001, Arg in TCL1-009, Lys in TCL1-016, and Asp in TCL1-008). It is noteworthy that the net charge of each HCDR3/LCDR3 pair was relatively neutral, either because of the pairing of two segments with pI values at different ends of the isoelectric spectrum or with similar, more neutral pI values (Table 2). The presence of charged amino acids in HCDR3 is seen more often in treatment-resistant U-CLL.
Finally, nine HCDR3s were enriched in Tyr (Y), Gly (G), and Ser (S) primarily due to the use of DFL segments (Table 2, HCDR3). This clustering of uncharged amino acids is reminiscent of human B-CLL where these amino acids are seen in rearrangements by using VH1-69 (38) and VH4-39 (13), genes associated with aggressive U-CLL.
Stereotypic VHDJH rearrangements.
Leukemic clones of some patients with B-CLL have HCDR3s with strikingly homologous amino acid sequences and, in many cases, these HCDR3 associate with identical VH and occasionally VL segments, thereby creating sets of patients with quasi-identical antigen-binding sites (7, 10, 13–17, 41). Furthermore, when protein databases are searched for antibodies with HCDR3s of appreciable amino acid sequence similarity (≥60%) with B-CLL mAb, virtually only rearrangements from B-CLL patients are found, with rearrangements from other B cell lymphoproliferative disorders, autoantibody-expressing B cells, and normal B cells identified much less frequently.
Among our 20 TCL1-derived leukemias, we found 10 that comprise five sets of clones defined by ≥80% amino acid similarity in HCDR3 (set I, TCL1-001 and -002; II, TCL1-003 and -004; III, TCL1-005 and -006; IV, TCL1-007 and -008; and V, TCL1-009 and -010; Table 2, HCDR3). The members of set II have identical HCDR3 sequences. Furthermore, the members of set I differ by only 1 aa, and those of sets II, IV, and V by 2 aa. For sets I, II, III, and V, the same VH gene is associated with structurally very similar HCDR3s. Furthermore, the members of set II that have identical VHDJH segments also use the same Vκ and Jκ segments to create LCDR3s that differ by only 1 aa. Finally, the VHDJH rearrangements of set I pair with the same Vκ (Table 2, LCDR3). Thus, the presence of stereotypic H and L chain rearrangements seen in human B-CLL, and most often in unmutated cases, is replicated in these TCL1-derived leukemias.
(ii) Comparison of Rearranged VHDJH Segments with Those Present in Public Databases.
Amino acid sequences of H and L chain rearrangements in human B-CLL cells often resemble autoantigen-reactive human and murine mAb (13, 14). To check for possible autoreactivity, we compared the amino acid sequences of the TCL1 HCDR3s with those found in the GenBank and IMGT databases. Significant similarity was found for two TCL-1 sequences. The sequences of TCL1-001 and -002 (Set I), which are comprised of identical VH, D, JH, Vκ, and Jκ segments of almost complete amino acid identity, were virtually identical to the rearranged H and L chain sequences previously determined from murine autoantibodies reactive with phosphatidylcholine (PtC) and with erythrocytes digested by bromelain, a process that makes PtC determinants more accessible to antibody binding (Table 3). In addition, TCL1-007 and -008 were very similar in HCDR3 (but not in VH) to anti-microbial (anti-Brucella abortus and anti-phosphocholine) antibodies (Fig. 2, which is published as supporting information on the PNAS web site). Human B-CLL cases with markedly similar HCDR3s associated with different VH genes belonging to the same VH clan have been reported (14).
Table 3.
Amino acid sequence comparison of TCL1 clones with other clones of known specificity having >75% HCDR3 similarity
| Heavy chain 1 |
Light chain |
|||||||
|---|---|---|---|---|---|---|---|---|
| VH NC1-A7 | HCDR3 | FR4 | Vκ4-91 | LCDR3 | FR4 | Specificity | ||
| TCL1-001 | TAMYYC | AGDRRGYWYFDV | WG | TCL1-001 | VATYYC | QQGGGIPLT | PG | |
| TCL1-002 | …... | …..T…... | .. | TCL1-002 | …... | ….…R. | .. | |
| X53339* | …... | …..D…... | .. | X53348* | …... | ….…R. | .. | Anti-PtC |
| X53338† | …... | …..S…... | .. | X53349† | …... | ….….. | .. | Anti-PtC |
| X53348‡ | …... | …..D…... | .. | X53350‡ | …... | .X….… | .. | Anti-PtC |
| U54362§ | …... | …..Y…... | .. | U54505§ | …... | ….…R. | .. | Anti-PtC |
| U64364¶ | …... | …..Y…... | .. | U54507¶ | …... | ….…R. | .. | Anti-PtC |
| U64367* | …... | … YY…... | .. | U54509* | …... | .X…..R. | .. | Anti-PtC |
| M22439** | …... | … YD…... | .. | M34591** | …... | ….…R. | .. | Anti-BrMRBC from B-1 cells |
The IMGT and VBase databases of Ig V region sequences were searched for HCDR3 and VHDJH rearrangements exhibiting ≥75% similarity with the TCL-1 HCDR3s. Fifteen were found, 7 with comparison VLJL rearrangements very similar to those of TCL1 mAb and 8 with VLJL using different gene segments. HCDR3 and LCDR3 of the four mAb are shown; these CDR3s are also identical in nucleotide sequence in both VH and VL. The eight antibodies that used different VL genes were AY172889, an antibody from B-1 cells; AF118985, AF118934, AF118992, U64382, U64371, and AF119029, anti-phosphatidylcholine antibodies; and AY437060, an anti-nuclear antibody. ·, sequence identity; x, unknown amino acid.
*Sequences for anti-PtC antibody clone NC5-A11 (42).
†Sequences for anti-PtC antibody clone NC1-A7 (42).
‡Sequences for anti-PtC antibody clone NC6-C12 (42).
§Sequences for anti-PtC antibody clone AE0017 (42).
¶Sequences for anti-PtC antibody clone AE0022 (42).
‖Sequences for anti-PtC antibody clone AE0045 (42).
**Sequences for anti-BrMRBC from B-1 cells (43).
(iii) Confirmation That Sequence Similarity Extends to Antigen Binding.
To confirm that the amino acid sequence similarity between the TCL1 mAb and anti-autoantigen and anti-microbial mAb identified above translated to binding of similar antigens, we produced hybridomas from selected TCL1 clones to generate soluble mAb and test their binding with a number of antigens. Because the database similarities mentioned above indicated that the TCL1 mAb resembled those made by murine B-1 cells that often bind phospholipids, nucleic acids, and cell surface antigens, we included phosphorylcholine, dextran, low-density lipoprotein (LDL), ss- and dsDNA, cardiolipin, bacterial lipopolysaccharide, pneumococcal polysaccharide, mouse IgG2a mAb, and mouse erythrocytes digested with bromelain in the test panel. Also, because many B-1 cells react with carbohydrate epitopes, the Consortium for Functional Genomics screened all mAb for reactivity with an extensive array of glycans.
Monoclonal antibody from TCL1-002 was poly- and autoreactive, binding in a dose-dependent manner to low-density lipoprotein (LDL) and Br-treated erythrocytes (Fig. 3 A and B, which is published as supporting information on the PNAS web site). The TCL1-020 mAb also bound Br-treated RBCs, albeit to a lesser degree (Fig. 3B). Finally, mAb from nos. 003 and 004 (Set II) that share VHDJH/VLJL rearrangements with only 1-aa difference in LCDR3 (Tables 1 and 2) also bound blood group H type 1 antigens, based on reactivity with the following polysaccharides: Fucα1-2Galb1-3GalNAcb1-3Gala1-4Galb1-4Glcb-Sp9, Fucα1-2Galβ1-3GalNAcα-Sp8, Fucα1-2Galb1-3GalNAcb1-4(Neu5Aca2-3)Galb1-4Glcb-Sp0, and Fucα 1-2Galβ1-3GalNAcβ1-4(Neu5Acα 2-3)Galβ1-4Glcβ-Sp9 (Fig. 3 C and D; detailed data accessible at www.functionalglycomics.org/static/consortium/main.shtml).
Thus, all of the mAb selected for analysis were poly- and/or autoreactive, binding to a glycerophospholipid (PtC based on reactivity with Br-treated RBCs), a lipoprotein (low-density lipoprotein), or polysaccharides (Fucα1-linkages). Although reactivity of human B-CLL cells with nonprotein antigens has been suggested (3, 5, 13, 14), formal proof of this hypothesis has yet to be obtained.
Discussion
Our data indicate that the TCL1 Tg mouse model replicates the Ig V-region rearrangements characteristic of the aggressive, treatment-resistant form of human B-CLL. BCRs comprised of genes differing minimally, if at all, from the germ-line sequence are a hallmark of poor-outcome human B-CLL, and the BCRs in the TCL1-mediated B cell lymphoproliferative disease display this feature (Table 1).
Another cardinal feature of human B-CLL is nonstochastic use and association of individual V region segments, with biases differing between U-CLL and mutated B-CLL (3–5). Based on the presently known numbers of murine segments, certain VH, D, JH, and VL genes appear overexpressed in the TCL1-derived B cell leukemias. This assumption applies particularly for rearrangements involving VH11, VH12, and VH4, because these VH families contain only one (VH11 and 12) or two (VH4) genes, a very minor fraction of the entire murine VH repertoire. More extensive studies of TCL1 leukemias will need to confirm the apparent overuse of these and other VH (V165.1) genes and certain D, JH, and VL segments (Tables 1 and 2).
Human B-CLL cells also exhibit unique HCDR3 features that characterize and differentiate aggressive U-CLL from indolent mutated B-CLL. Specifically, unmutated poor outcome cases frequently exhibit long HCDR3s containing multiple neutral tyrosine and serine residues (7, 38) that may confer CDR3 flexibility and favor polyreactivity. This characteristic is especially true for human leukemic rearrangements using VH1-69 and 4-39, two genes virtually always associated with rapid downhill clinical courses (8, 9, 13). The HCDR3s in TCL1 mice also contain many of these same amino acids (Table 2). Furthermore, HCDR3s of human B-CLL cells often contain charged amino acids, frequently not coded by germ-line D and JH segments. In the TCL1 leukemias, positively and negatively charged residues are seen frequently (55%) at or adjacent to the VH–D and D–JH junctions, deriving from simple and complex rearrangement events including insertion of nontemplated nucleotides (Table 2; see also Table 4, which is published as supporting information on the PNAS web site).
The startling discovery in human B-CLL of sets of patients with remarkably similar BCRs comprised of identical V[D]J segments (7, 10–14, 16, 17, 38), seen more often in poor outcome U-CLL, is also found in the TCL1 model. Although the cohort of leukemias we studied is modest (n = 20), half of these clones fit into five distinct sets defined by >80% HCRD3 amino acid similarity (Table 2). Furthermore, the members of four of these groups express identical VH genes. Finally, for two of the sets, the associated Vκ is identical and, in one set, has recombined with the same Jκ (Tables 1 and 2); for the other set where the expressed JL segment differs (Jκ 5 vs. Jκ 2), the LCDR3 amino acid sequences remain 92% similar.
In the human disease, findings such as these led to the hypothesis that B-CLL derives from a subset of B cells with BCRs of restricted structure (13) that is selected by specific antigen(s) or a class of antigens, in particular, a combination of autoantigens and microbial antigens, for clonal expansion and eventual transformation (3–5). Human B-CLL BCR sequences resembling closely those of known autoantibodies (13, 14) and auto- and poly-reactive antigen-binding (41, 44–46), enriched in aggressive U-CLL (41), further support this model. The TCL1 leukemias also display these two properties, because our comparisons with murine antibodies of known structure and antigen-binding specificity indicate. In particular, TCL1-001 and -002, which are virtually identical to a series of mAb from B-1 cells reactive with PtC and Br-treated erythrocytes (Table 3), and TCL1-020 bind PtC displayed on Br-treated RBCs (Fig. 3B). In addition, TCL1-005 and -006 exhibit VHDJH rearrangements comprised of VH11 and distinct D and JH segments typical of anti-PtC antibodies (Fig. 2A; refs. 47–49), although the TCL1 mAb do not use a Vκ 9 gene. Finally, two mAb from nos. 003 and 004 that share VHDJH/VLJL rearrangements bind polysaccharides found in blood group H type 1 antigens. The identification of these specific antigens in the TCL1 model, in particular one that is a component of all mammalian and certain bacterial membranes, may provide valuable clues for the human disease.
The model of ongoing autoantigen-mediated leukemogenesis predicts a growth and survival advantage for clones with BCRs reactive with antigens frequently encountered in vivo. In this regard, we noted nine animals that concomitantly displayed minor B cell expansions in their spleens (Table 4 and Table 5, which are published as supporting information on the PNAS web site). These clones expressed VH or VL genes with significant levels of V gene mutations (3.1%, 3.5%, and 2.1%). The clonal evolution that occurred in two of these TCL1 spleen cell suspensions (nos. 001 and 012) favored unmutated clones, which outstripped the mutated, minor B cell clones over time in vivo (Fig. 4 A and B, which is published as supporting information on the PNAS web site). Thus, the murine leukemia may develop from a selective B cell pool with unmutated BCRs, driven by repetitive interactions with (auto)antigens and stimulated by the overexpression of TCL1 that enhances activation of the Akt kinase, a key survival molecule in the BCR-signaling pathway in human B-CLL (50), that can protect from apoptosis (51–53) and augment surface receptor-mediated signaling.
Based on the apparent biases in VH (VH11, 12, and 4) and JH (JH1) gene use and association and the antigen specificities described herein, the TCL1 clones likely derive from the B-1a subset (54), consistent with finding the initial, preleukemic clonal expansions in the peritoneal cavity (21, 55). The paucity of N nucleotide insertion also favors a B-1a lineage (Fig. 1).
A key unanswered question in human B-CLL is finding the normal B cell counterpart of the leukemic cell. Because B-CLL cells uniformly express CD5, a human B cell equivalent for the murine B-1a cell is considered the most likely candidate for the leukemic precursor (56–58). However, it has been difficult to identify a B-1 counterpart in man, because the poor response to T cell independent antigens of circulating and follicular mantle resident human CD5+ B cells (59) and their dearth of poly- and autoreactivity (41) suggest that these CD5+ cells are not the human B-1a equivalent. It has been proposed that U-CLL cells might derive from marginal zone (MZ) B cells because of the similarity between human B-CLL mAb and mAb reactive with autoantigens and, to a lesser degree, to microbial antigens (3, 5). Because the murine B-1 and MZ compartments may intersect (60, 61) and both compartments contain autoreactive B cells (62–64), if the overexpression of TCL1 causes the human disease, the present data, in particular the poly/autoreactivity of TCL1 clones with lipoprotein, phospholipid, and polysaccharides could support a derivation of human B-CLL from a B-1a equivalent or from an MZ equivalent.
Materials and Methods
Animals.
Spleens, lymph nodes, and blood were collected from Eμ-TCL1 Tg mice when they developed extensive B-CLL-like disease (Table 6, which is published as supporting information on the PNAS web site). In compliance with Federal and Institutional Animal Care and Use Committee requirements, animals were euthanized when symptoms became disabling.
Isolation of DNA and RNA and Synthesis of cDNA.
DNA was purified from cell lysates by using DNeasy spin columns by following the manufacturer’s protocol (DNeasy Tissue Kit; Qiagen, Valencia, CA). RNA was extracted by using TRIzol Reagent (Invitrogen, Carlsbad, CA). One microgram of RNA was reverse transcribed to cDNA with 200 units of SuperScript II Reverse Transcriptase (Invitrogen)/1 unit of RNase inhibitor (Eppendorf, Hamburg, Germany)/20 pmol oligo dT primer (Roche Applied Science, Indianapolis) in a 20-μl volume. Reactants were incubated at 42°C for 1 h, heated at 65°C for 10 min, and then diluted to a final volume of 100 μl.
PCR Amplification of VH and Vκ Transcripts.
VHDJH and Vκ Jκ cDNAs were amplified by using consensus sense and antisense primers (Data Set 1, which is published as supporting information on the PNAS web site) and Platinum TaqDNA Polymerase High Fidelity (Invitrogen) for 35 cycles, each consisting of 1-min denaturation at 94°C, 1-min annealing at 52°C, and 1-min extension at 68°C. Amplification was completed by an additional 10-min extension at 68°C. To determine isotypes, antisense primers specific for murine μ, γ, and α CH or κ CL regions were paired with appropriate sense VH or VL family primers.
cDNA Sequencing and Mutational Analysis.
PCR products were ligated into pCRII vectors and cloned into Top10 cells (Invitrogen). cDNAs were amplified with M13 forward and reverse primers, and their DNA sequences were determined on both strands by using an automated sequenator (PerkinElmer Instruments, Norwalk, CT).
To confirm that nucleotide differences identified by database comparisons were true somatic mutations and not polymorphic or previously undescribed VH germ-line genes, DNA from splenocytes of non-Tg mice was amplified by using primer pairs specific for the intron 5′ of FR1 of the most similar VH gene and for 3′ recombination signal sequence (Data Set 2, which is published as supporting information on the PNAS web site). PCR products were cloned into TOPO TA vectors (Invitrogen) and up to 60 individual colonies sequenced. Three previously undescribed germ-line genes were identified and reported to GenBank (accession nos. DQ093183, XE1; DQ093184, XE2; DQ093185, XE3).
Sequence Comparison to Germ-Line Genes in Public Databases.
Nucleotide and aa sequences of V region segments from all major and minor clones were compared with each other and to those contained in public databases by using Ig-blast and IMGT/V-QUEST. HCDR3 was considered as the region immediately after the 3′ VH-encoded conserved Cys (C; TGT) and the 5′ JH-encoded Trp (TGG). LCDR3 was considered as the region after the 3′ VL-encoded conserved Cys and the 5′JH-encoded Phe (F; TTC).
Determining Reactivity of Soluble TCL1 mAb.
Hybridomas producing TCL1 mAb were produced by fusion of leukemic cells with SP2.0 myeloma cells and cloned by limiting dilution; those clones expressing the appropriate leukemic Ig rearrangements were identified by RT-PCR and nucleotide sequence analyses. The mAb concentration in supernatants from cultured clones ranged from 40 to 120 μg/ml. Reactivity with NP-BSA, NP-Ficoll, phosphorylcholine-hydroxyphenylacetic-BSA (Biosearch Technologies, Novato, CA), cardiolipin, LPS, calf thymus DNA (Sigma–Aldrich, St. Louis, MO), low-density lipoprotein (Biomedical Technologies, Stoughton, MA), pneumococcal polysaccharide (Statens Serum Institut, Copenhagen, Denmark), dextran (Amersham Pharmacia), and murine IgG2a mAb was tested by ELISA by using antigen-coated plates and anti-mouse IgM-HRP (Southern Biotechnology Associates) for detection. Reactivity with bromelain-treated RBCs was investigated as described in ref. 65. Data ≥10 times the negative controls were considered positive. The Consortium for Functional Glycomics, Protein–Carbohydrate Interaction Core H (Richard Alvarez, Director, and Angela Lee, Glycan Specialist) screened the TCL1 mAb for reactivity with a large battery of carbohydrate epitopes by using a glycan array (www.functionalglycomics.org/static/consortium/main.shtml).
Supplementary Material
Acknowledgments
We thank Richard Alvarez, Director, and Angela Lee, Glycan Specialist, of the Protein–Carbohydrate Interaction Core H of the Consortium for Functional Glycomics for their advice and assistance in performing the glycan array analysis and Drs. Charles C. Chu and Thomas L. Rothstein for helpful discussions and suggestions. These studies were supported in part by the Jean Walton Fund for Lymphoma and Myeloma Research (N.C.), the Joseph Eletto Leukemia Research Fund (N.C.), the Leukemia and Lymphoma Society Translational Research Program Grant 6043-06 (to D.G.E.), and a Sidney Kimmel Cancer Research Foundation Grant (to N.Z.). The Consortium for Functional Glycomics is funded by National Institute of General Medical Sciences Grant GM62116.
Abbreviations
- B-CLL
B cell chronic lymphocytic leukemia
- BCR
B cell antigen receptor
- PtC
phosphatidylcholine
- Tg
transgenic
- U-CLL
unmutated B-CLL.
Footnotes
References
- 1.Rai K. R., Patel D. V. In: Hematology: Basic Principles and Practice. Hoffman R., Benz E., Shattil S., Furie B., Cohen H., Silberstein L., editors. New York: Churchill Livingstone; 1995. pp. 1308–1321. [Google Scholar]
- 2.Travade P., Chastang C., Dighiero G., Binet J. L. Blood Cells. 1987;12:485–502. [PubMed] [Google Scholar]
- 3.Chiorazzi N., Ferrarini M. Annu. Rev. Immunol. 2003;21:841–894. doi: 10.1146/annurev.immunol.21.120601.141018. [DOI] [PubMed] [Google Scholar]
- 4.Stevenson F. K., Caligaris-Cappio F. Blood. 2004;103:4389–4395. doi: 10.1182/blood-2003-12-4312. [DOI] [PubMed] [Google Scholar]
- 5.Chiorazzi N., Rai K. R., Ferrarini M. N. Engl. J. Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder H. W., Jr., Dighiero G. Immunol. Today. 1994;15:288–294. doi: 10.1016/0167-5699(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 7.Fais F., Ghiotto F., Hashimoto S., Sellars B., Valetto A., Allen S. L., Schulman P., Vinciguerra V. P., Rai K., Rassenti L. Z., et al. J. Clin. Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damle R. N., Wasil T., Fais F., Ghiotto F., Valetto A., Allen S. L., Buchbinder A., Budman D., Dittmar K., Kolitz J., et al. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 9.Hamblin T. J., Davis Z., Gardiner A., Oscier D. G., Stevenson F. K. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 10.Hashimoto S., Dono M., Wakai M., Allen S. L., Lichtman S. M., Schulman P., Vinciguerra V. P., Ferrarini M., Silver J., Chiorazzi N. J. Exp. Med. 1995;181:1507–1517. doi: 10.1084/jem.181.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobin G., Thunberg U., Johnson A., Thorn I., Soderberg O., Hultdin M., Botling J., Enblad G., Sallstrom J., Sundstrom C., et al. Blood. 2002;99:2262–2264. doi: 10.1182/blood.v99.6.2262. [DOI] [PubMed] [Google Scholar]
- 12.Tobin G., Thunberg U., Johnson A., Eriksson I., Soderberg O., Karlsson K., Merup M., Juliusson G., Vilpo J., Enblad G., et al. Blood. 2003;101:4952–4957. doi: 10.1182/blood-2002-11-3485. [DOI] [PubMed] [Google Scholar]
- 13.Ghiotto F., Fais F., Valetto A., Albesiano E., Hashimoto S., Dono M., Ikematsu H., Allen S. L., Kolitz J., Rai K. R., et al. J. Clin. Invest. 2004;113:1008–1016. doi: 10.1172/JCI19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messmer B. T., Albesiano E., Efremov D. G., Ghiotto F., Allen S. L., Kolitz J., Foa R., Damle R. N., Fais F., Messmer D., et al. J. Exp. Med. 2004;200:519–525. doi: 10.1084/jem.20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tobin G., Thunberg U., Karlsson K., Murray F., Laurell A., Willander K., Enblad G., Merup M., Vilpo J., Juliusson G., et al. Blood. 2004;104:2879–2885. doi: 10.1182/blood-2004-01-0132. [DOI] [PubMed] [Google Scholar]
- 16.Widhopf G. F., 2nd, Rassenti L. Z., Toy T. L., Gribben J. G., Wierda W. G., Kipps T. J. Blood. 2004;104:2499–2504. doi: 10.1182/blood-2004-03-0818. [DOI] [PubMed] [Google Scholar]
- 17.Ghia P., Stamatopoulos K., Belessi C., Moreno C., Stella S., Guida G., Michel A., Crespo M., Laoutaris N., Montserrat E., et al. Blood. 2005;105:1678–1685. doi: 10.1182/blood-2004-07-2606. [DOI] [PubMed] [Google Scholar]
- 18.Dohner H., Stilgenbauer S., Benner A., Leupolt E., Krober A., Bullinger L., Dohner K., Bentz M., Lichter P. N. Engl. J. Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 19.Phillips J. A., Mehta K., Fernandez C., Raveche E. S. Cancer Res. 1992;52:437–443. [PubMed] [Google Scholar]
- 20.Zapata J. M., Krajewska M., Morse H. C., 3rd, Choi Y., Reed J. C. Proc. Natl. Acad. Sci. USA. 2004;101:16600–16605. doi: 10.1073/pnas.0407541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bichi R., Shinton S. A., Martin E. S., Koval A., Calin G. A., Cesari R., Russo G., Hardy R. R., Croce C. M. Proc. Natl. Acad. Sci. USA. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Virgilio L., Narducci M. G., Isobe M., Billips L. G., Cooper M. D., Croce C. M., Russo G. Proc. Natl. Acad. Sci. USA. 1994;91:12530–12534. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narducci M. G., Virgilio L., Engiles J. B., Buchberg A. M., Billips L., Facchiano A., Croce C. M., Russo G., Rothstein J. L. Oncogene. 1997;15:919–926. doi: 10.1038/sj.onc.1201246. [DOI] [PubMed] [Google Scholar]
- 24.Herling M., Patel K. A., Khalili J., Schlette E., Kobayashi R., Medeiros L. J., Jones D. Leukemia. 2006;20:280–285. doi: 10.1038/sj.leu.2404017. [DOI] [PubMed] [Google Scholar]
- 25.Zanesi N., Aqeilan R., Drusco A., Kaou M., Sevignani C., Costinean S., Bortesi L., La Rocca G., Koldovsky P., Volinia S., et al. Cancer Res. 2006;66:915–920. doi: 10.1158/0008-5472.CAN-05-3426. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda F., Ishii K., Bourvagnet P., Kuma K., Hayashida H., Miyata T., Honjo T. J. Exp. Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meindl A., Klobeck H. G., Ohnheiser R., Zachau H. G. Eur. J. Immunol. 1990;20:1855–1863. doi: 10.1002/eji.1830200834. [DOI] [PubMed] [Google Scholar]
- 28.Huber C., Schable K. F., Huber E., Klein R., Meindl A., Thiebe R., Lamm R., Zachau H. G. Eur. J. Immunol. 1993;23:2868–2875. doi: 10.1002/eji.1830231121. [DOI] [PubMed] [Google Scholar]
- 29.Williams S. C., Frippiat J. P., Tomlinson I. M., Ignatovich O., Lefranc M. P., Winter G. J. Mol. Biol. 1996;264:220–232. doi: 10.1006/jmbi.1996.0636. [DOI] [PubMed] [Google Scholar]
- 30.Mainville C. A., Sheehan K. M., Klaman L. D., Giorgetti C. A., Press J. L., Brodeur P. H. J. Immunol. 1996;156:1038–1046. [PubMed] [Google Scholar]
- 31.Thiebe R., Schable K. F., Bensch A., Brensing-Kuppers J., Heim V., Kirschbaum T., Mitlohner H., Ohnrich M., Pourrajabi S., Roschenthaler F., et al. Eur. J. Immunol. 1999;29:2072–2081. doi: 10.1002/(SICI)1521-4141(199907)29:07<2072::AID-IMMU2072>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 32.Johnston C. M., Wood A. L., Bolland D. J., Corcoran A. E. J. Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 33.Roschenthaler F., Kirschbaum T., Heim V., Kirschbaum V., Schable K. F., Schwendinger J., Zocher I., Zachau H. G. Eur. J. Immunol. 1999;29:2065–2071. doi: 10.1002/(SICI)1521-4141(199907)29:07<2065::AID-IMMU2065>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 34.Kirschbaum T., Roschenthaler F., Bensch A., Holscher B., Lautner-Rieske A., Ohnrich M., Pourrajabi S., Schwendinger J., Zocher I., Zachau H. G. Eur. J. Immunol. 1999;29:2057–2064. doi: 10.1002/(SICI)1521-4141(199907)29:07<2057::AID-IMMU2057>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 35.Chang S., Mohan C. Mol. Immunol. 2005;42:1293–1301. doi: 10.1016/j.molimm.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Ivanov I. I., Schelonka R. L., Zhuang Y., Gartland G. L., Zemlin M., Schroeder H. W., Jr. J. Immunol. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- 37.Stamatopoulos K., Belessi C., Hadzidimitriou A., Smilevska T., Kalagiakou E., Hatzi K., Stavroyianni N., Athanasiadou A., Tsompanakou A., Papadaki T., et al. Blood. 2005;106:3575–3583. doi: 10.1182/blood-2005-04-1511. [DOI] [PubMed] [Google Scholar]
- 38.Johnson T. A., Rassenti L. Z., Kipps T. J. J. Immunol. 1997;158:235–246. [PubMed] [Google Scholar]
- 39.Widhopf G. F., 2nd, Kipps T. J. J. Immunol. 2001;166:95–102. doi: 10.4049/jimmunol.166.1.95. [DOI] [PubMed] [Google Scholar]
- 40.Zemlin M., Klinger M., Link J., Zemlin C., Bauer K., Engler J. A., Schroeder H. W., Jr., Kirkham P. M. J. Mol. Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Herve M., Xu K., Ng Y.-S., Wardemann H., Albesiano E., Messmer B., Chiorazzi N., Meffre E. J. Clin. Invest. 2005;115:1636–1643. doi: 10.1172/JCI24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seidl K. J., MacKenzie J. D., Wang D., Kantor A. B., Kabat E. A., Herzenberg L. A. Int. Immunol. 1997;9:689–702. doi: 10.1093/intimm/9.5.689. [DOI] [PubMed] [Google Scholar]
- 43.Hardy R. R., Carmack C. E., Shinton S. A., Riblet R. J., Hayakawa K. J. Immunol. 1989;142:3643–3651. [PubMed] [Google Scholar]
- 44.Broker B. M., Klajman A., Youinou P., Jouquan J., Worman C. P., Murphy J., Mackenzie L., Quartey-Papafio R., Blaschek M., Collins P., et al. J. Autoimmun. 1988;1:469–481. doi: 10.1016/0896-8411(88)90068-6. [DOI] [PubMed] [Google Scholar]
- 45.Sthoeger Z. M., Wakai M., Tse D. B., Vinciguerra V. P., Allen S. L., Budman D. R., Lichtman S. M., Schulman P., Weiselberg L. R., Chiorazzi N. J. Exp. Med. 1989;169:255–268. doi: 10.1084/jem.169.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borche L., Lim A., Binet J. L., Dighiero G. Blood. 1990;76:562–569. [PubMed] [Google Scholar]
- 47.Hardy R. R., Wei C. J., Hayakawa K. Immunol. Rev. 2004;197:60–74. doi: 10.1111/j.0105-2896.2004.0100.x. [DOI] [PubMed] [Google Scholar]
- 48.Wang H., Clarke S. H. Curr. Opin. Immunol. 2004;16:246–250. doi: 10.1016/j.coi.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Arnold L. W., Spencer D. H., Clarke S. H., Haughton G. Int. Immunol. 1993;5:1365–1373. doi: 10.1093/intimm/5.11.1365. [DOI] [PubMed] [Google Scholar]
- 50.Petlickovski A., Laurenti L., Li X., Marietti S., Chiusolo P., Sica S., Leone G., Efremov D. G. Blood. 2005;105:4820–4827. doi: 10.1182/blood-2004-07-2669. [DOI] [PubMed] [Google Scholar]
- 51.Datta S. R., Brunet A., Greenberg M. E. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 52.Pekarsky Y., Koval A., Hallas C., Bichi R., Tresini M., Malstrom S., Russo G., Tsichlis P., Croce C. M. Proc. Natl. Acad. Sci. USA. 2000;97:3028–3033. doi: 10.1073/pnas.040557697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pekarsky Y., Hallas C., Croce C. M. Am. J. Pharmacogenomics. 2003;3:31–36. doi: 10.2165/00129785-200303010-00005. [DOI] [PubMed] [Google Scholar]
- 54.Hayakawa K., Hardy R. R., Parks D. R., Herzenberg L. A. J. Exp. Med. 1983;157:202–218. doi: 10.1084/jem.157.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pekarsky Y., Calin G. A., Aqeilan R. Curr. Top Microbiol. Immunol. 2005;294:51–70. doi: 10.1007/3-540-29933-5_4. [DOI] [PubMed] [Google Scholar]
- 56.Rajewsky K., Gu H., Vieira P., Forster I. Cold Spring Harbor Symp. Quant. Biol. 1989;54:209–217. doi: 10.1101/sqb.1989.054.01.026. [DOI] [PubMed] [Google Scholar]
- 57.Gu H., Forster I., Rajewsky K. EMBO J. 1990;9:2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kipps T. J. Adv. Immunol. 1989;47:117–185. doi: 10.1016/s0065-2776(08)60663-x. [DOI] [PubMed] [Google Scholar]
- 59.Zupo S., Dono M., Azzoni L., Chiorazzi N., Ferrarini M. Eur. J. Immunol. 1991;21:351–359. doi: 10.1002/eji.1830210216. [DOI] [PubMed] [Google Scholar]
- 60.Martin F., Kearney J. F. Immunol. Rev. 2000;175:70–79. [PubMed] [Google Scholar]
- 61.Martin F., Oliver A. M., Kearney J. F. Immunity. 2001;14:617–629. doi: 10.1016/s1074-7613(01)00129-7. [DOI] [PubMed] [Google Scholar]
- 62.Chen X., Martin F., Forbush K. A., Perlmutter R. M., Kearney J. F. Int. Immunol. 1997;9:27–41. doi: 10.1093/intimm/9.1.27. [DOI] [PubMed] [Google Scholar]
- 63.Bendelac A., Bonneville M., Kearney J. F. Nat. Rev. Immunol. 2001;1:177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 64.Li Y., Li H., Weigert M. J. Exp. Med. 2002;195:181–188. doi: 10.1084/jem.20011453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yurino H., Ishikawa S., Sato T., Akadegawa K., Ito T., Ueha S., Inadera H., Matsushima K. Toxicol. Sci. 2004;81:139–147. doi: 10.1093/toxsci/kfh179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.