Parkinson disease (PD) is the second most common progressive neurodegenerative disorder, with a prevalence of approximately 1% at the age of 65, increasing to 4–5% by the age of 85 (1, 2). PD patients suffer from bradykinesia, tremor, cogwheel rigidity, and postural instability. PD is due to the relatively selective loss of dopaminergic neurons in the substantia nigra pars compacta, which leads to a profound reduction in striatal dopamine (DA). Lewy bodies and dystrophic neurites (Lewy neurites)accompany the loss of dopaminergic neurons (3). Lewy bodies are a pathologic hallmark of PDand classically are round eosinophilic inclusions composed of a halo of radiating fibrils and a less defined core. Both Lewy bodies and Lewy neurites are comprised of cytoplasmic accumulations of aggregated proteins (3). PD represents a heterogeneous disorder with common clinical manifestations and, for the most part, common neuropathologic findings. The majority of cases of PD appear to be sporadic in nature (4); however, there may be genetic risk factors that increase the likelihood of developing PD, much in the same way that the apoE4 allele increases the risk of developing Alzheimer disease (AD) (5). Familial PD with specific genetic defects may account for fewer than 10% of all cases of PD (4); however, the identification of these rare genes and their functions has provided tremendous insight into the pathogenesis of PD and opened up new areas of investigation (6, 7).
Familial-linked PD genes
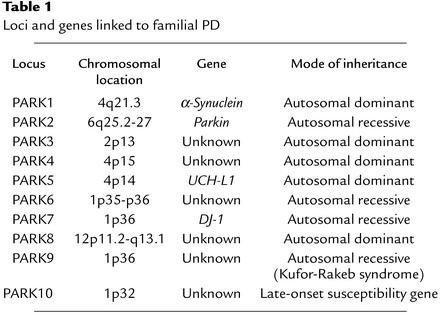
Three genes have been clearly linked to PD, and a number of other genes or genetic linkages have been identified that may cause PD (Table 1). The first “PD gene” to be identified, PARK1, was the gene encoding the presynaptic protein α-synuclein (8, 9). The second PD gene, PARK2, is caused by mutations in the gene for parkin (10), and it leads to autosomal recessive juvenile parkinsonism (AR-JP). The third PD gene, PARK7, results from mutations in DJ-1 (11). Mutations in α-synuclein, parkin, and DJ-1 definitely cause PD (see below). A mutation in the gene (PARK5) encoding ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) in two family members of a small German kindred with autosomal dominant PD has been described (9). Whether it represents a disease-causing mutation is unclear, as linkage analysis was not used to identify the mutation. Furthermore, comprehensive analysis of several PD family kindreds has not identified this mutation in any other family, and no other potential pathogenic mutations of this gene have been identified in humans (4). At this time, it is not possible to prove its pathogenic role in PD. However, recent functional studies suggest that it may be causally related to PD (12).
Table 1.
Loci and genes linked to familial PD
A locus located on chromosome 2p13 (PARK3) has been described in a subset of families with autosomal dominant inheritance and typical Lewy body pathology (13). The penetrance of the mutation on chromosome 2p13 was estimated to be 40% based on the occurrence of the affected haplotype in clinically asymptomatic members of the linked families. PARK3-linked families also show signs of dementia, and neuropathology revealed, in addition to neuronal loss in the substantia nigra and typical brainstem Lewy bodies, the presence of neurofibrillary tangles and Alzheimer plaques (4). PARK4 is linked to the short arm of chromosome 4 (4p15) (14). The PARK4 locus appears to segregate with both PD and postural tremor with an autosomal dominant inheritance pattern. Furthermore, affected family members with PD have several atypical features, including early weight loss, dysautonomia, and dementia (15). An autosomal recessive locus on chromosome 1, PARK6, has recently been described in a large Sicilian family and is linked to chromosome 1 (1p35-p36) (16). Linkage analysis in a consanguineous family from the southwest Netherlands revealed another locus (PARK7) on chromosome 1p36, which is genetically and clinically distinct from PARK6 (17). PARK8 is linked to chromosome 12p11.2-q13.1 and is inherited in an autosomal dominant fashion with partial penetrance (18). Neuropathologic examination of four patients revealed nigral degeneration without Lewy bodies (18). Kufor-Rakeb syndrome, an autosomal recessive nigro-striatal-pallidal-pyramidal neurodegenerative disorder, has been mapped to a 9-cM region of chromosome 1p36 and designated PARK9 (19). A recent Icelandic study suggests that genetic variability is a major contributor to PD in this population (20), and the locus was recently localized to a region on chromosome 1p32 and designated PARK10 (21). It is likely that there are other gene loci, as not all familial PD has been linked to the current loci. The identification of the genes for PARK1 (α-synuclein), PARK2 (parkin), and PARK7 (DJ-1) has led to new insights and direction in PD research and pathogenesis (see below). The identification of the other genes will undoubtedly shed much additional light on the molecular mechanisms of PD pathogenesis.
Besides identifying genetic mutations that cause PD, recent genomic screens have also identi-fied genetic factors that may be important in its de-velopment (20). In particular, linkage and mutation analysis indicates that the parkin gene, in addition to being a direct cause of PD, is influential in the de-velopment of early-onset PD (22). Multiple genetic factors appear to be important in the development of idiopathic late-onset PD. Four single-nucleotide polymorphisms in the tau gene are significantly associated with an increased risk of developing PD (23). Thus, the association of PD with the haplotype of tau and the evidence for linkage to that region of chromosome 17q suggest that tau, or a gene in linkage disequilibrium with tau, is a genetic risk factor for PD. Three large case-series studies also established a significant association between polymorphism of the tau gene and PD (24–26). Frontotemporal dementia with parkinsonism (FTDP) is caused, in part, by mutations in tau (27). Chromosome 9q also seems to be a region that incurs a genetic risk factor for PD (23). Other suggestive linkages have been identified on chromosomes 1, 3q, 5q, 8p, 10, and 16 (22, 28). Genes influencing the age of onset of PD may be linked to chromosome 1p and chromosomes 6 and 10 (29). Interestingly, the linkage on chromosome 10 also influences the age of onset of AD (29). Other work has suggested that environmental factors also play a role in the development of PD (30). It is likely that genetic susceptibility coupled with environmental exposure contributes to the development and manifestation of sporadic PD.
α-Synuclein mutations
Mutations in the α-synuclein gene are clearly a very rare cause of PD (8). The first mutation identified was an A53T mutation resulting from a G→A transition at position 209 (9). This mutation was originally found in the Contursi kindred and was also identified in several Greek kindreds. Recent haplotype analyses suggest that the Contursi kindred and the Greek kindreds share a common ancestor (31). Another mutation (A30P) resulting from a G→C transition at position 88 was identified in a small German kindred (32). Furthermore, genetic variability in the α-synuclein gene is a risk factor for the development of PD (33). Affected individuals with α-synuclein mutations have typical idiopathic PD, including levodopa responsiveness and Lewy bodies, although the age of onset is somewhat lower and progression appears to be more rapid (31). α-Synuclein is a 140–amino acid protein that contains repetitive imperfect repeats of KTKEGV in the amino-terminal half, a hydrophobic region, and an acidic carboxy-terminal region (34) (Figure 1). In humans, there are at least three different synuclein family members, designated α-, β-, and γ-synuclein, and they are expressed from three different genes (34). Synucleins are abundant brain proteins whose physiologic functions are poorly understood. α-Synuclein has been shown to bind to a number of proteins as well as lipid membranes. The physiologic significance of these interactions are not known. Whether these interactions contribute to disease pathogenesis is not clear, as disease-causing mutations in α-synuclein, for the most part, do not have dramatic effects on these interactions (35). It has been suggested that α-synuclein may play some role in the modulation of synaptic vesicle turnover and synaptic plasticity (34). α-Synuclein knockout mice are viable and fertile and exhibit normal brain structure and a normal complement of dopaminergic cell bodies, fibers, and synapses. Thus, a loss of function in α-synuclein is unlikely to cause PD, and mutations in α-synuclein that cause PD are likely to be gain-of-function mutations (36). α-Synuclein knockout mice have increased DA release following paired stimuli and an attenuation of DA-dependent locomotor responses to amphetamine, which suggest that α-synuclein may be an essential presynaptic, activity-dependent negative regulator of dopaminergic neu-rotransmission (36). α-Synuclein appears to be the primary component of the Lewy body (35). It has the ability to polymerize into approximately 10-nm fibrils in vitro, and bundles of these fibrils are the major component of Lewy bodies and Lewy neurites (35).
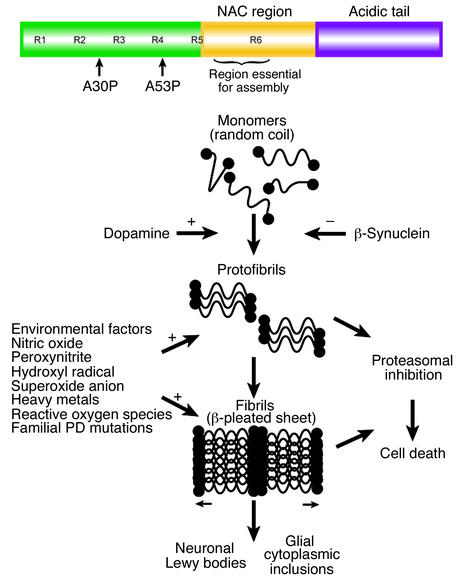
Figure 1.
Structure of α-synuclein and a model of α-synuclein aggregation and toxicity. The modular structure of α-synuclein, illustrating the location of familial-associated mutations and other key features of α-synuclein, including the imperfect (KTKEGV) repeats (R), is depicted in the top panel. The bottom panel shows a model of α-synuclein aggregation and toxicity and the proposed factors that enhance (+) or inhibit (–) the formation of toxic aggregated forms of α-synuclein. DA enhances the formation of the protofibrillar form of α-synuclein and prevents it from aggregating into the fibrillar form (not shown).
Overexpression of human wild-type α-synuclein in mice using the PDGF promoter yielded mice with selective decrements in DA nerve terminals in the striatum, with a concomitant reduction in tyrosine hydroxylase catalytic activity (37). A variety of transgenic mice overexpressing wild-type or mutant forms of α-synuclein have been described with varying degrees of pathology and α-synuclein abnormalities. None of the mammalian transgenic models fully recapitulate PD, but they have proved useful for studying synucleinopathy-induced neurodegeneration (38–44). α-Synuclein–dependent neurodegeneration is associated with abnormal accumulation of detergent-insoluble α-synuclein, and abnormal proteolytic processing of α-synuclein and the A53T α-synuclein mutant appears to cause significantly greater in vivo toxicity as compared with the other α-synuclein variants (7, 40). β-Synuclein, the nonamyloidogenic homologue of α-synuclein, is an inhibitor of aggregation of α-synuclein and rescues the motor deficits, neurodegenerative alterations, and neuronal α-synuclein accumulations seen in human PDGF-promoter α-synuclein transgenic mice (45). Thus, β-synuclein might be a neutral negative regulator of α-synuclein aggregation, and the antiamyloidogenic property of β-synuclein might provide a novel strategy for the treatment of neurodegenerative disorders. In addition to the mammalian models, Drosophila models have been developed (46). When normal and mutant forms of α-synuclein are overexpressed in Drosophila, the flies develop an adult-onset (midlife) progressive loss of DA neurons and filamentous interneuronal inclusions that contain α-synuclein. Overexpression of heat-shock protein HSP70 rescues the motoric and neuropathologic features of transgenic flies expressing normal and mutant forms of α-synuclein (47). Chaperones may play a role in PD, as Lewy bodies in human postmortem tissue immunostain for chaperones (47). The transgenic mouse and Drosophila models are persuasive in implicating α-synuclein in the pathogenesis of PD (6).
How derangements in α-synuclein lead to neuronal dysfunction and death is not known. However, aggregation and fibrillation are thought to play a central role (35), perhaps leading to a dysfunction in protein handling by inhibiting the proteasome (48) (Figure 1). It is not clear how mutations in α-synuclein cause selective degeneration of dopaminergic neurons, since α-synuclein is a ubiquitously expressed protein. However, oxidative ligation of DA to α-synuclein leads to the accumulation of the α-synuclein protofibril, which may be the toxic α-synuclein moiety (49). Although α-synuclein can form adducts with other catecholamines related to DA, it has been suggested that adduct formation provides an explanation for the dopaminergic selectivity of α-synuclein–associated neurotoxicity in PD (49). α-Synuclein toxicity in human dopaminergic neurons requires endogenous DA production and is mediated by reactive oxygen species. It does not seem to involve DA adduct formation but, instead, is mediated by 54- to 83-kDa soluble protein complexes that contain α-synuclein and 14-3-3 protein (50).
Parkin mutations
Linkage analysis of 13 families with AR-JP mapped the localization of the AR-JP gene to chromosome 6q25.2-27 (51). The identification of the AR-JP gene was facilitated by the discovery of a microdeletion in a family with AR-JP. The gene causing AR-JP was designated parkin and encodes a protein of 465 amino acids, with moderate similarity to ubiquitin at its amino-terminus and a RING-finger motif at the carboxy-terminus (10) (Figure 2). In the patient with a microdeletion, exons 3–7 were deleted, and four other AR-JP patients from three unrelated families also had a deletion affecting exon 4 (10). Since the initial discovery of the parkin gene, many groups have identified mutations in parkin, including exonic deletions, insertions, and several missense mutations (52) (Figure 2). So far, most of the point mutations reside in the RING-IBR-RING domains of parkin, suggesting that this region is key to parkin function. Mutations in parkin appear to be a major cause of autosomal recessive PD (53). Indeed, parkin mutations are now considered to be one of the major causes of familial PD.
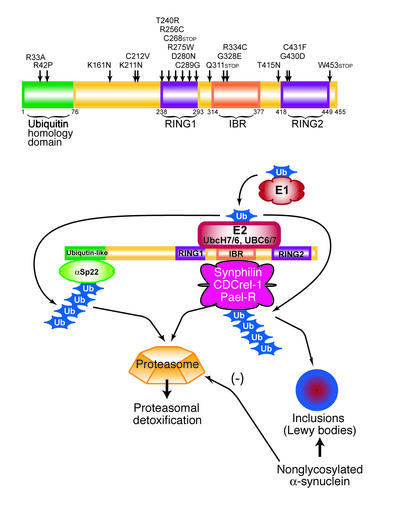
Figure 2.
Structure of parkin and a model of parkin-mediated ubiquitination and its substrates. In the top panel, the modular architecture of parkin is depicted, and the locations of the reported familial-associated disease-causing missense mutations are indicated. Disease-causing deletions, insertions, and frameshifts are not depicted (see text for details). The bottom panel shows a model of parkin-mediated ubiquitination and Lewy body formation. Parkin may play key roles in both ubiquitination and Lewy body formation in conjunction with the E1 ubiquitin-activating enzyme and the E2s UbcH7, UbcH8, UBC6, and UBC7. Nonglycosylated α-synuclein accumulates in Lewy bodies through mechanisms that are not clear, but it may inhibit (–) proteasomal function. Ub, ubiquitin.
The modular architecture of parkin led to insight into its function, as several other proteins had similar modular structures (54). In particular, a few proteins with RING-finger motifs were shown to be involved in E2-dependent ubiquitination (55). Proteins are targeted for degradation in the 20S proteasome by covalent attachment of ubiquitin (56). The 26S proteasome recognizes multiubiquitin chains that are formed by linkage through the lysine residue at position 48 in the ubiquitin protein. Polyubiquitination occurs by the cooperation of several sequentially acting enzymes. The ubiquitin-activating enzyme E1 activates the ubiquitin in an ATP-dependent manner; then ubiquitin is transferred to a ubi-quitin-conjugating enzyme, E2. A final step requires an E3 ubiquitin-protein ligase, which facilitates the transfer of ubiquitin to the target protein. Substrate specificity of the ubiquitin system is largely conferred by the E3 ubiquitin-protein ligase. Parkin was shown to be an E2-dependent E3 ubiquitin-protein ligase (55, 57, 58). It appears to use both UbcH7 and UbcH8 as its E2s, and it also utilizes the ER-associated E2s UBC6 and UBC7 (56). Familial-associated mutations in parkin impair the binding to either UbcH7 or UbcH8 and are defective in E3 ubiquitin-protein ligase activity, which suggests that disruption of the E3 ubiquitin-protein ligase activity of parkin is probably the cause of autosomal recessive PD. Since the loss of the E3 ligase activity of parkin may cause autosomal recessive PD, it is of great importance to identify the protein substrates of parkin. It is conceivable that dysfunction of the proteasomal processing of one or more of these proteins leads to dopaminergic dysfunction. Several potential substrates for parkin have recently been identified. The first substrate identified was the synaptic vesicle–associated protein CDCrel-1 (55). CDCrel-1 belongs to a family of septin GTPases, and it has been suggested that it regulates synaptic vesicle release in the nervous system (59). Whether CDCrel-1 is involved in the release of DA is not yet known, but it is possible that mutations in parkin affect CDCrel-1 modulation of DA release, which ultimately contributes to the parkinsonian state.
Synphilin-1 is also a substrate for parkin-targeted ubiquitination (60). The function of synphilin-1 is unknown, but it was identified and cloned as an α-synuclein–interacting protein (61). It is also a synaptic vesicle–enriched protein, and it is present in Lewy bodies (61, 62). Coexpression of synphilin-1 with α-synuclein in cultured cells results in the formation of Lewy body–like aggregates containing both proteins; and in the presence of parkin, a significant percentage of these aggregates become ubiquitinated (60). The observation that patients with mutations in parkin do not have Lewy bodies has led to the speculation that pathogenic mechanisms caused by mutations in parkin are different from those that occur in sporadic PD and in PD due to mutations in α-synuclein (54). On the other hand, the interactions of synphilin-1 with both parkin and α-synuclein suggest a common link between the different causes of PD and connect the pathogenesis of PD caused by mutations in parkin with that of PD caused by alterations in α-synuclein. Parkin may be intimately involved in the ubiquitination of Lewy body–associated proteins, such as synphilin-1. Furthermore, the sequestration of parkin in inclusions may contribute to its loss of function. In the absence of parkin, Lewy body–associated proteins would not be ubiquitinated and the formation of Lewy bodies would be impaired (54, 56). Consistent with this notion is the observation that familial-associated mutations of parkin fail to ubiquitinate synphilin-1. Recently, a patient with an R275W mutation in one allele of parkin and a 40-bp exon 3 deletion in the other allele revealed the presence of Lewy body pathology in regions typically affected in PD (63). Interestingly, the R275W parkin mutation reduces the catalytic activity of parkin, but it still has substantial enzyme activity (60). Thus, this mutation appears to be the exception that proves the rule and indicates that parkin is required for the formation of Lewy pathology. Lewy pathology may contribute, in part, to neuronal cell death by the sequestration of the function of parkin, which serves to degrade specific proteins (56). Using immunological methods in the normal human brain, Shimura et al. identified an O-glycosylated isoform of α-synuclein (αSp22) that contains complex monosaccharide chains (64). Familial-associated parkin mutants failed to bind αSp22, and, in an in vitro ubiquitination assay, αSpp22 was ubiquitinated by normal, but not by mutant, parkin. Whether or not αSp22 is present in the brain is controversial, since most laboratories have not been able to detect it and this type of complex glycosylation is highly unusual for a cytoplasmic protein (7). Nonglycosylated α-synuclein, the major species in the brain, does not appear to be a parkin substrate in vitro or in the brain, as parkin fails to interact with nonglycosylated α-synuclein and fails to ubiquitinate nonglycosylated α-synuclein (60, 64).
A more general role for parkin in the ubiquitin proteasomal-degradation pathway is suggested by the recent observations that parkin is upregulated by unfolded-protein stress. Parkin has been found to suppress unfolded protein-stress induced toxicity (58). Parkin may function in the unfolded-protein response, as it is localized to the microsomal fraction as well as to the cytosol and Golgi fractions (52). The unfolded-protein response regulates multiple ER and secretory pathway genes, and it is conceivable that mutations or deletions of the parkin gene could result in the accumulation of misfolded substrate proteins in the ER, leading to DA cell death in AR-JP. Recently, an unfolded putative G protein–coupled transmembrane receptor, the parkin-associated endothelial-like receptor (Pael-R), was found to be a parkin substrate (65). When overexpressed, Pael-R tends to become unfolded and insoluble and causes unfolded protein–induced cell death. Parkin ubiquitinates Pael-R, and coexpression of parkin results in protection against Pael-R–induced cell toxicity. Pael-R accumulates in the brains of AR-JP patients and thus may be an important parkin substrate. In the brain, Pael-R is expressed predominantly in oligodendrocytes, but it is also expressed at exceptionally high levels in neurons containing tyrosine hydroxylase. Thus, Pael-R is an attractive parkin substrate whose accumulation may account for the loss of DA neurons in AR-JP.
DJ-1 mutations
In genetically isolated communities in the Netherlands, Bonifati and colleagues, using an RT-PCR strategy, identified a deletion in exons 1AB to 5 of the DJ-1 gene that showed complete cosegregation with PD and the disease allele in a Dutch family (11). In addition, a T→C transition at position 497 from the open reading frame start in the cDNA of DJ-1 resulting in the substitution of a highly conserved leucine at position 166 of the DJ-1 protein by a proline was identified that shows complete cosegregation with the disease allele in an Italian family (11). In the Dutch family the DJ-1 protein is absent, and in the Italian family DJ-1 appears to be functionally inactive. Thus, mutations in the DJ-1 gene definitely cause PD, probably through a loss of function. It is difficult at this juncture to fully appreciate how mutations in the DJ-1 gene cause PD, as its function is largely unknown (11). However, DJ-1 was identified as a hydroperoxide-responsive protein that becomes more acidic following oxidative stress, suggesting that it may function as an antioxidant protein (66). Furthermore, DJ-1 is sumoylated through binding to the SUMO-1 ligase PIAS, suggesting that it might be involved in the regulation of transcription (67). Other putative functions of DJ-1 have been raised (11), but how a loss of function of DJ-1 leads to loss of DA neurons in PD awaits further study.
Summary
Ultimately it would be of great interest to link all the multiple genes and the sporadic causes of PD into a common pathogenic biochemical pathway — much in the same way that, in AD, the metabolism of the amyloid precursor protein has been shown to be central to the degenerative process. Derangements in protein handling seem to be central in the pathogenesis of PD. Aggregation, fibrillation, and proteasom-al dysfunction seem central to the pathogenesis of α-synuclein–induced injury. Mutations in parkin lead to dysfunction in the proteasomal-degradation pathway. It will be of interest to determine whether parkin is involved in the ubiquitination and elimination of proteins whose proteasomal degradation is impaired by derangements in α-synuclein. The major nonglycosylated form of α-synuclein is linked to parkin through their mutual interactions with synphilin-1, and the minor glycosylated α-synuclein species may be directly linked to parkin, providing further evidence for a common biochemical pathway. How the unfolded-protein stress response fits into a common pathway is not clear. Oxidative stress seems to play a prominent role in sporadic PD, and oxidative stress leads to synuclein aggregation and/or proteasomal dysfunction. Recent data suggest that there may be selective derangements in the protea-somal system in the substantia nigra of sporadic PD patients (68). Thus, protein mishandling may be central to the pathogenesis of PD. Another possible common pathogenic mechanism is the disruption of synaptic function, as α-synuclein, synphilin-1, CDCrel-1, and parkin are synaptically enriched proteins, but this awaits further clarification and investigation. How DJ-1 fits into a common pathogenic pathway is not clear, but its potential role in oxidative stress would fit with the “oxidative stress” hypothesis. The fact the DJ-1 is sumoylated, a ubiquitin-like protein modification, suggests that it might be a target of parkin, tying it into the ubiquitin/proteasomal pathway. Only the future can tell whether DJ-1 and the other yet-to-be-identified PD genes will fit into one of these pathways or whether they will reveal new pathways. Surprises are undoubtedly in store as the identification of these genes leads to a better understanding of the pathogenesis of PD.
Acknowledgments
The authors thank Weza Cotman for secretarial assistance. This work was supported by grants from the US Public Health Service (NS38377) and the Edward D. and Anna Mitchell Family Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Parkinson disease (PD); dopamine (DA); Alzheimer disease (AD); autosomal recessive juvenile parkinsonism (AR-JP); parkin-associated endothelial-like receptor (Pael-R).
References
- 1.Lang AE, Lozano AM. Parkinson’s disease. Second of two parts. N. Engl. J. Med. 1998;339:1130–1143. doi: 10.1056/NEJM199810153391607. [DOI] [PubMed] [Google Scholar]
- 2.Lang AE, Lozano AM. Parkinson’s disease. First of two parts. N. Engl. J. Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- 3.Forno LS. Neuropathology of Parkinson’s disease. J. Neuropathol. Exp. Neurol. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Gasser T. Genetics of Parkinson’s disease. J. Neurol. 2001;248:833–840. doi: 10.1007/s004150170066. [DOI] [PubMed] [Google Scholar]
- 5.Roses AD. Apolipoprotein E and Alzheimer’s disease. The tip of the susceptibility iceberg. Ann. NY Acad. Sci. 1998;855:738–743. doi: 10.1111/j.1749-6632.1998.tb10653.x. [DOI] [PubMed] [Google Scholar]
- 6.Dawson T, Mandir A, Lee M. Animal models of PD: pieces of the same puzzle? Neuron. 2002;35:219–222. doi: 10.1016/s0896-6273(02)00780-8. [DOI] [PubMed] [Google Scholar]
- 7.Giasson BI, Lee VM. Parkin and the molecular pathways of Parkinson’s disease. Neuron. 2001;31:885–888. doi: 10.1016/s0896-6273(01)00439-1. [DOI] [PubMed] [Google Scholar]
- 8.Polymeropoulos MH, et al. Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science. 1996;274:1197–1199. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- 9.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 10.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 11.Bonifati, V., et al. 2002. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. doi:10.1126/science.1077209. [DOI] [PubMed]
- 12.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 13.Gasser T, et al. A susceptibility locus for Parkinson’s disease maps to chromosome 2p13. Nat. Genet. 1998;18:262–265. doi: 10.1038/ng0398-262. [DOI] [PubMed] [Google Scholar]
- 14.Farrer M, et al. A chromosome 4p haplotype segregating with Parkinson’s disease and postural tremor. Hum. Mol. Genet. 1999;8:81–85. doi: 10.1093/hmg/8.1.81. [DOI] [PubMed] [Google Scholar]
- 15.Muenter MD, et al. Hereditary form of parkinsonism--dementia. Ann. Neurol. 1998;43:768–781. doi: 10.1002/ana.410430612. [DOI] [PubMed] [Google Scholar]
- 16.Valente EM, et al. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am. J. Hum. Genet. 2001;68:895–900. doi: 10.1086/319522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Duijn CM, et al. Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am. J. Hum. Genet. 2001;69:629–634. doi: 10.1086/322996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funayama M, et al. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann. Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 19.Hampshire DJ, et al. Kufor-Rakeb syndrome, pallido-pyramidal degeneration with supranuclear upgaze paresis and dementia, maps to 1p36. J. Med. Genet. 2001;38:680–682. doi: 10.1136/jmg.38.10.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sveinbjornsdottir S, et al. Familial aggregation of Parkinson’s disease in Iceland. N. Engl. J. Med. 2000;343:1765–1770. doi: 10.1056/NEJM200012143432404. [DOI] [PubMed] [Google Scholar]
- 21.Hicks AA, et al. A susceptibility gene for late-onset idiopathic Parkinson’s disease. Ann. Neurol. 2002;52:549–555. doi: 10.1002/ana.10324. [DOI] [PubMed] [Google Scholar]
- 22.Scott WK, et al. Complete genomic screen in Parkinson disease: evidence for multiple genes. JAMA. 2001;286:2239–2244. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- 23.Martin ER, et al. Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. JAMA. 2001;286:2245–2250. doi: 10.1001/jama.286.18.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maraganore DM, et al. Case-control study of the extended tau gene haplotype in Parkinson’s disease. Ann. Neurol. 2001;50:658–661. doi: 10.1002/ana.1228. [DOI] [PubMed] [Google Scholar]
- 25.Golbe LI, et al. The tau A0 allele in Parkinson’s disease. Mov. Disord. 2001;16:442–447. doi: 10.1002/mds.1087. [DOI] [PubMed] [Google Scholar]
- 26.Pastor P, et al. Familial atypical progressive supranuclear palsy associated with homozigosity for the delN296 mutation in the tau gene. Ann. Neurol. 2001;49:263–267. doi: 10.1002/1531-8249(20010201)49:2<263::aid-ana50>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 27.Hutton M, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 28.DeStefano AL, et al. Genome-wide scan for Parkinson’s disease: the GenePD Study. Neurology. 2001;57:1124–1126. doi: 10.1212/wnl.57.6.1124. [DOI] [PubMed] [Google Scholar]
- 29.Li YJ, et al. Age at onset in two common neurodegenerative diseases is genetically controlled. Am. J. Hum. Genet. 2002;70:985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanner CM, et al. Parkinson disease in twins: an etiologic study. JAMA. 1999;281:341–346. doi: 10.1001/jama.281.4.341. [DOI] [PubMed] [Google Scholar]
- 31.Papadimitriou A, et al. Mutated alpha-synuclein gene in two Greek kindreds with familial PD: incomplete penetrance? Neurology. 1999;52:651–654. doi: 10.1212/wnl.52.3.651. [DOI] [PubMed] [Google Scholar]
- 32.Kruger R, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat. Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 33.Farrer M, et al. alpha-Synuclein gene haplotypes are associated with Parkinson’s disease. Hum. Mol. Genet. 2001;10:1847–1851. doi: 10.1093/hmg/10.17.1847. [DOI] [PubMed] [Google Scholar]
- 34.Clayton DF, George JM. The synucleins: a family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci. 1998;21:249–254. doi: 10.1016/s0166-2236(97)01213-7. [DOI] [PubMed] [Google Scholar]
- 35.Murray IVJ, Lee VM, Trojanowski JQ. Synucleinopathies: a pathological and molecular review. Clinical Neuroscience Research. 2001;1:445–455. [Google Scholar]
- 36.Abeliovich A, et al. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 37.Masliah E, et al. Dopaminergic loss and inclusion body formation in alpha-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287:1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- 38.Giasson BI, et al. Neuronal α-synucleinopathy with severe movement disorder in mice expressing A53T human α-synuclein. Neuron. 2002;34:521–533. doi: 10.1016/s0896-6273(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 39.Lee MK, Price DL. Advances in genetic models of Parkinson’s Disease. Clinical Neuroscience Research. 2001;1:456–466. [Google Scholar]
- 40.Lee MK, et al. Human alpha-synuclein-harboring familial Parkinson’s disease-linked Ala-53 right-arrow Thr mutation causes neurodegenerative disease with alpha-synuclein aggregation in transgenic mice. Proc. Natl. Acad. Sci. USA. 2002;99:8968–8973. doi: 10.1073/pnas.132197599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahle PJ, et al. Subcellular localization of wild-type and Parkinson’s disease-associated mutant alpha-synuclein in human and transgenic mouse brain. J. Neurosci. 2000;20:6365–6373. doi: 10.1523/JNEUROSCI.20-17-06365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuoka Y, et al. Lack of nigral pathology in transgenic mice expressing human alpha-synuclein driven by the tyrosine hydroxylase promoter. Neurobiol. Dis. 2001;8:535–539. doi: 10.1006/nbdi.2001.0392. [DOI] [PubMed] [Google Scholar]
- 43.Richfield EK, et al. Behavioral and neurochemical effects of wild-type and mutated human alpha-synuclein in transgenic mice. Exp. Neurol. 2002;175:35–48. doi: 10.1006/exnr.2002.7882. [DOI] [PubMed] [Google Scholar]
- 44.van der Putten H, et al. Neuropathology in mice expressing human alpha-synuclein. J. Neurosci. 2000;20:6021–6029. doi: 10.1523/JNEUROSCI.20-16-06021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimoto M, Rockenstein E, Mante M, Mallory M, Masliah E. β-Synuclein inhibits α-synuclein aggregation: a possible role as an anti-parkinsonian factor. Neuron. 2001;32:213–223. doi: 10.1016/s0896-6273(01)00462-7. [DOI] [PubMed] [Google Scholar]
- 46.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 47.Auluck PK, Chan HYE, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2001;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka Y, et al. Inducible expression of mutant alpha-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum. Mol. Genet. 2001;10:919–926. doi: 10.1093/hmg/10.9.919. [DOI] [PubMed] [Google Scholar]
- 49.Conway KA, Rochet JC, Bieganski RM, Lansbury PT., Jr Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 50.Xu J, et al. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat. Med. 2002;8:600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- 51.Matsumine H, et al. Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2-27. Am. J. Hum. Genet. 1997;60:588–596. [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Dawson VL, Dawson TM. Parkin: clinical aspects and neurobiology. Clinical Neuroscience Research. 2001;1:467–482. [Google Scholar]
- 53.Lucking CB, et al. Association between early-onset Parkinson’s disease and mutations in the parkin gene. French Parkinson’s Disease Genetics Study Group. N. Engl. J. Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 54.Dawson TM. New animal models for Parkinson’s disease. Cell. 2000;101:115–118. doi: 10.1016/S0092-8674(00)80629-7. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung KK, Dawson VL, Dawson TM. The role of the ubiquitin-proteasomal pathway in Parkinson’s disease and other neurodegenerative disorders. Trends Neurosci. 2001;24(Suppl.):S7–S14. doi: 10.1016/s0166-2236(00)01998-6. [DOI] [PubMed] [Google Scholar]
- 57.Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 58.Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 2000;275:35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- 59.Beites CL, Xie H, Bowser R, Trimble WS. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- 60.Chung KK, et al. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat. Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 61.Engelender S, et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat. Genet. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]
- 62.Wakabayashi K, et al. Synphilin-1 is present in Lewy bodies in Parkinson’s disease. Ann. Neurol. 2000;47:521–523. [PubMed] [Google Scholar]
- 63.Farrer M, et al. Lewy bodies and parkinsonism in families with parkin mutations. Ann. Neurol. 2001;50:293–300. doi: 10.1002/ana.1132. [DOI] [PubMed] [Google Scholar]
- 64.Shimura H, et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson’s disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 65.Imai Y, et al. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 66.Mitsumoto A, Nakagawa Y. DJ-1 is an indicator for endogenous reactive oxygen species elicited by endotoxin. Free Radic. Res. 2001;35:885–893. doi: 10.1080/10715760100301381. [DOI] [PubMed] [Google Scholar]
- 67.Takahashi K, et al. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J. Biol. Chem. 2001;276:37556–37563. doi: 10.1074/jbc.M101730200. [DOI] [PubMed] [Google Scholar]
- 68.McNaught KS, Olanow CW, Halliwell B, Isacson O, Jenner P. Failure of the ubiquitin-proteasome system in Parkinson’s disease. Nat. Rev. Neurosci. 2001;2:589–594. doi: 10.1038/35086067. [DOI] [PubMed] [Google Scholar]