Abstract
Modeling of disease pathogenesis and immunity often is carried out in large animals that are natural targets for pathogens of human or economic relevance. Although murine mAbs are a valuable tool in identifying certain host/pathogen interactions, progress in comparative immunology would be enhanced by the use of mAbs isolated from the host species. Such antibodies would reflect an authentic host immune response to infection or vaccination, and as they are host derived, would allow the application of in vivo experiments that previously have been unrealizable in large animals because of induction of an antispecies immune response. The advent of antibody phage display technology provides a way of producing host-derived mAbs in animals where the molecular genetics of Ig formation are known. Exploiting recent advances in the molecular immunology of cattle, we report here the design of an optimized phage display vector, pComBov, for the construction of combinatorial libraries of bovine Ig antigen-binding fragments (Fab) of native sequence. By using this system, we initially have generated and characterized a panel of bovine mAbs against a model antigen glutathione S-transferase. The isolated mAbs showed features typical of bovine Igs and recognized glutathione S-transferase with high specificity in ELISA and by Western blotting. The pComBov expression system can be readily adapted for the preparation of libraries from related ruminant species and advances the use of monoclonal reagents derived in this way for comparative studies in animals of economic importance.
The study of host-pathogen interactions in an outbred population of the natural target species provides valuable insights into the molecular basis of pathogenesis and immunity. With the studies often performed in large domesticated animals, progress in this area frequently is constrained by a lack of reagents and by the limited applicability of advances pioneered in immunologically well-characterized species such as the mouse. There therefore has been a trend to model disease processes in animals that are more immunologically amenable than the true target species. One area that typifies this difficulty is that of mAb technology, originally described for the production of murine Igs through the generation of hybridoma cells (1). Although attempts to produce monospecific human antibodies by other immortalization methods, for example by transformation with Epstein–Barr virus (2), have met with success, it has proved difficult to apply this strategy to large domesticated animals of economic significance, especially cattle, where there have been only isolated reports of the generation of mAbs through the formation of heterohybridoma lines of uncertain stability (3, 4). The advent of antibody phage-display technology, where specific mAbs are generated by using molecular cloning techniques (5, 6), offers the opportunity to redress this constraint, because the methods that ultimately yield mAbs are founded on an understanding of the molecular immunology of the species under study, rather than the availability of stable transformed cell lines or methods to immortalize B lymphocytes. This technology has been used successfully to isolate human mAbs against viruses, self-antigens, and tumor antigens (5, 7). We have sought to apply this technology to cattle to generate a panel of bovine mAbs, specifically to further our work with a bovine model of papillomavirus (PV)-induced mucosal epithelial tumors (8). However, in principle, the methods are applicable to any species for which there is a comparable understanding of the genetic basis of Ig formation.
mAbs derived from the host species offer several advantages over conventional murine hybridoma technology in the field of comparative immunology. First, antigenic determinants recognized by the natural host immune system may differ from those induced by vaccination in experimental animals. For example, a T helper cell epitope of human PV-16 identified in several strains of mice by vaccination does not appear to be recognized by the natural human host (9). Therefore, host-derived mAbs may more accurately reflect genuine host immune responses to a given antigen. In addition, if these antibodies exhibited a high binding affinity against antigen, they would become reagents of choice in experimental and diagnostic assays traditionally performed with murine mAbs. Although human mAbs isolated by this methodology have been shown to be of variable affinity, a number of techniques now exist that increase the binding affinity of highly specific mAbs with low affinities (10). Second, a common procedure in experimental mouse models is the in vivo administration of specific mAbs to authenticate in vitro observations. It previously has not been possible to perform this type of experiment by using murine mAbs in experimental animals other than mice, because of an antispecies recognition of the antibodies and rapid clearance by the host immune system. The ability to perform in vivo studies in large animals would represent a major advance in the field of comparative immunology.
Like many domesticated species, cattle predominantly express Ig λ light chains over κ chains (11). In addition, despite the apparent complexity of the bovine λ locus (12), our work (13) and that of others (14) have shown that the light chain repertoire is dominated by expression of a single family of Vλ segments. Conveniently, the Ig heavy chain repertoire is also founded on expression of a single gene family comprising up to 15 near-identical members contributing to all bovine heavy chains characterized to date (15–18). This relative molecular simplicity is not unique to cattle. Similar processes operate in chickens, rabbits, pigs, goats, and sheep (11); however, it is an advantage in the production of recombinant antibodies, as fewer sets of oligonucleotide primers are required to recover the bovine Ig repertoire by PCR amplification. This report describes the construction of a phage-display vector pComBov for expression of fully bovine antibodies as antigen-binding antibody fragments (Fabs), the generation of a combinatorial Ig library from bovine lymph node tissue, and the isolation of bovine antibodies against a model antigen, glutathione S-transferase (GST).
MATERIALS AND METHODS
Vector Construction.
Initially, the vector pComb3H, a derivative of pComb3 (19) optimized for expression of human fragments, was modified to encode bovine framework (FR) 1 residues upstream of the SstI (SacI) cloning site for light chain amplicons (12, 13, 15). Five hundred picomoles of partially overlapping oligonucleotide pairs (5′ P-Lmod1/2 and Hmod1/2) (Fig. 1) were annealed then extended in Klenow fragment in the presence of dNTPs and purified by acrylamide gel electrophoresis. The Lmod1 fragment was ligated into NruI/SstI cut pComb3H. Because the bovine λ constant region (CL) carries a SacI site (12, 13, 15), this sequence was amplified from bovine genomic DNA by PCR using Pfu polymerase (Stratagene) and the mut3/4 primers (Fig. 1). To remove the SacI site from the CL sequence, two fragments were PCR-amplified by using the mut1/4 and mut2/3 oligonucleotides. The resulting products were combined, and an additional amplification was carried out by using the mut3/4 primers. The modified CL fragment was digested with XbaI and SstI and ligated into the light chain FR1-modified plasmid. To complete vector construction, the double-stranded Hmod fragment encoding bovine heavy chain FR1 sequences was digested with XhoI and NcoI and ligated into the plasmid. In the resulting phage display vector pComBov, light chain (VL) and heavy chain (VH) variable region amplicons are cloned as SstI/BstEII and RsrII/SpeI fragments, respectively.
Figure 1.
Oligonucleotide primers used in expression vector and library construction. Primer sequences are written 5′ to 3′. Restriction endonuclease recognition sites are underlined. Degenerate nucleotides are indicated as follows: S = C or G; W = A or T; M = A or C; Y = C or T.
Nucleic Acid Sequencing.
All plasmid sequence modifications and complete nucleotide sequences of the isolated antibodies were determined by using oligonucleotide primers as shown in Fig. 1 and performed on an automated Applied Biosystems sequencer.
RNA Isolation and PCR Amplification of Antibody Chains.
A female Simmental calf was injected i.m. with 300–400 μg of GST/bovine PV-4 L2a fusion protein in Alhydrogel (Intervet Laboratories, Cambridgeshire, U.K.) (20) four times over a 3-month period. Five days after the last injection, the animal was sacrificed, and the popliteal and iliac lymph nodes were removed. Total RNA was isolated by using RNAzol (Biogenesis, Bournemouth, U.K.), and 15 μg was reverse-transcribed to cDNA by using an oligo(dT) primer according to GeneAmp RNA PCR kit protocols (Perkin–Elmer). VH (including the amino-terminal heavy chain constant region CH1) and VL regions were amplified by PCR using Taq polymerase and VH1/2 and VL1/2 oligonucleotide primers (Fig. 1). PCR products were purified by gel electrophoresis then digested with excess restriction enzymes as detailed below.
Library Construction.
The method of library construction was essentially as described (21) with the following modifications: pComBov was digested with an excess of SstI and BstEII, treated with phosphatase, and then purified by agarose gel electrophoresis. Two micrograms of plasmid was ligated to 330 ng of SstI/BstEII-digested VL fragments with 20 units of T4 DNA ligase. Ligated DNA was electroporated into Escherichia coli XL1Blue by using a Bio-Rad Gene Pulser and amplified as described (21). The size of the light chain library was determined by plating aliquots of the culture on Luria–Bertani agar plates containing 100 μg/ml of carbenicillin (22). Phagemid DNA containing the light chain library was prepared by using Maxiprep columns (Qiagen, Crawley, U.K.), then digested with an excess of RsrII and SpeI and purified as above. Two micrograms of the VL library was ligated to 660 ng of RsrII/SpeI-cut VH PCR product, transformed into XL1Blue cells, and amplified and titered as above. To produce recombinant phage, 1012 plaque-forming units of VCSM13 helper phage was added, and the cultures were amplified overnight and phage-precipitated as described (21). To determine the efficiency of insertion of light and heavy chain fragments, DNA from single clones was amplified by PCR using the Lseq/mut2 (VL) or Hseq/gIII (VH) oligonucleotides (Fig. 1). To assess the diversity of the amplified sequences, the resulting products were digested with EcoRII (BstNI) and electrophoresed on an 8% polyacrylamide gel to determine restriction patterns.
Panning of Library.
If required, library phage stocks were reamplified as follows: 20 μl of stored phage was added to 10 ml of log-phase XL1Blue cells, incubated at 20°C for 15 min, before adding 10 ml Superbroth (SB) (22) containing 10 μg/ml of tetracycline and 20 μg/ml of carbenicillin (SB/tet10/carb20). The culture was shaken for 1 hr at 37°C, then the concentration of carbenicillin was increased to 50 μg/ml. Additional incubations, addition of VCSM13, overnight incubation, and phage precipitation were as for the VH insertion above (21). Panning of the library was performed essentially as described (21). Microwell plates were coated with 1 μg of GST in carbonate buffer (15 mM Na2CO3/35 mM NaHCO3, pH 9.6), before blocking in Tris-buffered saline (TBS)/3% BSA. Fifty microliters of fresh phage suspension containing 1011–1012 phage/well was bound for 2 hr at 37°C, then the wells were washed in TBS/0.5% Tween 20 for 5 min. Wells were washed twice in round 1, five times in round 2, and 10 times in round 3 and any subsequent rounds. Phage were eluted in 0.1 M HCl, pH 2.2/1 mg/ml BSA for 10 min, followed by vigorous pipetting. Eluted phage were neutralized immediately with 6 μl of 2 M Tris base, then added to 2 ml of fresh log-phase XL1Blue cells for 15 min, and amplified as described (21). Phage output was determined by plating aliquots of eluted phage on Luria–Bertani agar/100 μg/ml of carbenicillin. The phage input at each round was determined by infection of 100-μl aliquots of XL1Blue cells with dilutions of the phage suspension and plating as above. Panning was repeated for 3–5 rounds over consecutive days.
Characterization of Phage Clones.
ELISA. Output clones from the final round of panning were picked into 200 μl of Superbroth/tet10/carb50/1% glucose in 96-well round bottom plates and grown at 37°C overnight. Five microliters of each culture was added to 200 μl of medium as above but containing 0.1% glucose and grown for an additional 2–4 hr, after which 109 VCSM13 was added. After 15 min at 20°C, the cultures were grown for an additional 2 hr at 37°C. Finally, kanamycin was added to a final concentration of 70 μg/ml, and the plates were incubated at 30°C overnight. Culture supernatants were added to microwell plates coated with 1 μg of GST or BSA and blocked in TBS/3% BSA. The phage were bound for 2 hr at 37°C, followed by extensive washing of the wells in TBS/0.1% Tween 20. Bound phage were detected with biotin-linked anti-fd bacteriophage antibody, followed by ExtrAvidin-alkaline phosphatase (Sigma). Enzyme substrate BluePhos (Dynatech) was added to the wells, and absorbance was read at 630 nm.
Restriction mapping and sequencing.
Individual clones were grown in Superbroth/carb50/1% glucose overnight at 37°C, and plasmid DNA was isolated by using Qiaprep spin miniprep columns (Qiagen). DNA was digested with EcoRII and analyzed by electrophoresis on an 8% polyacrylamide gel. Individual restriction digestion patterns were visualized by staining in ethidium bromide. Plasmid DNA from selected clones was sequenced at the VH and VL regions by using primers shown in Fig. 1.
Expression and Detection of Soluble Fab.
Plasmid DNA from selected clones was digested with NheI and SpeI, then religated and electroporated into XL1Blue cells. For ELISA, clones were grown overnight at 37°C in 2YT (22)/carb50/1% glucose in 96-well plates. Ten microliters of culture was added to 200 μl of 2YT/carb50/0.1% glucose and grown at 37°C until OD600 ≈ 0.8, then induced with 1 mM isopropyl β-d-thiogalactopyranoside and incubated overnight at 30°C. Culture supernatants containing Fab were added to antigen-coated and blocked ELISA plates as above for 2 hr at 37°C. In addition to the antigens above, 1 μg of the following proteins was coated to the wells: keyhole limpet hemocyanin, hen egg lysozyme, chicken egg ovalbumin, and fluorescein isothiocyanate-BSA conjugate (Sigma). Bound Fab was detected with alkaline-phosphatase-conjugated F(ab′)2-specific anti-bovine IgG antibody (Jackson ImmunoResearch), by using BluePhos substrate. For Western blot, Fab clones were grown as above but in 5-ml vol. One microgram of GST or BSA was separated on a 10% SDS/PAGE gel, transferred to nitrocellulose, then blocked in TBS/5% skim milk powder/0.1% Tween 20. Blots were incubated for 1 hr in culture supernatants containing Fab, washed in TBS/0.1% Tween 20 then incubated with horseradish peroxidase (HRP)-conjugated F(ab′)2-specific anti-bovine IgG (Jackson ImmunoResearch) for an additional hour. Blots were developed by using ECL substrate (Amersham Pharmacia) and exposed to x-ray film. Anti-GST mAb (Autogen, Wiltshire, U.K.) followed by HRP-conjugated anti-mouse IgG (New England Biolabs) was used as a positive control.
RESULTS
Vector Construction.
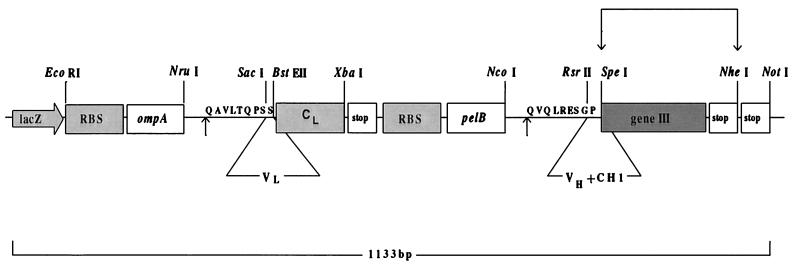
Changes were introduced into the phage display vector pComb3H to optimize the system for expression of bovine Igs of native sequence. In the first of these, DNA downstream of the unique NruI site, located in the bacterial leader, was replaced, retaining the ompA reading frame and adding codons for QAVLTQPSS, the native amino terminus of FR1 of the bovine λ light chain (12, 13), downstream of the predicted point of cleavage by signal peptidase. Codons for the last three amino acids were chosen so as to create a SacI (SstI) restriction site for insertion of light chain amplicons. Because the bovine λ constant domain carries a SacI site (12, 13, 15), we amplified this region from lymphocyte genomic DNA and engineered silent mutations by overlap PCR. At the same time, a BstEII site was introduced to span codons for serine, valine, and threonine (SVT), residues at the amino-terminal end of the λ constant region (12, 13, 15). This modification enabled the insertion of light chains as SacI/BstEII fragments, the remainder of the λ constant domain being encoded by the modified vector. Finally, sequences were optimized upstream of the site of insertion of heavy chain amplicons. A small NcoI/XhoI fragment was replaced with synthetic DNA encoding the amino-terminal residues of bovine heavy chain FR1 (QVQLRESGPS; refs. 15–18). Codons were selected to provide an RsrII site spanning the last three codons. Selection of appropriate primers then allowed insertion of heavy chain amplicons as RsrII/SpeI fragments into the reading frames for the upstream pelB leader and the product of phage gene III, the minor coat protein that enables display of expressed Fabs. A diagram of the modified region of the vector, pComBov, is shown in Fig. 2. Expression of bovine Fabs as soluble protein is possible through excision of geneIII on a SpeI/NheI fragment. At this stage, an affinity tag (24) also can be inserted to simplify purification (P.M.O., unpublished work).
Figure 2.
pComBov expression cassette. Modified from pComb3H phagemid (19) on a pBluescript background (23). The CL was cloned from bovine genomic DNA. Digestion with SpeI and NheI removes the geneIII sequence, allowing expression of soluble Fab. Arrows indicate the position of cleavage of secretory sequences (pelB or ompA) during periplasmic expression of Fab. Amino acid resides (single-letter code) immediately after the cleavage site are the optimized bovine amino-terminal Ig sequences. RBS, ribosome binding site.
Immunization, Library Construction, and Panning.
After four vaccinations, the immunized animal was shown to have a high serum antibody titer against the GST-L2a fusion protein, with a lower reactivity against GST alone (not shown). Five days after the final injection, the animal was sacrificed. On examination, the draining popliteal and iliac lymph nodes were found to be very enlarged. After RNA isolation, Ig VH and VL sequences were amplified by PCR and used to construct a library in pComBov via a standard two-stage protocol. The initial light chain library comprised more than 107 clones with an insertion frequency of about 92% (not shown). This library formed the basis for insertion of heavy chains, yielding a final library of more than 2 × 107 clones, of which approximately 100% carried VH inserts as judged by PCR. Both libraries showed diversity of cloned VH and VL inserts after restriction digestion with EcoRII (BstN1), which cuts at a high frequency in Ig variable regions (25). To confirm that the reading frames were intact and that recombinant bovine Fabs could be expressed in a stable form, the geneIII sequence was removed from randomly selected individual clones and Fab-expressing culture supernatants were separated by SDS/PAGE. Bovine Fab were detected by Western blot by using anti-bovine IgG F(ab′)2 mAb (not shown). Having established that the expression system was functional, we proceeded to pan the library against GST as a model antigen.
The number of GST-specific phage clones increased with each successive round of panning, as determined by phage ELISA against GST or BSA (Table 1). After three rounds of panning, almost 100% of the output clones expressed Fab reactive with GST. However, after a fourth round of panning, specificity appeared to decline.
Table 1.
Enrichment of anti-GST recombinant phage over four rounds of panning
| Panning round | Input no. of phage | Output no. of phage | % Recovery | No. GST positive phage/no. tested |
|---|---|---|---|---|
| 1 | 6.5 × 1011 | 1 × 104 | 0.000015 | NT |
| 2 | 5.3 × 1011 | 2.5 × 106 | 0.00047 | 5/30 |
| 3 | 7 × 1011 | 2.5 × 107 | 0.0036 | 29/30 |
| 4 | 1.2 × 1012 | 2.7 × 108 | 0.023 | 21/27 |
NT, not tested.
Characterization of Selected Phage Clones.
Of the 30 clones analyzed by phage ELISA after round 3, 23 with apparent GST specificity were randomly chosen for further characterization. Initially, plasmid DNA was purified for digestion with EcoRII and analysis on polyacrylamide gels. This process revealed that of the 23 clones, all but three possessed a common restriction pattern, indicating close similarity, and were judged dominant. These clones represented approximately 87% (20/23) of the anti-GST phage clones isolated after three rounds of panning. Of the 20 dominant clones, eight were randomly chosen for sequencing along with the three that exhibited more rare restriction patterns. Three VL sequences emerged from the dominant group of clones (Fig. 3). From the length of complementarity-determining region (CDR) 1, the fifth, seventh, 10th, and 11th amino acids encoded in this region and features of CDR2, it is clear that these light chain V regions are derived from the Vλ1a subgroup (13).
Figure 3.
(A) Alignment of predicted amino acid sequences for the selected anti-GST clones. The nucleotide sequences of the VH and VL inserts of eight dominant and three minority clones were determined and translated into protein sequence (single-letter code). Unique VL (i–iii) and VH (i–iv) are shown for the dominant clones. Each minority clone expressed unique VL and VH sequence patterns. The sequences are segregated into CDR and FR regions based on homology to known bovine sequences (11–18). A dash (–) indicates identity to the uppermost sequence; ∗ indicates no amino acid at this position. (B) VL and VH chain usage in the dominant selected anti-GST clones. Three VL (i–iii) and four VH (i–iv) sequences were found in various combinations in the eight dominant clones. Clones G67, G72, G74, and G76 expressed the same VL and VH combination and are therefore identical. Clone G63 differs by only two VH FR residues from clone G77.
Four VH sequences were expressed by clones in the dominant group (Fig. 3). Although they appear more diverse as a group than the light chains with which they are paired, overall these VH sequences are closely related, supporting the notion that these Fabs recognize common or closely related GST epitopes. Given the frequency with which extended VH CDR3 sequences have been reported for bovine Igs in the literature (15–18), it is the modest length of CDR3 (5 aa) that is the most striking feature of this group.
It is considered that the antigen specificity of antibodies is conferred by the heavy chain, and that this specificity can be maintained despite pairing with a number of different light chains—the concept of chain promiscuity (26). This promiscuity was demonstrated in the isolated Fabs: clones G65 and G77 possessed common VL sequences but were associated with heavy chains that differ in CDRs 1 and 2; clones G65 and G81 shared common VH sequences but with divergent light chains (Fig. 3). It is not known whether chain promiscuity is a result of the random nature of combinatorial library construction or whether the isolated Fabs represent original pairings of VL and VH chains selected in vivo.
The three minority clones sequenced on the basis of their uncommon EcoRII restriction patterns also expressed light chains derived from the Vλ1a subgroup but possessed quite distinct features in the CDRs (Fig. 3). The heavy chains were generally more typical of those previously characterized from cattle in that CDR3 ranged in length from 17 to 22 aa.
Expression of Soluble Fab.
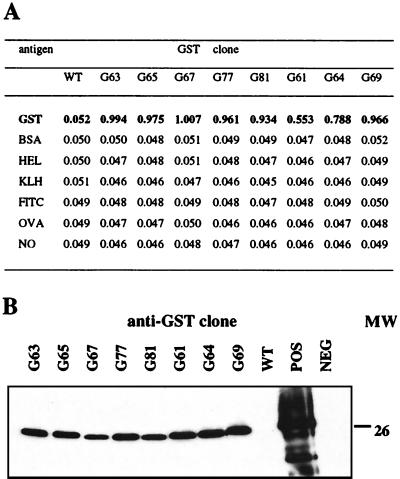
The geneIII sequence was removed from four of the dominant clones that express different VH chains (G63, G65, G67, and G77) and from the three minority clones. Culture supernatants or periplasmic extracts containing soluble Fab were tested for antigen specificity against a variety of proteins. All of the clones showed absolute specificity for GST by ELISA (Fig. 4). Relative affinity measurements of each individual Fab were not calculated, and therefore a comparison of ELISA titers between clones is not possible. The Fab supernatants also were tested for antigenic specificity by Western blot. As shown by ELISA, the bovine Fabs exhibited a strong and specific signal against GST (Fig. 4B) but did not detect BSA (not shown).
Figure 4.
Antigen specificity of anti-GST Fab. (A) ELISA. Fab supernatants were diluted 1:2 and tested for reactivity against a range of protein antigens. Optical densities were read at 630 nm after 15 min; data are results from one of three similar experiments. Reactivity against GST is indicated in bold. WT, pComBov (no VH or VL sequences); HEL, hen egg lysozyme; KLH, keyhole limpet hemocyanin; FITC, fluorescein isothiocyanate-BSA conjugate; OVA, chicken ovalbumin; NO, no coating antigen. (B) Western blot GST and BSA were separated by SDS/PAGE and transferred to nitrocellulose, then incubated with supernatants containing anti-GST Fab. Fab recognized GST (26 kDa) but not BSA (not shown). WT, as in A; POS, positive control anti-GST mouse mAb; NEG, anti-bovine Ig-horseradish peroxidase only; MW, molecular weight (kDa).
DISCUSSION
In principle, antibody phage display allows the isolation of mAb from any species for which the molecular basis of Ig formation is understood. This technology is well advanced in human immunology, where mAbs generated by phage display are being applied in both research and immunotherapy. We sought to apply antibody phage display to large domesticated animals of economic importance, which often are used in comparative immunology as models of disease. Our model of bovine PV-4-induced tumor development in mucosal epithelium is one such example, in which we want to further define the bovine host response to a prophylactic vaccine (8). Although murine mAbs could be used to delineate certain host responses in vitro and determine authentic bovine Ig responses to vaccination, and have the ability to perform future in vivo experiments, such as transfer of passive immunity, we considered it appropriate to attempt to isolate bovine mAbs.
We chose to produce antibodies as Fab fragments, by using an optimized vector based on that developed by Barbas et al. (19), pComb3H. Although there are now reports of the production of recombinant antibodies from a range of other nonhuman, nonlaboratory species, namely chickens (27, 28), rabbits (29–31), and camels (32, 33), the authors either have engineered single-chain variable region proteins differing from native Igs in both primary amino acid sequence and conformation or have used phage display vectors that yield Fab fragments but do not encode optimized FR1 sequences to match those of the host. One unique feature of the system that we report here is that it yields Fab fragments that are entirely matched to bovine FR sequences (12–18). We anticipate that this feature will prove beneficial in future in vivo studies, as the native sequence of these proteins should fail to provoke an immune response in the bovine host. A similar premise has been demonstrated by the successful use of fully human mAbs in immunotherapy, an area in which widespread use of murine mAb previously has been hampered by an antispecies immune response (10). Although the antibodies described here are Fab molecules and their half-life in vivo may differ from that of full-length Igs, it would be a relatively simple undertaking to add the remaining bovine CH regions to synthesize complete bovine antibodies in an appropriate expression vector (34).
We constructed a Ig combinatorial library in pComBov from the lymphoid cells of a GST-bovine PV-4 fusion protein-vaccinated animal. We chose to use immune B cells rather than those from a naive animal for two reasons. Immune libraries are highly biased toward the expression of V-genes specific for the immunizing protein, an advantage when an analysis of humoral immune response to infection is required. Additionally, as the Igs have undergone somatic mutation in vivo, this approach has the potential to produce antibodies of greater specificity and affinity than those that would be isolated from a similar size library constructed from naive lymphocytes. Although the nature of the combinatorial library construction does not necessarily mean that native pairings of heavy and light chains are generated, an increase in the frequency of antibody-secreting cells induced by vaccination should correspondingly increase the likelihood of isolating natural pairings.
Initially, we constructed a library from peripheral B cells and attempted to isolate Fabs against GST, which often is used as a fusion partner for our recombinant PV proteins. This approach was unsuccessful in spite of a moderate anti-GST serum titer of Ig present in the donor animal (not shown). However, peripheral blood contains a limited number of B cells secreting antibody and may not be an optimum source of lymphocytes for the generation of combinatorial libraries (35). When we constructed a library from lymph nodes of the same animal, we successfully and rapidly isolated a range of antibodies against GST. After the success of our model antigen, we are now isolating bovine mAbs against the immunizing fusion partner bovine PV-4 L2a, which will be reported separately. It should be noted that the isolation of human mAbs against human PV-16 E4 (36) and E7 (37) recently has been reported.
Almost all of the isolated anti-GST Fabs appeared to be very closely related, as predicted by restriction mapping and demonstrated by sequence analysis, and it is likely that the Fabs in the major group that we identified react with common or closely related immunodominant GST epitopes. Although the anti-GST Fab clones represented less commonly were more divergent in sequence, especially in the CDRs, their relationship to the dominant clones is nevertheless close. These clones may recognize other epitopes of GST or have a different affinity for the same epitope recognized by the dominant clones. It is striking that the antigen-specific Fabs that we have characterized are so polarized in the length of the heavy chain CDR3 (predominantly 5 aa, or less commonly 22 residues). It is therefore evident that while long CDR3 sequences may be a common feature of the bovine Ig heavy chain (15–18), binding to antigen is equally possible with a more modest area of interaction.
This paper demonstrates that antibody phage display technology can be successfully applied to cattle, and we would expect that a similar approach could be formulated for other large animals where the molecular structure of Ig is known. Indeed, the substantial sequence similarity between ovine and bovine Igs (11) also should enable expression of sheep Fabs after minor modification to pComBov. Given the methods that now exist to increase the size and diversity of combinatorial libraries, and the affinity of the generated mAbs (10), we anticipate that the production of recombinant fully bovine mAbs in pComBov or its derivatives should advance aspects of comparative immunology, studies of infectious disease, and animal productivity via immunomodulation.
Acknowledgments
We thank D. Burton and C. Barbas for providing the pComb3H expression plasmid. This work is supported by the Medical Research Council. M.S.C. is a Cancer Research Campaign Life Fellow.
ABBREVIATIONS
- CDR
complementarity-determining region
- CL
light chain constant region
- Fab
antigen-binding antibody fragment
- FR
framework region
- GST
glutathione S-transferase
- PV
papillomavirus
- TBS
Tris-buffered saline
- VH
heavy chain variable region
- VL
light chain variable region
Footnotes
References
- 1.Kohler G, Milstein C. Nature (London) 1975;25:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 2.Cole S P, Campling B G, Atlaw T, Kozbor D, Roder J D. Mol Cell Biochem. 1984;62:109–120. doi: 10.1007/BF00223301. [DOI] [PubMed] [Google Scholar]
- 3.Groves D J, Tucker E M. Vet Immunol Immunopathol. 1989;23:1–14. doi: 10.1016/0165-2427(89)90105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suter M. Vet Immunol Immunopathol. 1992;33:285–300. doi: 10.1016/0165-2427(92)90001-7. [DOI] [PubMed] [Google Scholar]
- 5.Burton D R, Barbas C F., III Adv Immunol. 1994;57:191–280. doi: 10.1016/s0065-2776(08)60674-4. [DOI] [PubMed] [Google Scholar]
- 6.Winter G, Griffiths A D, Hawkins R E, Hoogenboom H R. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 7.Nissim A, Hoogenboom H R, Tomlinson I M, Flynn G, Midgley C, Lane D, Winter G. EMBO J. 1994;13:692–698. doi: 10.1002/j.1460-2075.1994.tb06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campo M S. Vet J. 1997;154:175–188. doi: 10.1016/s1090-0233(97)80019-6. [DOI] [PubMed] [Google Scholar]
- 9.Frazer I H, Tindle R W. Papillomavirus Rep. 1992;3:53–58. [Google Scholar]
- 10.Vaughan T J, Osbourn J K, Tempest P R. Nat Biotechnol. 1998;16:535–539. doi: 10.1038/nbt0698-535. [DOI] [PubMed] [Google Scholar]
- 11.Pastoret P P, Griebel P, Bazin H, Govaerts A, editors. Handbook of Vertebrate Immunology. London: Academic; 1998. [Google Scholar]
- 12.Parng C L, Hansal S, Goldsby R A, Osborne B A. J Immunol. 1996;157:5478–5486. [PubMed] [Google Scholar]
- 13.Sinclair M C, Gilchrist J, Aitken R. J Immunol. 1995;155:3068–3078. [PubMed] [Google Scholar]
- 14.Armour K L, Tempest P R, Fawcett P H, Fernie M L, King S I, White P, Taylor G, Harris W J. Mol Immunol. 1994;31:1369–1372. doi: 10.1016/0161-5890(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 15.Jackson T, Morris B A, Sanders P G. Mol Immunol. 1992;29:667–676. doi: 10.1016/0161-5890(92)90204-b. [DOI] [PubMed] [Google Scholar]
- 16.Sinclair M C, Gilchrist J, Aitken R. J Immunol. 1997;159:3883–3889. [PubMed] [Google Scholar]
- 17.Berens S J, Wylie D E, Lopez O J. Int Immunol. 1997;9:189–199. doi: 10.1093/intimm/9.1.189. [DOI] [PubMed] [Google Scholar]
- 18.Saini S S, Hein W R, Kaushik A. Mol Immunol. 1997;34:641–651. doi: 10.1016/s0161-5890(97)00055-2. [DOI] [PubMed] [Google Scholar]
- 19.Barbas C F, III, Kang A S, Lerner R A, Benkovic S J. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrachud L M, Grindlay G J, McGarvie G M, O’Neil B W, Wagner E R, Jarrett W F H, Campo M S. Virology. 1995;211:204–208. doi: 10.1006/viro.1995.1392. [DOI] [PubMed] [Google Scholar]
- 21.Burton D R, Barbas C F, III, Persson M A A, Koenig S, Chanock R M, Lerner R A. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 1. New York: Wiley; 1994–1998. [Google Scholar]
- 23.Short J M, Fernandez J M, Sorge J A, Huse W D. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoogenboom H R, Griffiths A D, Johnson K S, Chiswell D J, Hudson P, Winter G. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marks J D, Hoogenboom H R, Bonnert T P, McCafferty J, Griffiths A D, Winter G. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 26.Collet T A, Roben P, O’Kennedy R, Barbas C F, III, Burton D R, Lerner R A. Proc Natl Acad Sci USA. 1992;89:10026–10030. doi: 10.1073/pnas.89.21.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies E L, Smith J S, Birkett C R, Manser J M, Anderson-Dear D V, Young J R. J Immunol Methods. 1995;186:125–135. doi: 10.1016/0022-1759(95)00143-x. [DOI] [PubMed] [Google Scholar]
- 28.Yamanaka H I, Inoue R, Ikeda T O. J Immunol. 1996;157:1156–1162. [PubMed] [Google Scholar]
- 29.Ridder R, Schmitz R, Legay F, Gram H. Biotechnology. 1995;13:255–260. doi: 10.1038/nbt0395-255. [DOI] [PubMed] [Google Scholar]
- 30.Lang I M, Barbas C F, III, Schleef R R. Gene. 1996;172:295–298. doi: 10.1016/0378-1119(96)00021-2. [DOI] [PubMed] [Google Scholar]
- 31.Foti M, Granucci F, Ricciardi-Castagnoli P, Spreafico A, Ackermann M, Suter M. J Immunol Methods. 1998;213:201–212. doi: 10.1016/s0022-1759(98)00029-5. [DOI] [PubMed] [Google Scholar]
- 32.Arabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. FEBS Lett. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 33.Lauwereys M, Arabi Ghahroudi M, Desmyter A, Kinne J, Holzer W, De Genst E, Wyns L, Muyldermans S. EMBO J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter P, Merchant A M. Curr Opin Biotechnol. 1997;8:449–454. doi: 10.1016/s0958-1669(97)80067-5. [DOI] [PubMed] [Google Scholar]
- 35.Yip Y L, Hawkins N J, Clark M A, Ward R L. Immunotechnology. 1997;3:195–203. doi: 10.1016/s1380-2933(97)00013-4. [DOI] [PubMed] [Google Scholar]
- 36.Doorbar J, Foo C, Coleman N, Medcalf L, Hartley O, Prospero T, Napthine S, Sterling J, Winter G, Griffin H. Virology. 1997;238:40–52. doi: 10.1006/viro.1997.8768. [DOI] [PubMed] [Google Scholar]
- 37.Wang-Johanning F, Gillespie G Y, Grim J, Rancourt C, Alvarez R D, Siegal G P, Curiel D T. Cancer Res. 1998;58:1893–1900. [PubMed] [Google Scholar]