Abstract
To elucidate the function of PPARγ in leptin-deficient mouse (ob/ob) liver, a PPARγ liver-null mouse on an ob/ob background, ob/ob-PPARγ(fl/fl)AlbCre+, was produced using a floxed PPARγ allele, PPARγ(fl/fl), and Cre recombinase under control of the albumin promoter (AlbCre). The liver of ob/ob-PPARγ(fl/fl)AlbCre+ mice had a deletion of exon 2 and a corresponding loss of full-length PPARγ mRNA and protein. The PPARγ-deficient liver in ob/ob mice was smaller and had a dramatically decreased triglyceride (TG) content compared with equivalent mice lacking the AlbCre transgene (ob/ob-PPARγ(fl/fl)AlbCre–). Messenger RNA levels of the hepatic lipogenic genes, fatty acid synthase, acetyl-CoA carboxylase, and stearoyl-CoA desaturase-1, were reduced in ob/ob-PPARγ(fl/fl)AlbCre+ mice, and the levels of serum TG and FFA in ob/ob-PPARγ(fl/fl)AlbCre+ mice were significantly higher than in the control ob/ob-PPARγ(fl/fl)AlbCre– mice. Rosiglitazone treatment exacerbated the fatty liver in ob/ob-PPARγ(fl/fl)AlbCre– mice compared with livers from nonobese Cre– mice; there was no effect of rosiglitazone in ob/ob-PPARγ(fl/fl)AlbCre+ mice. The deficiency of hepatic PPARγ further aggravated the severity of diabetes in ob/ob mice due to decreased insulin sensitivity in muscle and fat. These data indicate that hepatic PPARγ plays a critical role in the regulation of TG content and in the homeostasis of blood glucose and insulin resistance in steatotic diabetic mice.
Introduction
PPARγ is a nuclear receptor that heterodimerizes with retinoid X receptor α and activates genes involved in lipid storage and metabolism. It is required for lipid homeostasis (1, 2). Immortalized fibroblasts lacking PPARγ lose the potential for differentiation to mature adipocytes (3), indicating that PPARγ is absolutely required for differentiation of preadipocytes to mature adipocytes. PPARγ also plays a critical role in the regulation of cholesterol homeostasis in the macrophage (4, 5). PPARγ–/– embryos die at embryonic day 9.5–10 due to placental dysfunction (6). Therefore, determination of the physiological function of PPARγ in mice has been limited to the study of heterozygous PPARγ+/– animals (7, 8).
PPARγ is expressed at the highest level in adipose tissue (9, 10), colon epithelium (11–13), and macrophages (14, 15). In contrast to these tissues or cells, the expression of PPARγ in liver is very low (10, 15, 16). PPARγ is normally expressed in both human and murine liver at only 10–30% of the level in adipose tissue (10, 15, 16). The function of PPARγ in liver is not clear. However, it is noteworthy that PPARγ is expressed at elevated levels in the liver of a number of murine models of diabetes or obesity, including aP2/DTA (17), A-ZIP/F1 (18), ob/ob (19, 20), db/db (19), KKA (21), and 5-HT2cR (19) mutant mice. Levels of hepatic PPARγ were elevated by seven- to ninefold in ob/ob and db/db mice compared with wild-type mice. The elevation in expression of the UCP2 and CD36 genes, known target genes of PPARγ, suggests that PPARγ in ob/ob liver is functional (19). However, the physiological role of hepatic PPARγ remains to be clarified.
To determine the role of hepatic PPARγ in a diabetic mouse model and to circumvent the embryonic lethality of a standard gene knockout model, conditional-null mice were created using the Cre-loxP strategy and a Cre transgene derived from the interferon α/β promoter (MxCre) (4). The resultant PPARγ(fl/fl)MxCre mice lacked expression of PPARγ in liver, but also in other tissues, including spleen and kidney. To produce a liver-specific PPARγ-null mouse model, a liver-specific albumin promoter–driven Cre transgene, AlbCre, was used to generate PPARγ(fl/fl)AlbCre mice. These mice were bred with ob/ob mice to obtain liver-specific disruption of PPARγ in ob/ob mice. Here we show that ob/ob mice with a liver-specific disruption of PPARγ exhibited a dramatic improvement in fatty liver but had exacerbated hyperglycemia and insulin resistance.
Methods
Generation of liver-specific PPARγ conditional-null mice.
PPARγ(fl/fl) mice, produced as described (4), were bred with a mouse containing the AlbCre transgene (22), kindly provided by Derek LeRoith of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH. This transgene was used in an earlier study to create an HNF4α liver-null mouse (23). Heterozygous (fl/+) animals carrying one copy of AlbCre were then interbred with fl/+ littermates lacking Cre to generate liver-specific PPARγ conditional-null mice and littermate control mice. PPARγ(fl/fl)AlbCre+ or PPARγ(fl/fl)AlbCre– mice were intercrossed with heterozygotic C57BL/6J-Lepob mice obtained from The Jackson Laboratory (Bar Harbor, Maine) to generate double heterozygotes (PPARγ fl/+, OB/ob). Since mouse PPARγ and leptin are both on chromosome 6, the double heterozygotes were crossed until recombination occurred to generate OB/ob-PPARγ(fl/fl) genotype mice. The OB/ob-PPARγ(fl/fl)AlbCre+ or OB/ob-PPARγ(fl/fl)AlbCre– mice were then crossed to generate ob/ob-PPARγ(fl/fl)AlbCre+ or ob/ob-PPARγ(fl/fl)AlbCre– mice. Mice were reared on a 12-hour light/dark cycle and fed water and a pellet chow diet (NIH-07) ad libitum. For rosiglitazone (SmithKline Beecham Pharmaceuticals, West Sussex, United Kingdom) treatment, a powdered diet (AIN-93G; Dyets Inc., Bethlehem, Pennsylvania, USA) was blended with the drug and administered for 3 weeks at approximately 3 mg/kg/day. The National Cancer Institute Animal Care and Use Committee approved all animal studies, which were carried out in accordance with Institute of Laboratory Animal Resources (ILAR) guidelines.
DNA and RNA analysis.
Southern and Northern blot analysis and RNase protection assays were performed as previously described (4). The cDNA probes used for Northern blotting were described in previous reports (4, 23) except for the probes indicated below. cDNA probes for malic enzyme (MAL), ATP-citrate lyase (ACL), and glycerol-3-phosphate acyltransferase (GPAT) were amplified by PCR from a mouse liver cDNA library using gene-specific primers and cloned into pGEM-T Easy Vector (Promega Corp., Madison, Wisconsin, USA). The primers used for PCR were as follows. MAL 5′, CGATGATAAGGTCTTCCTCACCAC and 3′, TCCTATGGAGTGTTTGGGTTCG. ACL 5′, GGTCAATCTCTCTCTGGATGGAGT and 3′, GGATGGTCTTGGCATAGTCATAGG. GPAT 5′, CGAAGGTCACTACAATGGCGAAC and 3′, GGTCTCTTTGAAAACCCCGATG. The identities of the probes were confirmed by nucleotide sequencing.
Measurement of lipids, lipoprotein lipase activity, VLDL export, and insulin.
Serum lipid levels and lipoproteins were analyzed as previously described (4). The clearance rate of exogenous triglyceride (TG) was measured in mice fasted for 4 hours and then gavaged with 400 μl of olive oil. Blood was taken at 0, 60, 90, 270, and 390 minutes after administration of oil, and plasma TGs were measured. For liver TG and cholesterol concentrations, total lipids were extracted from 100 mg liver as previously described (24) and the extracts were used for the measurement of each lipid class. Postheparin lipoprotein and hepatic lipase activities were assayed in triplicate using 14C-labeled triolein substrate as previously described (25). Postheparin plasma was collected as previously described (25). VLDL export rates were measured according to an earlier method (26). Plasma insulin was measured with a radioimmunoassay kit (Linco Research Inc., St. Charles, Missouri, USA).
Glucose levels and glucose tolerance tests.
Glucose tolerance tests, performed on conscious mice following a 6-hour fast, were done by intraperitoneal administration of glucose (2 mg/g). Blood samples were taken at 0, 30, 60, 90, and 120 minutes from the tail vein and were analyzed for glucose concentrations using a Glucometer Elite (Bayer Corp., Elkhart, Indiana, USA).
Histology.
Livers from 7- to 8-week-old representative mice were fixed in 10% neutral buffered formalin and embedded in paraffin. Sections were cut at a thickness of 4–6 μm and stained with H&E.
Hyperinsulinemic-euglycemic clamp.
The clamp studies were performed using 5-week-old male ob/ob mice using the protocol previously described (27).
Results
Production of a liver-specific PPARγ-null mouse on an ob/ob background.
The conditional floxed allele of the PPARγ gene (Figure 1a) was described in a previous report (4). No difference in phenotype was noted between 15-week-old PPARγ(fl/fl)AlbCre– and PPARγ(fl/fl)AlbCre+ mice, probably reflecting the low level of PPARγ expression in liver of nonobese wild-type mice (10, 15, 28). To elucidate the role of PPARγ in liver, PPARγ(fl/fl)AlbCre mice were crossed with ob/ob mice. The ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ mice thus obtained had a distribution of offspring genotypes that followed the predicted mendelian frequencies. These crosses also generated lean mice, wild type for the leptin gene, from the same litter, designated OB/OB-PPARγ(fl/fl)AlbCre– and OB/OB-PPARγ(fl/fl)AlbCre+.
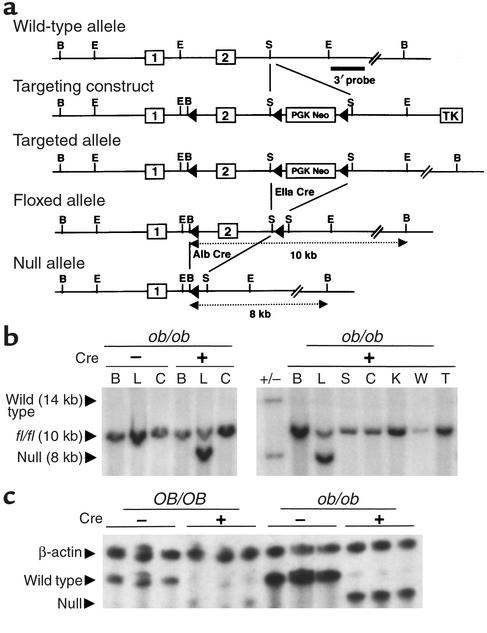
Figure 1.
Gene targeting and conditional deletion of exon 2 of the PPARγ gene. (a) Restriction maps of the wild-type allele, targeting vector, targeted allele, floxed allele, and null allele. The indicated 3′ probe was used to assess recombination events by Southern blot analysis. Open boxes represent exons and are numbered as indicated. PGK neomycin (PGK Neo) and thymidine kinase (TK) are positive and negative selection cassettes, respectively. Restriction sites: B, BamHI; E, EcoRI; S, SacI. (b) Southern blot analysis of BamHI-digested genomic DNA isolated from brain (B), liver (L), colon (C), spleen (S), kidney (K), white adipose (W), and tail (T) in ob/ob-PPARγ(fl/fl)AlbCre+ or ob/ob-PPARγ(fl/fl)AlbCre– mice. Fragments hybridizing with 3′ probe from the wild-type, floxed, and deleted alleles migrate at approximately 14, 10, and 8 kb, respectively. (c) RNase protection analysis of PPARγ mRNA in OB/OB- or ob/ob-PPARγ(fl/fl)AlbCre mouse livers. Total RNA from three separate mouse livers in each genotype were hybridized with riboprobes for β-actin and PPARγ. The products were then separated on a 5.0% polyacrylamide gel. The size of the protected mRNA fragments for PPARγ and β-actin is as follows; wild-type PPARγ, 195 nt; null PPARγ, 165 nt; and β-actin; 250 nt.
To examine liver-specific deletion of the floxed exon 2 of the PPARγ gene, the recombination event was analyzed by Southern blotting of genomic DNA isolated from various tissues using a 3′ probe (Figure 1a). Deletion of exon 2 was found to occur in liver in a Cre+-specific manner, as indicated by the presence of an 8-kb hybridizing band that represents the PPARγ allele lacking exon 2. However, in addition to the deleted band, the intact floxed exon 2 allele, represented by a 10-kb band, was also detected in the PPARγ(fl/fl)AlbCre+ mice (Figure 1b). The incomplete deletion is probably the result of nonparenchymal cells such as Kupffer, endothelial, and stellate cells that do not express the albumin promoter, as noted in previous reports (22, 29). The loss of full-length PPARγ mRNA and the presence of a truncated PPARγ mRNA product was confirmed in ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ liver using RNase protection assays (Figure 1c). The truncated PPARγ transcripts were detected only in Cre+ mice. Using an β-actin riboprobe to normalize for the amount of total RNA, the mRNA levels of PPARγ in ob/ob-PPARγ(fl/fl)AlbCre– mice were shown to be markedly higher than in OB/OB-PPARγ(fl/fl)AlbCre– mice. In addition, PPARγ(fl/fl)AlbCre+-derived mRNA levels in both OB/OB and ob/ob mice were significantly lower than native PPARγ transcripts, suggesting the possibility that the truncated mRNA is inherently less stable than wild-type PPARγ mRNA (4) or that PPARγ expression is self-regulated (30).
The deficiency of liver-specific PPARγ improves fatty liver in ob/ob mice.
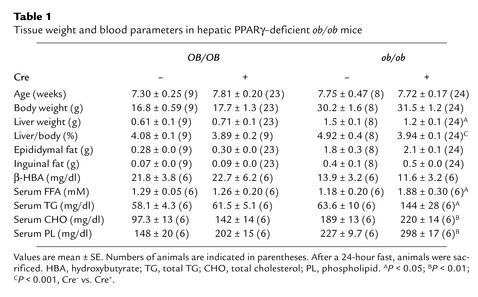
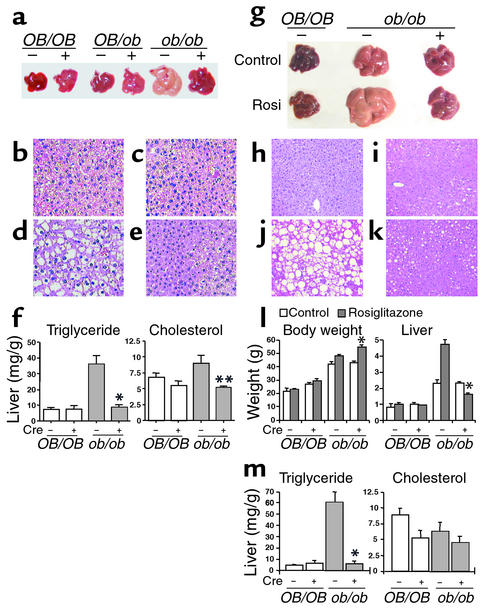
The ob/ob genetic background causes severe obesity and a fatty liver. To assess the potential effects of liver-specific PPARγ deficiency, body and tissue weights were measured. For a period of 8 weeks after birth, no significant difference in body and white adipose weight was observed between ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ mice (Table 1). However, the liver weight of ob/ob-PPARγ(fl/fl)AlbCre+ mice was significantly lower than that of ob/ob-PPARγ(fl/fl)AlbCre– mice; the OB/OB-PPARγ(fl/fl)AlbCre– and OB/OB-PPARγ(fl/fl)AlbCre+ livers were not different. These results suggest that PPARγ has a physiological function in the liver of ob/ob mice. Livers in the ob/ob-PPARγ(fl/fl)AlbCre– mice were significantly enlarged relative to those of OB/OB-PPARγ(fl/fl)AlbCre– mice and were yellowish in appearance, typical of fatty liver (Figure 2a). However, ob/ob-PPARγ(fl/fl)AlbCre+ mice had a dramatically improved fatty liver. Therefore, the elevated PPARγ expression in ob/ob liver appears to be a pathophysiological response to the state of severe obesity and diabetes. At this time, no phenotypic differences have been seen between OB/OB-PPARγ(fl/fl)AlbCre– and OB/OB-PPARγ(fl/fl)AlbCre+ mice. Histological analysis of the liver from each genotyped mouse revealed the presence of numerous hepatocyte vacuoles in the ob/ob-PPARγ(fl/fl)AlbCre– liver; hepatocyte vacuoles observed in the ob/ob-PPARγ(fl/fl)AlbCre+ liver were much smaller and less numerous than those seen in ob/ob-PPARγ(fl/fl)AlbCre– liver (Figure 2, b–e). These vacuoles were positive for the presence of lipid as revealed by oil red O staining (data not shown) or measurement of hepatic TG content. The hepatic TG content of ob/ob-PPARγ(fl/fl)AlbCre+ mice was also significantly lower (75% lower) than that of ob/ob-PPARγ(fl/fl)AlbCre– mice (Figure 2f). Rosiglitazone treatment of ob/ob-PPARγ(fl/fl)AlbCre– mice caused a marked increase in hepatic TG content and size compared with ob/ob-PPARγ(fl/fl)AlbCre+ mice (Figure 2g). Histological data also revealed numerous large hepatocyte vacuoles (Figure 2, h–k). However, these effects of rosiglitazone on liver were not observed in Cre+ mice. The TG content and liver weight in ob/ob-PPARγ(fl/fl)AlbCre+ mice were 8.5% and 33% that of rosiglitazone-treated ob/ob-PPARγ(fl/fl)AlbCre– mice, respectively (Figure 2, l and m). These results strongly suggest that hepatic PPARγ is involved in the development of fatty liver in ob/ob mice.
Table 1.
Tissue weight and blood parameters in hepatic PPARγ–deficient ob/ob mice
Figure 2.
Effect of hepatic PPARγ deficiency in ob/ob and OB/OB mice. (a–f) Nontreatment mice. (a) Liver of littermates. Livers of 8-week-old mice were used. (b–e) Histology of livers from OB/OB-PPARγ(fl/fl)AlbCre– (b) and ob/ob-PPARγ(fl/fl)AlbCre– (d) or OB/OB-PPARγ(fl/fl)AlbCre+ (c) and ob/ob-PPARγ(fl/fl)AlbCre+ mice (e). H&E staining was performed for liver sections (original magnification, ×100) from each genotyped mouse. (f) Total cholesterol and TG content in OB/OB-PPARγ(fl/fl)AlbCre– (n = 5: 2 males and 3 females), OB/OB-PPARγ(fl/fl)AlbCre+ (n = 4: 2 males and 2 females), ob/ob-PPARγ(fl/fl)AlbCre– (n = 6: 3 males and 3 females), and ob/ob-PPARγ(fl/fl)AlbCre+ (n = 15: 8 males and 7 females) mice. (g–m) Rosiglitazone-treated mice. (g) Rosiglitazone-treated livers. (h–k) H&E staining of livers from rosiglitazone-treated OB/OB- PPARγ(fl/fl)AlbCre– (h) and ob/ob-PPARγ(fl/fl)AlbCre– (j) or OB/OB- PPARγ(fl/fl)AlbCre+ (i) and ob/ob-PPARγ(fl/fl)AlbCre+ mice (k). (l) Body and liver weight in rosiglitazone-treated and control mice. For control groups: OB/OB-PPARγ(fl/fl)AlbCre–, n = 4 (2 males and 2 females); OB/OB-PPARγ(fl/fl)AlbCre+ (n = 3, males); ob/ob-PPARγ(fl/fl)AlbCre–, n = 7 (3 males and 4 females); and ob/ob-PPARγ(fl/fl)AlbCre+, n = 5 (3 males and 2 females). Rosiglitazone groups: OB/OB-PPARγ(fl/fl)AlbCre–, n = 5 (2 males and 3 females); OB/OB-PPARγ(fl/fl)AlbCre+, n = 4 (3 males and 1 female); ob/ob-PPARγ(fl/fl)AlbCre–, n = 10 (6 males and 4 females); and ob/ob-PPARγ(fl/fl)AlbCre+, n = 9 (5 males and 4 females). (m) Total cholesterol and TG content in rosiglitazone-treated mice. The mouse number for each genotype was described in l. Data are mean ± SE. *P < 0.001, **P < 0.01 compared with Cre– mice. Rosi, rosiglitazone.
Hepatic PPARγ controls the expression of lipogenic genes in ob/ob mice.
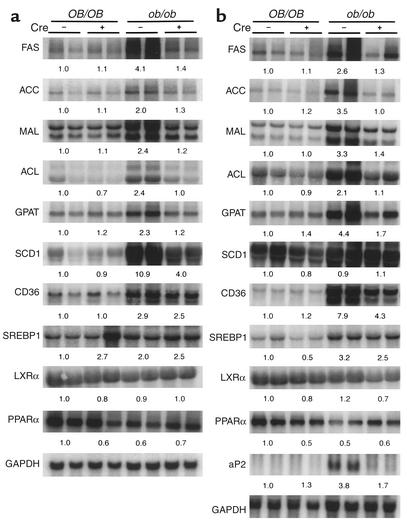
The PPARγ-mediated transactivation of several genes involved in lipid metabolism and transport has been demonstrated (4, 10). However, these studies analyzed adipose tissue or macrophages that highly express PPARγ. PPARγ-regulated gene expression in liver remains unclear. To uncover genes regulated by hepatic PPARγ and to determine the mechanism of the decrease in hepatic TGs in ob/ob mice lacking expression of PPARγ, mRNA from livers of untreated (Figure 3a) and rosiglitazone-treated (Figure 3b) mice was analyzed. The mRNAlevels of the fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC), ATP-citrate lyase (ACL), malic enzyme (MAL), glycerol-3-phosphate acyltransferase (GPAT), and stearoyl-CoA desaturase-1 (SCD1) genes in ob/ob-PPARγ(fl/fl)AlbCre+ mice were clearly lower than that of ob/ob-PPARγ(fl/fl)AlbCre– mice. However, their expression levels were unchanged in OB/OB-PPARγ(fl/fl)AlbCre– or OB/OB-PPARγ(fl/fl)AlbCre+ mice. Rosiglitazone treatment additively induced the expression of ACC, MAL, GPAT, CD36, and aP2 mRNA in ob/ob-PPARγ(fl/fl)AlbCre– mice but not in Cre+ mice (Figure 3b). The aP2 gene was not expressed in ob/ob-PPARγ(fl/fl)AlbCre– livers of untreated mice (data not shown). The mRNA levels of genes associated with glucose metabolism, apolipoprotein receptors, and apolipoproteins were unchanged by loss of PPARγ (data not shown). Since the lipogenic genes that were induced in ob/ob-PPARγ(fl/fl)AlbCre– mice are directly or indirectly involved in fatty acid and TG synthesis, these results suggest that hepatic PPARγ in ob/ob mice regulates TG content by controlling lipid synthesis. Further, other transcription factors that regulate the expression of lipogenic genes were analyzed. However, no difference in expression between ob/ob-PPARγ(fl/fl)AlbCre– and Cre+ mice was observed for the SREBP, liver X receptor-α (LXRα), or PPARα genes (Figure 3, a and b).
Figure 3.
Northern blot analysis to assess the effect of PPARγ deficiency on hepatic gene expression in untreated and rosiglitazone-treated ob/ob mice. Total RNA was isolated from nonfasting male mice and 20 μg was subjected to electrophoresis on a 1.2% agarose gel, transferred to a nylon membrane, and hybridized with the indicated 32P-labeled cDNA probes. (a) Northern blot of untreated ob/ob mice liver. (b) Northern blots of rosiglitazone-treated ob/ob mice liver. Quantitation of the bands was performed using the PhosphorImager from Molecular Dynamics and are expressed as the fold change, after correction for GAPDH levels, relative to OB/OB-PPARγ(fl/fl)AlbCre– mice. Values are averages obtained from two animals.
The deficiency of liver-specific PPARγ causes accumulation of VLDL and an elevation of FFA content in the ob/ob mouse.
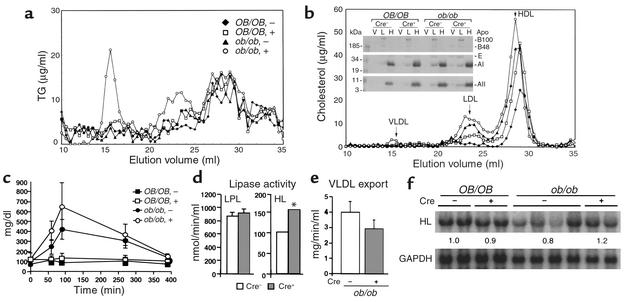
The serum lipid contents of ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ mice are summarized in Table 1. Levels of all lipid classes (TG, FFA, cholesterol, and phospholipids), of ob/ob-PPARγ(fl/fl)AlbCre+ mice were significantly higher than those of ob/ob-PPARγ(fl/fl)AlbCre– mice after 24 hours of fasting. Contrary to this result, no significant difference was observed in the two OB/OB-PPARγ(fl/fl)AlbCre mouse groups. To more precisely characterize the nature of the serum lipids and their associated lipoproteins in ob/ob-PPARγ(fl/fl)AlbCre+ mice, FPLC analysis was performed on 24-hour fasting serum. The elution profile by TG content revealed that the majority of the serum TG in ob/ob-PPARγ(fl/fl)AlbCre+ mice results from VLDL (Figure 4a). Consistent with these data, Western blot analysis of the apolipoprotein content of the VLDL, LDL, and HDL fractions revealed that ob/ob-PPARγ(fl/fl)AlbCre+ mice have increased apoB, apoE, and apoA in the VLDL fractions compared with ob/ob-PPARγ(fl/fl)AlbCre– mice (Figure 4b, inset). Elution profiles determined by cholesterol content revealed that LDL and HDL are also elevated in ob/ob-PPARγ(fl/fl)AlbCre+ mice (Figure 4b). The elevation of these lipoproteins appears to explain the elevation of cholesterol and phospholipid content in ob/ob-PPARγ(fl/fl)AlbCre+ mice (Table 1). The accumulation of VLDL in ob/ob-PPARγ(fl/fl)AlbCre+ mice raised questions about the potential for catabolism of exogenous TG-rich apolipoproteins as chylomicrons. To investigate this possibility, mice were gavaged with olive oil, and serum TG was sequentially measured (Figure 4c). The elevated rate of serum TG in ob/ob-PPARγ(fl/fl)AlbCre+ mice showed a tendency toward slower clearance than in Cre– mice, although this difference did not reach statistical significance. These results suggest that the deficiency of hepatic PPARγ causes an impairment of TG-rich lipoprotein clearance. Furthermore, to elucidate potential mechanisms for increased serum TG and FFA in ob/ob-PPARγ(fl/fl)AlbCre+ mice, lipase activities and VLDL export rates were measured. However, no significant differences in the lipoprotein lipase (Figure 4d) and VLDL export rates (Figure 4e) were observed between ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ mice. However, hepatic lipase activity was significantly increased in ob/ob-PPARγ(fl/fl)AlbCre+ mice (Figure 4d). The result of Northern blotting showed that, with the exception of one mouse, hepatic lipase mRNA in ob/ob-PPARγ(fl/fl)AlbCre– liver is lower than in OB/OB-PPARγ(fl/fl)AlbCre– mice. This result is in agreement with an earlier report (31). Furthermore, the deficiency of hepatic PPARγ resulted in recovery of hepatic lipase expression to levels similar to those found in OB/OB-PPARγ(fl/fl)AlbCre– mice (Figure 4f), suggesting that the different activities between ob/ob-PPARγ(fl/fl)AlbCre– and Cre+ mice result from the differences in transcriptional levels.
Figure 4.
Effect of PPARγ deficiency on the catabolism of serum TG in the ob/ob mouse. Lipoproteins were separated from 60 μl of pooled mouse plasma samples (n = 6 for each genotype) by FPLC. The concentration of TG (a) and cholesterol (b) in each eluted fraction is indicated on the y axis. Inset: immunoblot analysis of apoB, apoE, apoA-I, and apoA-II contained within the VLDL (V), LDL (L), and HDL (H) top fractions from each mouse genotype. (c) Measurement of serum TG after gavage with oil. The clearance rate of exogenous TG was measured in mice fasted for 4 hours as described in Methods. (d and e) Measurement of plasma lipase activities (d) and VLDL export rates (e). In d, for ob/ob-PPARγ(fl/fl)AlbCre– mice, n = 7; ob/ob-PPARγ(fl/fl)AlbCre+, n = 10. In e, for ob/ob-PPARγ(fl/fl)AlbCre–, n = 7; ob/ob-PPARγ(fl/fl)AlbCre+, n = 6. Each assay was performed as described in Methods. LPL, lipoprotein lipase; HL, hepatic lipase. (f) Northern blotting for hepatic lipase. The Northern blot was performed as described in the legend to Figure 3. All data are mean ± SE.*P < 0.01, Cre– vs. Cre+ mice.
Deficiency of liver-specific PPARγ aggravates hyperglycemia and insulin resistance in ob/ob mice.
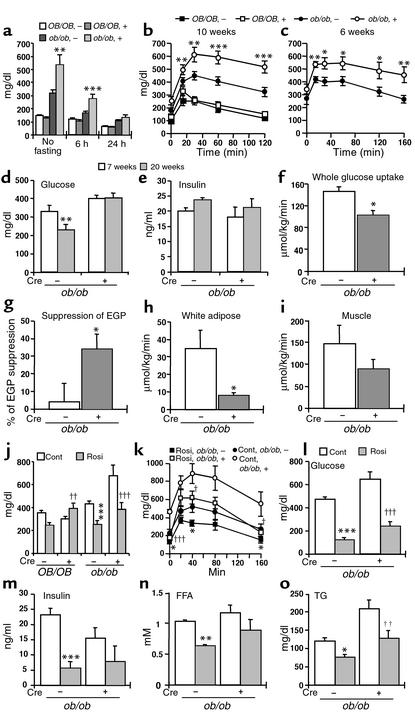
To assess the effects of deficiency of liver-specific PPARγ on glucose homeostasis, the level of blood glucose was measured (Figure 5a). Glucose levels of ob/ob-PPARγ(fl/fl)AlbCre+ mice were significantly higher than those of ob/ob-PPARγ(fl/fl)AlbCre– mice. To further characterize glucose metabolism, glucose tolerance tests were performed following an exogenous load of glucose. The glucose levels in ob/ob-PPARγ(fl/fl)AlbCre– mice were higher than those in the OB/OB-PPARγ(fl/fl)AlbCre– mice (Figure 5, b and c). Surprisingly, in ob/ob-PPARγ(fl/fl)AlbCre+ mice of both age groups, there was a significant elevation in blood glucose levels at all timepoints compared with ob/ob-PPARγ(fl/fl)AlbCre– mice. However, the elevated glucose levels in ob/ob-PPARγ(fl/fl)AlbCre+ mice were improved by treatment with rosiglitazone, suggesting that the effect of rosiglitazone on glucose levels does not depend on hepatic PPARγ (Figure 5, j and l).
Figure 5.
Effect of PPARγ deficiency on glucose homeostasis in the ob/ob mouse. (a) Blood glucose concentrations were measured in 10-week-old mice after no fasting and after 6 hours or 24 hours of fasting. For OB/OB-PPARγ(fl/fl)AlbCre– mice, n = 15; OB/OB-PPARγ(fl/fl)AlbCre+, n = 16; ob/ob-PPARγ(fl/fl)AlbCre–, n = 16; and ob/ob-PPARγ(fl/fl)AlbCre+, n = 11. (b and c) Glucose tolerance test. 10-week-old (b) and 6-week-old (c) mice were injected with glucose (2 mg/g). For 10-week-old mice, OB/OB-PPARγ(fl/fl)AlbCre– mice, n = 8; OB/OB-PPARγ(fl/fl)AlbCre+, n = 9; ob/ob-PPARγ(fl/fl)AlbCre–, n = 19; and ob/ob-PPARγ(fl/fl)AlbCre+, n = 15. For 6-week-old mice, ob/ob-PPARγ(fl/fl)AlbCre–, n = 9; and ob/ob-PPARγ(fl/fl)AlbCre+, n = 8. (d and e) Effect of rosiglitazone on glucose and insulin levels. Both of these measurements were performed using the same samples. For 20-week-old mice, ob/ob-PPARγ(fl/fl)AlbCre–, n = 13; ob/ob-PPARγ(fl/fl)AlbCre+, n = 8. For 7-week-old mice, ob/ob-PPARγ(fl/fl)AlbCre–, n = 11; ob/ob-PPARγ(fl/fl)AlbCre+, n = 6. (f–i) Hyperinsulinemic-euglycemic clamp. Measurements of (f) whole-body glucose uptake (g) suppression of basal endogenous glucose production (EGP) (h) white adipose glucose uptake, and (i) muscle glucose uptake. Male 5-week-old mice were used in this experiment. For ob/ob-PPARγ(fl/fl)AlbCre– mice, n = 4; ob/ob-PPARγ(fl/fl)AlbCre+, n = 6. All data are mean ± SE. *P < 0.05, Cre – vs. Cre+ mice. (j–o) Effect of rosiglitazone on glucose levels (j and l), glucose tolerance (k), insulin levels (m), FFA (n), and TG (o) in ob/ob mice. The legend to Figure 2l describes the conditions for rosiglitazone treatment in the glucose analysis studies (j). For rosiglitazone-treated mice: ob/ob-PPARγ(fl/fl)AlbCre–, n = 11; ob/ob-PPARγ(fl/fl)AlbCre+, n = 4. For control mice: ob/ob-PPARγ(fl/fl)AlbCre–, n = 10; ob/ob-PPARγ(fl/fl)AlbCre+, n = 4. All data are mean ± SE. *P < 0.05, **P < 0.01, ***P < 0.001, rosiglitazone-treated vs. control Cre– mice. †P < 0.05, ††P < 0.01, †††P < 0.001, rosiglitazone-treated vs. control Cre+ mice. Rosi, rosiglitazone; Cont, control.
No difference in insulin levels was observed between ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)Cre+ mice (Figure 5e), suggesting that exacerbated hyperglycemia and glucose intolerance of liver PPARγ–deficient mice may be caused by further impairment in insulin sensitivity. To directly measure the changes in insulin sensitivity in ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ mice, a hyperinsulinemic-euglycemic clamp study was performed (Figure 5, f–i). Whole-body glucose uptake (g) under hyperinsulinemic conditions is largely considered a measure of muscle insulin sensitivity. This variable was approximately 40% higher in ob/ob-PPARγAlbCre– mice than in ob/ob-PPARγ(fl/fl)AlbCre+ mice, indicating a worsening of muscle insulin sensitivity in ob/ob-PPARγ(fl/fl)AlbCre+ mice. The measurement of tissue glucose uptake revealed that white adipose (h) and muscle (i) glucose uptake was also lower in ob/ob-PPARγ(fl/fl)AlbCre+ mice than in ob/ob-PPARγ(fl/fl)AlbCre– mice. In contrast, ob/ob-PPARγ(fl/fl)AlbCre+ mice had more pronounced suppression of endogenous glucose production during the clamp, suggesting that liver insulin sensitivity is improved by the deficiency of hepatic PPARγ.
Rosiglitazone improves the plasma diabetic syndromes in hepatic PPARγ–deficient mice.
Finally, we examined the effect of rosiglitazone on symptoms of diabetes aggravated in hepatic PPARγ–deficient mice. The glucose and TG levels were elevated and the glucose intolerance observed in ob/ob-PPARγ(fl/fl)AlbCre+ was significantly improved by treating with rosiglitazone (Figure 5, j–l and o). Treating with rosiglitazone showed a tendency to lower insulin and FFA levels, though this did not reach statistical significance (Figure 5, m and n). These results suggest that the effect of rosiglitazone on the symptoms of diabetes aggravated in hepatic PPARγ–deficient mice does not solely depend on hepatic PPARγ.
Discussion
Deficiency of liver-specific PPARγ in ob/ob mice.
Southern blot analysis of tissue DNA from ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ mice revealed over 80% deletion in the Cre+ liver, with no indication of recombination in other tissues. The detection of alleles that were not recombined in PPARγ(fl/fl)AlbCre+ mice is similar to results of Southern blot analysis in AlbCre+ mice generated in other studies (22, 29). This failure to recombine is believed to result from the existence of nonparenchymal cell types in the liver that do not express the albumin promoter and hence do not express Cre recombinase (29). It is known that the content of nonparenchymal cells in rat liver is approximately 30% of total cells (32). Indeed, quantitation of Southern blot signals revealed that the band representing the unrecombined floxed allele band (10 kb) in ob/ob-PPARγ(fl/fl)AlbCre+ mouse liver represents 29.2% ± 1.1% (n = 5) of the total alleles (10 kb + 8 kb), a result that is in agreement with parenchymal cell–specific deletion mediated by the AlbCre transgene. RNase protection further revealed that PPARγ mRNA in PPARγ(fl/fl)AlbCre+ mice is less than 5% that found in livers of PPARγ(fl/fl)AlbCre– mice. Although truncated transcripts apparently derived from the recombined allele are detected, PPARγ proteins are not expected to be expressed since these transcripts have a reading frame that results in a new stop codon at amino acids 87 (PPARγ1) or 117 (PPARγ2). Bands corresponding to the expected translation products of PPARγ mRNA lacking exon 2, which are approximately 10 kDa (PPARγ1) and 13 kDa (PPARγ2), did not appear on Western blots, perhaps due to instability of the prematurely truncated proteins (4).
Hepatic PPARγ is critical for the development of fatty liver in ob/ob mice.
The mechanisms for hepatic steatosis in ob/ob mice are not well understood, but some evidence indicates that elevation of hepatic lipogenesis is involved (33). The expression of genes encoding enzymes involved in lipogenesis — FAS, ACC, MAL, SCD1, GPAT, and ACL — is increased in ob/ob liver (34). In fact, de novo fatty acid synthesis activity measured using 3H2O in ob/ob liver is eightfold higher than in lean mice (34).
In the present study, hepatic PPARγ was found to have a role in development of fatty liver in ob/ob mice. The results of Northern blot analysis showed that expression of the lipogenesis genes induced in ob/ob-PPARγ(fl/fl)AlbCre– mice was attenuated in ob/ob-PPARγ(fl/fl)AlbCre+ mice. The expression levels of these genes in ob/ob-PPARγ(fl/fl)AlbCre+ mice returned to the basal levels found in OB/OB-PPARγ(fl/fl)AlbCre– mice. These results indicate that hepatic PPARγ may modulate the amount of hepatic TG by regulating the expression levels of lipogenic genes. Rosiglitazone is representative of a class of antidiabetic agents that act by increasing insulin sensitivity (35); these drugs are also agonist ligands for PPARγ (36). Rosiglitazone treatment of ob/ob-PPARγ(fl/fl)AlbCre– mice caused exacerbation of the development of fatty liver and increased liver size; this was not observed in Cre+ mice, indicating a correlation between the activation of hepatic PPARγ and the development of fatty liver. Furthermore, rosiglitazone induced the expression of the ACC, GPAT, CD36, and aP2 genes in ob/ob-PPARγ(fl/fl)AlbCre– mice compared with untreated ob/ob-PPARγ(fl/fl)AlbCre– mice. Therefore, the elevation of fatty acid and TG synthesis, uptake of fatty acids, and lipid accumulation in liver may be included in possible mechanisms that result in more severe fatty liver in rosiglitazone-treated ob/ob-PPARγ(fl/fl)AlbCre– mice.
It should be noted that no obvious phenotypes were observed for OB/OB-PPARγ(fl/fl)AlbCre+ mice. However, the mice were not observed beyond 13 weeks of age. Therefore we cannot rule out the existence of a small, subtle phenotype yet undetected in these mice. Perhaps older mice or mice on special (high-fat) diets may cause more noticeable phenotypes to emerge.
SREBP1 proteins are basic helix-loop-helix leucine zipper family transcription factors that mainly regulate the expression of lipogenesis genes, including FAS, ACC, MAL, SCD1, GPAT, and ACL (37). Overexpression of SREBP1 in transgenic mice results in fatty liver. SREBP1-null mice on the ob/ob genetic background (lepob/ob × Srebp1–/– mice) were produced and fatty livers were markedly attenuated (38). Similar to ob/ob-PPARγ(fl/fl)AlbCre+ mice, the mRNA levels of lipogenic enzymes such as FAS, SCD1, GPAT, S14, and ACL were decreased in lepob/ob × Srebp1–/– mice. These results indicate that SREBP1 is also an important factor for the development of fatty liver in ob/ob mice.
Regulation of lipogenesis genes by hepatic PPARγ.
To our knowledge, no prior study has directly shown that PPARγ regulates the expression of the FAS, ACC, MAL, SCD1, GPAT, and ACL genes in vivo. It remains unclear whether PPARγ directly or indirectly regulates the transcription of these genes. It was shown that MAL (39) and SCD1 (40) are regulated by PPARα and have the peroxisome proliferator response element–like (PPRE-like) sequence in their promoter regions. Transfection analysis revealed a PPRE located at position –664 to –642 bp of the SCD1 promoter, suggesting that PPARγ can directly bind to a PPRE in the promoter of this gene. However, this raises questions about why PPARγ can specially regulate the expression of these genes when PPARα is predominantly expressed in liver of ob/ob-PPARγ(fl/fl)AlbCre+ and ob/ob-PPARγ(fl/fl)AlbCre– mice. Treatment of wild-type and PPARα-null mice with the highly selective PPARγ agonist rosiglitazone induces acyl-CoA oxidase, fatty acid–binding protein, and CYP4A mRNA’s, all known PPARα target genes (41). These results indicate that although expression of PPARγ in liver is lower than PPARα, it appears that residual PPARγ is capable of mimicking PPARα function with activation by potent agonist.
PPARγ is known to activate the expression of LXRα in macrophages (4, 5). Furthermore, SREBP1c expression is markedly increased in an LXR-dependent manner (42, 43). Therefore, Northern blotting was performed to elucidate whether LXRα, SREBP1, and PPARα are mediating some of the PPARγ-dependent phenotypes in liver. However, no difference in expression between ob/ob-PPARγ(fl/fl)AlbCre– and Cre+ mice was observed for the SREBP1, LXRα, or PPARα genes, suggesting that, at least at the mRNA level, these genes are not associated with the phenotypes obtained with the PPARγ liver-null ob/ob mice. Interestingly, SREBP1 mRNA in rosiglitazone-treated ob/ob mice were elevated in a PPARγ-independent manner. These results raise the possibility that rosiglitazone modulates the expression of these transcriptional factors through non–PPARγ-dependent pathways.
Elevation of serum TG and FFA levels in hepatic PPARγ–deficient mice.
Levels of TG and FFA in ob/ob-PPARγ(fl/fl)AlbCre+ mice were significantly higher than in ob/ob-PPARγ(fl/fl)AlbCre– mice. High levels of serum TG and FFA appear to be derived from elevated VLDL in blood. In addition, ob/ob-PPARγ(fl/fl)AlbCre+ mice have a tendency to accumulate not only VLDL but also chylomicrons as exogenous TG-rich lipoproteins.
The mechanism of elevated FFA and TG levels in hepatic PPARγ-null ob/ob mice remains to be defined. We believe the mechanism may be mediated by hormone-sensitive lipase in white adipose that results in decreased insulin sensitivity in hepatic PPARγ-null ob/ob mice. This severe insulin resistance would prevent the antilipolysis effect of insulin and may cause lipolysis in adipose, although high levels of TG are not accounted for by this mechanism.
In addition, VLDL and chylomicron accumulation in blood suggests that the hepatic PPARγ-null ob/ob mice have an impairment in TG-rich lipoprotein clearance. These results raise the possibility that the deficiency of hepatic PPARγ indirectly causes the decrease of lipoprotein lipase activity, which has a crucial role in the catabolism of TG-rich lipoproteins. However, no significant differences in lipoprotein lipase activity were observed, although hepatic lipase activity was significantly increased in ob/ob-PPARγ(fl/fl)AlbCre+ mice. Hepatic lipase elevation in ob/ob-PPARγ(fl/fl)AlbCre+ mice does not appear to significantly affect TG and FFA levels because hepatic lipase preferentially hydrolyzes TG from HDL and IDL/LDL, and not from VLDL (44). Even if hepatic lipase contributes to the catabolism of TG-rich lipoproteins, the increased activity will generate an opposite situation from the high TG level observed in ob/ob-PPARγ(fl/fl)AlbCre+ mice. Therefore, the low TG clearance rate may result from impaired uptake. We could not observe a difference in expression of LDL receptor, scavenger receptor class B type I, or apoE between ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ liver. However, we cannot rule out other factors that mediate the uptake of lipoproteins and their remnants, including TG. Further studies are needed to determine the mechanism for elevation of TG and FFA in ob/ob-PPARγ(fl/fl)AlbCre+ mice.
In lepob/ob × Srebp1–/– mice, plasma cholesterol levels were decreased (38), while the plasma cholesterol levels in ob/ob-PPARγ–null mice were increased compared with ob/ob mice. It is known that SREBP1 positively regulates expression of the LDL receptor through sterol regulatory element-1 in the promoter region (45). However, SREBP1-null mice had decreased plasma cholesterol levels (46) like SREBP1-deficient ob/ob mice. It is thought that SREBP2, which is increased by a deficiency in SREBP1, compensates for the loss of SREBP1 (46). Therefore, SREBP1-deficient mice achieve decreased cholesterol levels by mediating uptake through increased LDL receptor. The same mechanism could account for the phenotype of lepob/ob × Srebp1–/– mice. Contrary to the SREBP1 study, we did not observe a difference in expression of LDL receptor mRNA between ob/ob-PPARγ(fl/fl)AlbCre– and ob/ob-PPARγ(fl/fl)AlbCre+ mice (data not shown).
Deficiency of liver-specific PPARγ improves liver insulin sensitivity but further aggravates hyperglycemia and muscle and fat insulin resistance.
The ob/ob mouse is a valuable model for insulin resistance and type 2 diabetes. The relationship between PPARγ and insulin sensitivity is highly controversial (47, 48). In our studies, the deficiency of hepatic PPARγ aggravated the hyperglycemia and glucose intolerance in ob/ob mice. This result was surprising, since reduction in hepatic steatosis would be expected to increase liver insulin sensitivity and therefore improve glycemic control. The clamp study revealed that while liver insulin sensitivity in PPARγ-deficient ob/ob mice was relatively improved, fat and muscle insulin resistance was, to the contrary, further aggravated.
The mechanism of how hepatic PPARγ influences glucose clearance or insulin resistance is likely complex. One possible factor is high lipids in the blood. In agreement with a role for systemic FFA in the development of type 2 diabetes, it was shown that elevation of plasma FFA induces peripheral insulin resistance in humans and in rodent models within a few hours (49, 50). In addition, it was shown that FFA can have positive or negative effects on insulin secretion, depending on the experimental conditions used (51, 52). Thus, FFA might have a direct impact on glucose homeostasis via systemic insulin sensitivity and possibly through effects on insulin secretion.
The basal endogenous glucose production in PPARγ-null ob/ob mice was significantly higher than in ob/ob mice (92.75 ± 2.78 μmol/kg/min vs. 128.67 ± 6.36 μmol/kg/min, P < 0.01). However, endogenous glucose production does not always correlate well with liver insulin sensitivity as measured under standardized clamp conditions with comparable insulinemia and glycemia. PPARγ deletion “protects” liver against lipid overload, thus blunting the negative effects of circulating or hepatic lipids on liver insulin sensitivity. This leads to improvement in liver insulin sensitivity, i.e., relatively greater suppression of endogenous glucose production under clamp (hyperinsulinemic) conditions. This is in agreement with other studies showing that tissue lipid levels (53), rather than circulating lipid levels, is the principal determinant of tissue insulin sensitivity.
It was demonstrated that PPARγ+/– mice were protected from the development of high fat diet–induced insulin resistance (7). After 15 weeks on a high-fat diet, blood glucose levels tended to be lower in PPARγ+/– mice, and in a glucose tolerance test, the response of PPARγ+/– mice was lower than in the wild type. In the present report, deficiency of liver-specific PPARγ showed an opposite phenotype with respect to glucose metabolism. However, it should be noted that the mice generated in the present study and by earlier work (7) differ in at least two points. The ob/ob-PPARγ(fl/fl)AlbCre+ mice are deficient in liver-specific PPARγ and do not have functional leptin. Therefore, the data obtained with the two mouse lines are not directly comparable.
The mechanism of how hepatic PPARγ is induced in ob/ob mouse liver remains elusive. It is known that the expression of hepatic PPARγ is increased in some obese and diabetic model mice (17–21). These mice have phenotypes common to diabetic syndromes. This raises the possibility that longer periods of exposure to high insulin or glucose induces PPARγ in liver. In vitro studies have demonstrated that insulin induces PPARγ expression (54). Furthermore, a recent report revealed that PPARγ is not expressed in fatty livers induced by starvation or by choline-deficient diets (55). These observations clearly demonstrate that hepatic fatty change itself is not enough to induce PPARγ. Further experimentation is needed in order to determine the mechanism by which PPARγ is activated in ob/ob mouse liver.
In summary, liver-specific disruption of PPARγ in diabetic mice dramatically decreased hepatic TG and systemically aggravated insulin resistance. However, the effect of rosiglitazone on glucose levels did not depend on hepatic PPARγ. Results in this report and other reports (18) suggest that adipose tissue is required for the antihyperglycemic effect of rosiglitazone.
Acknowledgments
We are grateful to Christopher J. Nicol, Taro E. Akiyama, Linda G. Byrd, and Shioko Kimura for their helpful suggestions. K. Matsusue was supported by a postdoctoral fellowship from the Japanese Society for the Promotion of Science.
Footnotes
Martin Haluzik’s present address is: Department of Medicine, Faculty of Medicine, Charles University, Prague, Czech Republic.
Gilles Lambert’s present address is:INSERMU539, Nantes, France.
Marc L. Reitman’s present address is: Merck Research Laboratories, Rahway, New Jersey, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Cre recombinase under control of the albumin promoter (AlbCre); malic enzyme (MAL); ATP-citrate lyase (ACL); glycerol-3-phosphate acyltransferase (GPAT); triglyceride (TG); fatty acid synthase (FAS); acetyl-CoA carboxylase (ACC); stearoyl-CoA desaturase-1 (SCD1); liver X receptor (LXR); peroxisome proliferator response element (PPRE).
References
- 1.Hihi AK, Michalik L, Wahli W. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell. Mol. Life Sci. 2002;59:790–798. doi: 10.1007/s00018-002-8467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bocher V, Pineda-Torra I, Fruchart JC, Staels B. PPARs: transcription factors controlling lipid and lipoprotein metabolism. Ann. N. Y. Acad. Sci. 2002;967:7–18. doi: 10.1111/j.1749-6632.2002.tb04258.x. [DOI] [PubMed] [Google Scholar]
- 3.Rosen ED, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akiyama TE, et al. Conditional disruption of the peroxisome proliferator-activated receptor gamma gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol. Cell. Biol. 2002;22:2607–2619. doi: 10.1128/MCB.22.8.2607-2619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawla A, et al. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 6.Barak Y, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 7.Kubota N, et al. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 8.Rosen ED, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 9.Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 10.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre AM, et al. Activation of the peroxisome proliferator-activated receptor gamma promotes the development of colon tumors in C57BL/6J-APCMin/+ mice. Nat. Med. 1998;4:1053–1057. doi: 10.1038/2036. [DOI] [PubMed] [Google Scholar]
- 12.Saez E, et al. Activators of the nuclear receptor PPARgamma enhance colon polyp formation. Nat. Med. 1998;4:1058–1061. doi: 10.1038/2042. [DOI] [PubMed] [Google Scholar]
- 13.Seed B. PPARgamma and colorectal carcinoma: conflicts in a nuclear family. Nat. Med. 1998;4:1004–1005. doi: 10.1038/1990. [DOI] [PubMed] [Google Scholar]
- 14.Moore KJ, Fitzgerald ML, Freeman MW. Peroxisome proliferator-activated receptors in macrophage biology: friend or foe? Curr. Opin. Lipidol. 2001;12:519–527. doi: 10.1097/00041433-200110000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Fajas L, et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J. Biol. Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 16.Peters JM, Rusyn I, Rose ML, Gonzalez FJ, Thurman RG. Peroxisome proliferator-activated receptor alpha is restricted to hepatic parenchymal cells, not Kupffer cells: implications for the mechanism of action of peroxisome proliferators in hepatocarcinogenesis. Carcinogenesis. 2000;21:823–826. doi: 10.1093/carcin/21.4.823. [DOI] [PubMed] [Google Scholar]
- 17.Burant CF, et al. Troglitazone action is independent of adipose tissue. J. Clin. Invest. 1997;100:2900–2908. doi: 10.1172/JCI119839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao L, et al. Adipose tissue is required for the antidiabetic, but not for the hypolipidemic, effect of thiazolidinediones. J. Clin. Invest. 2000;106:1221–1228. doi: 10.1172/JCI11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memon RA, et al. Up-regulation of peroxisome proliferator-activated receptors (PPAR-alpha) and PPAR-gamma messenger ribonucleic acid expression in the liver in murine obesity: troglitazone induces expression of PPAR-gamma-responsive adipose tissue-specific genes in the liver of obese diabetic mice. Endocrinology. 2000;141:4021–4031. doi: 10.1210/endo.141.11.7771. [DOI] [PubMed] [Google Scholar]
- 20.Rahimian R, et al. Hepatic over-expression of peroxisome proliferator activated receptor gamma2 in the ob/ob mouse model of non-insulin dependent diabetes mellitus. Mol. Cell. Biochem. 2001;224:29–37. doi: 10.1023/a:1011927113563. [DOI] [PubMed] [Google Scholar]
- 21.Bedoucha M, Atzpodien E, Boelsterli UA. Diabetic KKAy mice exhibit increased hepatic PPARgamma1 gene expression and develop hepatic steatosis upon chronic treatment with antidiabetic thiazolidinediones. J. Hepatol. 2001;35:17–23. doi: 10.1016/s0168-8278(01)00066-6. [DOI] [PubMed] [Google Scholar]
- 22.Yakar S, et al. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Iverius PH, Brunzell JD. Human adipose tissue lipoprotein lipase: changes with feeding and relation to postheparin plasma enzyme. Am. J. Physiol. 1985;249:E107–E114. doi: 10.1152/ajpendo.1985.249.1.E107. [DOI] [PubMed] [Google Scholar]
- 26.Vaisman BL, et al. ABCA1 overexpression leads to hyperalphalipoproteinemia and increased biliary cholesterol excretion in transgenic mice. J. Clin. Invest. 2001;108:303–309. doi:10.1172/JCI200112517. doi: 10.1172/JCI12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haluzik M, et al. Adrenalectomy improves diabetes in A-ZIP/F-1 lipoatrophic mice by increasing both liver and muscle insulin sensitivity. Diabetes. 2002;51:2113–2118. doi: 10.2337/diabetes.51.7.2113. [DOI] [PubMed] [Google Scholar]
- 28.Peters JM, Rusyn I, Rose ML, Gonzalez FJ, Thurman RG. Peroxisome proliferator-activated receptor alpha is restricted to hepatic parenchymal cells, not Kupffer cells: implications for the mechanism of action of peroxisome proliferators in hepatocarcinogenesis. Carcinogenesis. 2000;21:823–826. doi: 10.1093/carcin/21.4.823. [DOI] [PubMed] [Google Scholar]
- 29.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 30.Davies GF, McFie PJ, Khandelwal RL, Roesler WJ. Unique ability of troglitazone to up-regulate peroxisome proliferator-activated receptor-gamma expression in hepatocytes. J. Pharmacol. Exp. Ther. 2002;300:72–77. doi: 10.1124/jpet.300.1.72. [DOI] [PubMed] [Google Scholar]
- 31.Liang CP, Tall AR. Transcriptional profiling reveals global defects in energy metabolism, lipoprotein, and bile acid synthesis and transport with reversal by leptin treatment in ob/ob mouse liver. J. Biol. Chem. 2001;276:49066–49076. doi: 10.1074/jbc.M107250200. [DOI] [PubMed] [Google Scholar]
- 32.Drochmans P, Wanson JC, Mosselmans R. Isolation and subfractionation on ficoll gradients of adult rat hepatocytes. Size, morphology, and biochemical characteristics of cell fractions. J. Cell Biol. 1975;66:1–22. doi: 10.1083/jcb.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishida K, et al. Enhancement of the aquaporin adipose gene expression by a peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 2001;276:48572–48579. doi: 10.1074/jbc.M108213200. [DOI] [PubMed] [Google Scholar]
- 34.Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 35.Zinman B. PPAR gamma agonists in type 2 diabetes: how far have we come in ‘preventing the inevitable’? A review of the metabolic effects of rosiglitazone. Diabetes Obes. Metab. 2001;3(Suppl. 1):S34–S43. [PubMed] [Google Scholar]
- 36.Lehmann JM, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J. Biol. Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 37.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi:10.1172/JCI200215593. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yahagi N, et al. Absence of sterol regulatory element-binding protein-1 (SREBP-1) ameliorates fatty livers but not obesity or insulin resistance in Lep(ob)/Lep(ob) mice. J. Biol. Chem. 2002;277:19353–19357. doi: 10.1074/jbc.M201584200. [DOI] [PubMed] [Google Scholar]
- 39.Castelein H, et al. The peroxisome proliferator activated receptor regulates malic enzyme gene expression. J. Biol. Chem. 1994;269:26754–26758. [PubMed] [Google Scholar]
- 40.Miller CW, Ntambi JM. Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9443–9448. doi: 10.1073/pnas.93.18.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLuca JG, et al. Evidence for peroxisome proliferator-activated receptor (PPAR)alpha-independent peroxisome proliferation: effects of PPARgamma/delta-specific agonists in PPARalpha-null mice. Mol. Pharmacol. 2000;58:470–476. doi: 10.1124/mol.58.3.470. [DOI] [PubMed] [Google Scholar]
- 42.Yoshikawa T, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jansen H, Verhoeven AJ, Sijbrands EJ. Hepatic lipase: a pro- or anti-atherogenic protein? J. Lipid Res. 2002;43:1352–1362. doi: 10.1194/jlr.r200008-jlr200. [DOI] [PubMed] [Google Scholar]
- 45.Streicher R, et al. SREBP-1 mediates activation of the low density lipoprotein receptor promoter by insulin and insulin-like growth factor-I. J. Biol. Chem. 1996;271:7128–7133. doi: 10.1074/jbc.271.12.7128. [DOI] [PubMed] [Google Scholar]
- 46.Shimano H, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebovitz HE, Banerji MA. Insulin resistance and its treatment by thiazolidinediones. Recent Prog. Horm. Res. 2001;56:265–294. doi: 10.1210/rp.56.1.265. [DOI] [PubMed] [Google Scholar]
- 48.Picard F, Auwerx J. PPAR(gamma) and glucose homeostasis. Annu. Rev. Nutr. 2002;22:167–197. doi: 10.1146/annurev.nutr.22.010402.102808. [DOI] [PubMed] [Google Scholar]
- 49.Dresner A, et al. Effects of free fatty acids on glucose transport and IRS-1-associated phosphatidylinositol 3-kinase activity. J. Clin. Invest. 1999;103:253–259. doi: 10.1172/JCI5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roden M, et al. Mechanism of free fatty acid-induced insulin resistance in humans. J. Clin. Invest. 1996;97:2859–2865. doi: 10.1172/JCI118742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGarry JD, Dobbins RL. Fatty acids, lipotoxicity and insulin secretion. Diabetologia. 1999;42:128–138. doi: 10.1007/s001250051130. [DOI] [PubMed] [Google Scholar]
- 52.Sako Y, Grill VE. A 48-hour lipid infusion in the rat time-dependently inhibits glucose-induced insulin secretion and B cell oxidation through a process likely coupled to fatty acid oxidation. Endocrinology. 1990;127:1580–1589. doi: 10.1210/endo-127-4-1580. [DOI] [PubMed] [Google Scholar]
- 53.Hajri T, Han XX, Bonen A, Abumrad NA. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J. Clin. Invest. 2002;109:1381–1389. doi:10.1172/JCI200214596. doi: 10.1172/JCI14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rieusset J, et al. Insulin acutely regulates the expression of the peroxisome proliferator-activated receptor-gamma in human adipocytes. Diabetes. 1999;48:699–705. doi: 10.2337/diabetes.48.4.699. [DOI] [PubMed] [Google Scholar]
- 55.Yu S, et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J. Biol. Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]