Abstract
Preadipocyte factor-1 (Pref-1) is a transmembrane protein highly expressed in preadipocytes. Pref-1 expression is, however, completely abolished in adipocytes. The extracellular domain of Pref-1 undergoes two proteolytic cleavage events that generate 50 and 25 kDa soluble products. To understand the function of Pref-1, we generated transgenic mice that express the full ectodomain corresponding to the large cleavage product of Pref-1 fused to human immunoglobulin-γ constant region. Mice expressing the Pref-1/hFc transgene in adipose tissue, driven by the adipocyte fatty acid–binding protein (aP2, also known as aFABP) promoter, showed a substantial decrease in total fat pad weight. Moreover, adipose tissue from transgenic mice showed reduced expression of adipocyte markers and adipocyte-secreted factors, including leptin and adiponectin, whereas the preadipocyte marker Pref-1 was increased. Pref-1 transgenic mice with a substantial, but not complete, loss of adipose tissue exhibited hypertriglyceridemia, impaired glucose tolerance, and decreased insulin sensitivity. Mice expressing the Pref-1/hFc transgene exclusively in liver under the control of the albumin promoter also showed a decrease in adipose mass and adipocyte marker expression, suggesting an endocrine mode of action of Pref-1. These findings demonstrate the inhibition of adipogenesis by Pref-1 in vivo and the resulting impairment of adipocyte function that leads to the development of metabolic abnormalities.
Introduction
Obesity is a prevalent health hazard in industrialized countries and is closely associated with a number of pathological disorders, including diabetes, hypertension, and cardiovascular diseases. Obesity arises from in-creased size of individual adipose cells due to lipid accumulation and from increased number of adipocytes arising from differentiation of adipose precursor cells to mature adipocytes under the appropriate nutritional and hormonal cues (1–3). Recently, studies of genes expressed during adipocyte differentiation established that PPARγ and C/EBPs play a central role in adipocyte gene expression and differentiation (4, 5). Use of exogenous factors with positive or negative effects on adipogenesis has indicated that expression of PPARγ and C/EBPs, and therefore adipose differentiation, requires a combination of hormones, growth factors, and proper ECM components (6, 7). Preadipocytes, therefore, must integrate signals from the extracellular environment for differentiation to occur. Little is known about soluble factors found specifically in preadipocytes that affect maintenance of the preadipose state or modulate conversion to adipocytes (8, 9). Previously, we have cloned preadipocyte factor-1 (Pref-1) as one such factor (10).
Pref-1, a transmembrane protein encoded by the pref-1/dlk1 gene, belongs to a family of EGF-like repeat-containing proteins that include Notch/Delta/Serrate, which are involved in cell fate determination. Pref-1 is highly expressed in 3T3-L1 preadipocytes, but its expression is abolished during differentiation into adipocytes (10–12). Pref-1 expression has also been shown to increase upon the loss of adipose tissue mass produced by leptin ad-ministration or in lipodystrophic mice overexpress-ing the nuclear form of SREBP-1c in adipose tissue (13, 14). Furthermore, constitutive expression of Pref-1 in 3T3-L1 cells inhibits adipocyte differentiation, where-as forced downregulation of Pref-1 by antisense ex-pression enhances adipogenesis (10, 15). These obser-vations suggest that Pref-1 may function in the mainte-nance of the preadipose state by inhibiting preadipocyte differentiation (10–12, 15). In this regard, glucocorticoids enhance adipose differentiation partly by downregulation of Pref-1 expression (15). Recently, we found that the transmembrane form of Pref-1 is cleaved at the juxtamembrane region to generate a 50-kDa large soluble form that contains the full ectodomain of Pref-1 (11). In addition, smaller fragments are also produced due to processing that occurs at a more N-terminal region. Together, these observations suggest that Pref-1 may function either in a juxtacrine/paracrine or in an endocrine manner to inhibit adipocyte differentiation.
Apart from preadipocytes, Pref-1 is also expressed in various tissues during embryogenesis, and its expression is abolished after birth (10, 16, 17). In this regard, the gene encoding for Pref-1 has been reported recently to be an imprinted gene that is paternally expressed due to differential methylation (18–20). Given the role of imprinted genes in fetal growth and development, in general, and the expression of Pref-1 in embryonic tissues, Pref-1 may have functions beyond the regulation of adipogenesis (18–22).
Here we show the inhibitory role of the large soluble form of Pref-1 on adipogenesis by generating transgenic mice expressing the full ectodomain of Pref-1 as a human immunoglobulin-γ constant (hFc) region fusion protein. The large soluble form of Pref-1 specifically expressed in adipose tissue caused a decrease in fat pad weight, with a decrease in average adipocyte cell size, as well as in the expression of adipocyte marker genes, indicating an impaired adipogenesis. Hepatic expression of the same Pref-1/hFc transgene also produced a decrease in adipose tissue mass, suggesting the endocrine mode of Pref-1 action. Transgenic mice with a substantial reduction in adipose tissue mass showed hypertriglyceridemia, impaired glucose tolerance, and reduced insulin sensitivity, demonstrating the significance of adipose tissue mass to the development of the diabetes-like syndrome.
Methods
Transgene constructs and generation of transgenic mice.
The transgene constructs were designed for either adipose- or liver-specific expression of the soluble Pref-1/hFc fusion protein under the control of the adipocyte fatty acid–binding protein (aP2) promoter or the albumin promoter, respectively. To generate a soluble Pref-1/hFc fusion protein, a PCR product encoding the large soluble form of Pref-1 (N-terminal 301 amino acids) with the Kozak sequence at the 5′ end and EagI site at the 3′ end was ligated in frame to the RT-PCR product of the human IgGγ-1 heavy chain Fc region sequence encoding the C-terminal 235 amino acids. Then, the Pref-1/hFc fusion gene was cloned into pBluescript containing either the 5.4-kb aP2 promoter or the 2.3-kb albumin promoter/enhancer (23, 24). The transgene fragments were released by digestion, excised from an agarose gel, and purified with a QIAquick gel extraction kit (QIAGEN Inc., Valencia, California, USA). The DNA was microinjected into fertilized eggs of C57BL/6 × FVB mice. For PCR analysis of both transgenic mice, primers specific for the 3′ end of the Pref-1 cDNA (5′-CAC GAG CTG CCT GTT CAG CAG CC-3′) and the 5′ end of the human Fc cDNA sequence (5′-CTT GAC CTC AGG GTC TTC GTG-3′) were used to amplify a 254-bp fragment. Transgenic mice and their wild-type control littermates were maintained under standard temperature and lighting.
Measurements for body weight, food intake, and total body fat.
All mice were weighed at 5-day intervals from 3 to 10 weeks of age. For food intake measurement, 10- to 12-week-old transgenic mice and their wild-type littermates were individually housed. Body weight and daily food consumption were measured for 8 days. Body fat and fecal fat content were determined by the modified method of Bligh and Dyer (25).
Western blot analysis.
Serum (3 μl) from 10-week-old wild-type and transgenic mice were subjected to 10% SDS-PAGE and transferred onto nitrocellulose membranes. Endogenous Pref-1 or Pref-1/hFc fusion protein was probed with a polyclonal rabbit anti-Pref-1 Ab (1:5,000) and a goat anti-rabbit IgG Ab conjugated with peroxidase (1:5,000) was used as a secondary Ab. Pref-1/hFc fusion protein was also probed with an anti-human IgG Ab conjugated with peroxidase (1:3,000). Protein (20 μg) isolated from extracts of 12-day-old whole embryos were used for Western blotting analysis.
Tissue collection and histology.
Mice were anesthetized in a CO2 chamber and euthanized by cervical dislocation. Inguinal, renal, and epididymal or parametrial depots of white adipose tissue and interscapular brown adipose tissue were collected from 10-week-old mice. For histology, renal fat tissue was excised and fixed in Bouin’s fluid, embedded in paraffin, and sliced into 8-μm sections. Sections were stained with hematoxylin and eosin. Images of adipose tissue sections were captured and adipocyte volumes were measured from at least 300 cells by NIH image software.
Northern blot analysis and RT-PCR.
Total RNA from tissues or whole embryos was prepared using TRIzol reagent (Invitrogen Corp., San Diego, California, USA). Total RNA (15 μg) was subjected to electrophoresis through a 1.2% formaldehyde-agarose gel in 2.2 M formaldehyde, 20 mM Mops, 1 mM EDTA, and transferred to a nylon membrane (Hybond N; Amersham Pharmacia Biotech, Piscataway, New Jersey, USA). After cross-linking by UV, the membranes were hybridized with 32P-labeled cDNA probes for aP2, FAS, SCD-1, UCP-1, C/EBPα, ADSF/resistin, adiponectin, and leptin in ExpressHyb solution (CLONTECH Laboratories Inc., Palo Alto, California, USA). For RT-PCR, total RNA from epididymal or parametrial fat pads from 10-week-old mice was reverse transcribed with SuperScript II (Invitrogen Corp.). The endogenous Pref-1 cDNA was amplified using primers (forward: 5′-GCC ATC GTC TTT CTC AAC AAG TG-3′; reverse: 5′-GTA AGC ATA GGC TTC ACT CGA TTC-3′) corresponding to the 3′ noncoding region, enabling detection of all forms of endogenous Pref-1 mRNA. β-actin primers (forward, 5′-TCC TAT GTG GGT GAC GAG GC-3′; reverse, 5′-CAT GG CTG GGG TGT TGA AGG-3′) were used as internal control.
Glucose and insulin tolerance tests.
Glucose and insulin tolerance tests were performed on 10- to 12-week-old transgenic mice, and their wild-type control littermates. For glucose-tolerance test, D-glucose (2 mg/g of body weight) was intraperitoneally injected into overnight fasted mice, and glucose levels were monitored at 0, 30, 60, and 120 min after injection using an Accu-Chek glucometer (Roche Diagnostics Corp., Indianapolis, Indiana, USA). Insulin-tolerance tests were performed on mice following a 5-hour fast. Animals were injected intraperitoneally with 0.5 U/kg body weight of insulin (Eli Lilly Co., Indianapolis, Indiana, USA). Tail-blood samples were taken at time 0, 30, 60, 90, and 120 min after injection for measurement of blood glucose levels.
Measurements of serum triglyceride and insulin levels.
Serum triglycerides were analyzed with triglyceride INT 10 kit (Sigma-Aldrich, St. Louis, Missouri, USA). Insulin levels after overnight fasting were measured using a Linco Rat RIA kit. Leptin and adiponectin levels in serum were determined by RIA (Linco Research Inc., St. Charles, Missouri, USA).
Skeletal preparations.
Embryos or adult mice were skinned, eviscerated, and fixed in 90% ethanol for at least 1 week. For cartilage staining, carcasses were incubated with 0.01% alcian blue in 80% ethanol and 20% glacial acetic acid for 3 days. Then samples were rehydrated with series of ethanol, 70, 40, and 15% for 2 hours. Subsequently, mice were incubated with alizarin red in 1% KOH for 5 days for bone staining. After rinsing with 1% KOH, samples were stored in 100% glycerol.
Statistical analysis.
Data are expressed as the mean ± SEM. The statistical significance of differences in mean values between transgenic and wild-type littermates was assessed by Student t test.
Results and Discussion
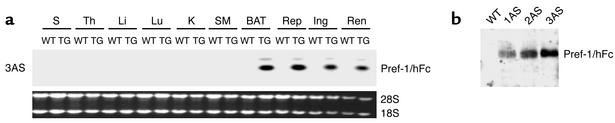
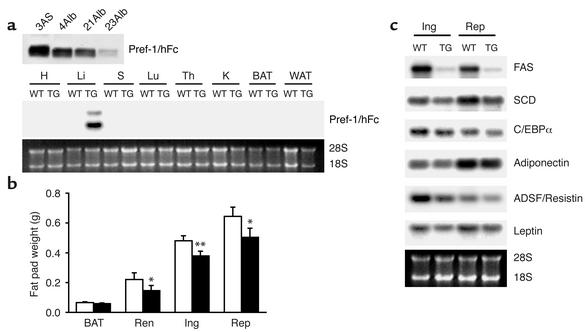
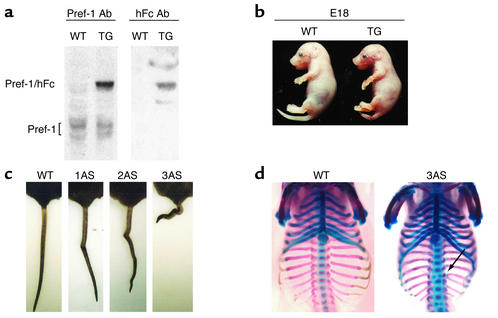
We employed the full Pref-1 ectodomain corresponding to the large soluble form of Pref-1 (10, 11, 26) as an Fc fusion protein to generate transgenic mice expressing the fusion gene in adipose tissue under the control of the aP2 promoter. Since Pref-1 has been known to function as a dimer, addition of human Fc to generate a Pref-1 fusion protein would enhance its dimerization and bioactivity (27, 28). The large soluble form of Pref-1 expressed in adipocytes in this manner may affect preadipocyte differentiation in a paracrine fashion as well as in a long-range endocrine action via the circulation. Five trans-genic founder lines expressing the transgene were generated. Northern blot analysis of various tissues from adult transgenic mice demonstrated that the aP2-Pref-1/hFc transgene transcript was expressed exclusively in adipose tissues, including inguinal, reproductive, and renal white adipose tissue, as well as in interscapular brown fat. The expression of the aP2-Pref-1/hFc transgene was not detected in any other tissue examined, including spleen, thymus, liver, lung, kidney, and muscle (Figure 1a). High levels of circulating Pref-1/hFc fusion protein were detected in three aP2-Pref-1/hFc transgenic lines (1AS, 2AS, and 3AS) (Figure 1b), whereas two other lines had barely detectable levels of plasma Pref-1/hFc. Wild-type mice, as expected, did not show any Pref-1/hFc in circulation as assessed by Western blot, using a specific Ab against hFc (Figure 1b).
Figure 1.
aP2-Pref-1/hFc transgene expression. (a) Analysis of transgene expression by Northern blot analysis in 3AS line of aP2-Pref-1/hFc mice. Total mRNA was extracted from the tissues of 10-week-old transgenic mice (TG) and wild-type littermates and probed with radiolabeled Pref-1 cDNA probe. S, spleen; Th, thymus; Li, liver; Lu, lung; K, kidney; SM, skeletal muscle; BAT, brown adipose tissue; Rep, reproductive fat pad; Ing, inguinal fat pad; Ren, renal fat pad. (b) Western blot analysis for Pref-1/hFc fusion protein in serum of three lines (1AS, 2AS, and 3AS) of aP2-Pref-1/hFc transgenic mice. The serum proteins were separated by SDS-PAGE and probed with human Fc antibody. The antibody detected the 75-kDa of Pref-1/hFc fusion protein in three founder lines of transgenic but not in wild-type mice.
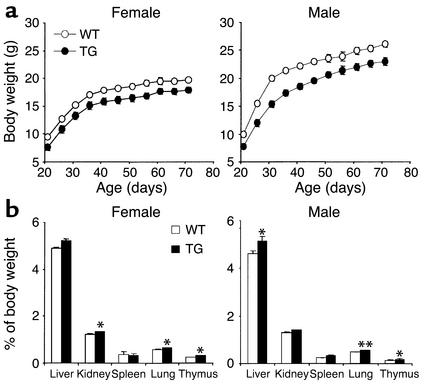
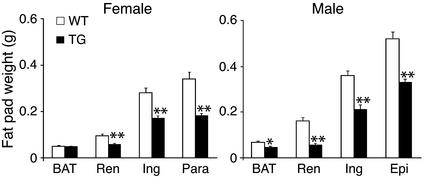
At 3 weeks of age, 3AS aP2-Pref-1/hFc transgenic mice, expressing the highest levels of circulating Pref-1/hFc, had lower body weight than their wild-type littermates (Figure 2a). The body weight of the 3AS aP2-Pref-1/hFc transgenic mice was lower than that of wild-type mice throughout life up to 10 weeks of age (P < 0.01) (Figure 2a). We observed no significant changes in food intake (relative to body weight) or in fat absorption as monitored by daily food consumption and fecal lipid content (data not shown). Despite the lower body weight, the mass of various organs, expressed as a percentage of body weight, was larger in the transgenic mice (Figure 2b). We then investigated whether the lower body weight of the transgenic mice was due to a decrease in fat pad mass. The 3AS transgenic mice had smaller inguinal, renal, and epididymal/parametrial fat pad weight at 10 weeks of age compared with wild-type littermates (in males, 42, 75, and 34%, respectively; in females 38, 39, and 50%, respectively; P < 0.01; Figure 3). In the midlevel expresser line (2AS), inguinal, renal, and reproductive fat pad weight was reduced to a lesser extent (in males, 26, 57, 25%, respectively, n = 5–6, P < 0.05; in females, 18, 27, and 38%, respectively, n = 5–8, P < 0.05). This indicates that the magnitude of decrease in white fat pad weight was positively correlated with the levels of transgene expression. We also observed a significant 36% reduction in interscapular brown adipose tissue in 3AS male transgenic mice, although the decrease was not obvious in the midlevel expresser line 2AS. Total body lipid content was also measured to examine whether the reduced proportion of fat pad mass was reflected in the total lipid content of the animals. The 3AS transgenic mice had less total body lipid (1.82 ± 0.08 g) than their wild-type littermates (2.86 ± 0.16 g, P < 0.01). The 36% reduction of total body lipid content in transgenic mice was similar to the decrease in fat pad weight described above, indicating that fat content in other tissues probably did not change significantly, and the decrease in total body fat can be attributed mainly to a reduction of adipose tissue mass.
Figure 2.
Growth curve and organ weight of 3AS aP2-Pref-1/hFc transgenic mice. (a) Growth curves for male and female wild-type (open circles) and transgenic (filled circles) (TG) mice fed a chow diet. Body weight of mice measured at 5-day intervals is shown; each point represents mean ± SEM from 6 to 13 mice. Body weight of Pref-1/hFc transgenic mice was significantly lower (P < 0.01) than those of wild-type mice at all ages. (b) Organ weights of 10-week-old mice are presented as percentage of body weight (n = 8–10 per group). Statistically significant differences between the groups are indicated as *P < 0.05 and **P < 0.01.
Figure 3.
Reduced fat pad weight in 3AS aP2-Pref-1/hFc transgenic mice. Fat depot weights from 10-week-old mice are presented (n = 6 per group). BAT, brown adipose tissue; Renal, renal fat pad; Ing, inguinal fat pad; Para, parametrial fat pad; Epi, epididymal fat pad. Statistically significant differences between the groups are indicated as *P < 0.05 and **P < 0.01.
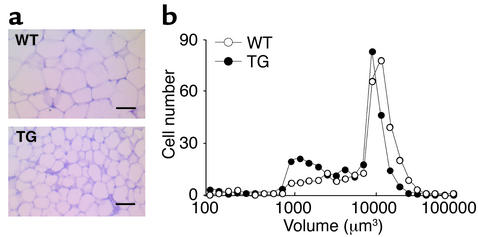
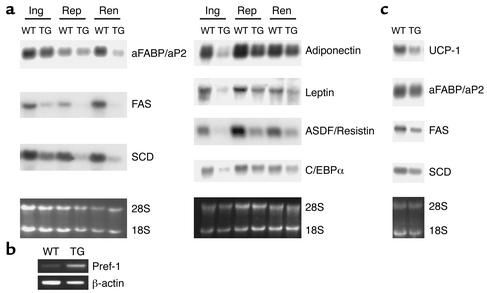
A decrease in adipose tissue mass can result from a reduction in adipocyte cell size and/or a decrease in adipocyte cell number due to an impairment of preadipocytes’ differentiation into adipocytes. We first measured the total amount of DNA in fat pads to establish whether decreased fat pad weight of transgenic mice was accompanied by a decreased number of cells. The total DNA content of adipose tissue did not differ between wild-type and transgenic mice (data not shown). Histological analysis of renal fat pads from the 3AS transgenic mice and wild-type littermates at 10 weeks of age revealed that adipocytes from transgenic mice were markedly smaller than those from wild-type mice (Figure 4a). Regarding the distribution of renal adipocyte volume, transgenic mice showed a shift of peaks down to smaller sizes, around 104 μm3 and 1 to 4 × 103 μm3 (Figure 4b). Adipose cell differentiation involves changes in expression of genes that result in the acquisition of fat cell phenotype and adipocyte function. Not only specialized in triglyceride storage, mature adipocytes are characterized by the ability to express and secret a wide range of molecules involved in many physiological processes, including energy homeostasis. Northern blot analysis showed that aP2-Pref-1/hFc transgenic mice had decreased expression of transcription factors known to be involved in adipogenesis, such as C/EBPα, as well as other adipocyte markers, such as aP2, FAS (fatty acid synthase), and SCD (stearoyl-CoA desaturase1), in the three different fat pads examined (Figure 5a). The expression of factors secreted by adipocytes, including leptin, adiponectin, and ADSF/resistin was also decreased. In contrast, expression of endogenous Pref-1, a preadipocyte marker (12, 13), was increased in Pref-1/hFc transgenic mice as detected by RT-PCR (Figure 5b). Moreover, interscapular brown adipose tissue also exhibited a decrease in expression of aP2, FAS, and SCD, as well as the brown adipocyte marker UCP-1 (uncoupling protein 1) (Figure 5c). Taken together, these observations suggest that the decrease in fat mass in aP2-Pref-1/hFc transgenic mice reflects an impairment of adipocyte differentiation by Pref-1/hFc in all adipose tissue pads, including brown fat. The inhibition of adipogenesis and the consequent decrease in fat mass in these transgenic mice agree with our previous in vitro studies on Pref-1 in which constitutive expression of Pref-1 leads to inhibition of 3T3-L1 adipocyte differentiation, whereas antisense Pref-1 enhances adipocyte differentiation (10, 15). Furthermore, the effectiveness of the large extracellular domain of Pref-1 in transgenic mice led us to conclude that the large soluble Pref-1 is sufficient to inhibit adipocyte differentiation. In agreement with these results, we have reported recently that only the large, soluble form of Pref-1 inhibits 3T3-L1 differentiation in vitro (29).
Figure 4.
Histological analysis of adipose tissue and cell size distribution. (a) Paraffin-embedded sections of renal white adipose tissue from 10-week-old male mice were stained with hematoxylin and eosin. Scale bar, 50 μm. (b) Distribution of the adipocyte volume in wild-type and aP2-Pref-1/hFc transgenic mice. The volume of at least 300 cells per sample (mean of four mice per group) was determined with the NIH image software.
Figure 5.
Gene expression in adipose tissue from aP2-Pref-1/hFc transgenic mice. (a) Northern blot analysis of adipocyte marker expression in white adipose tissue of wild-type and 3AS aP2-Pref-1/hFc transgenic mice. Total RNA from three different fat pads (Ing, inguinal; Rep, reproductive; Ren, renal) was probed with cDNA probes for different adipocyte markers. (b) Endogenous Pref-1 expression analyzed by RT-PCR. A primer set was designed to amplify only the endogenous Pref-1 message (see Methods). β-actin was used as an internal control. (c) Northern blot analysis for adipocyte markers in interscapular brown adipose tissue.
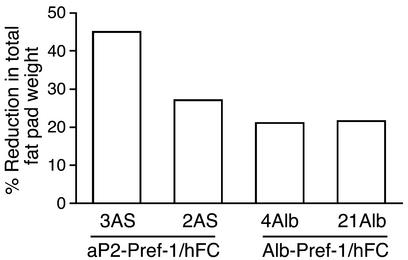
Since the aP2 promoter was employed to express the transgene in adipose tissue, the high levels of Pref-1/hFc fusion protein secreted in adipose tissue and the amounts present in the circulation of transgenic mice may have affected differentiation of preadi-pocytes to adipocytes in a paracrine or in an endo-crine manner, respectively. We therefore generated transgenic mice expressing the Pref-1/hFc fusion gene under the control of the albumin promoter. Three different Alb-Pref-1/hFc transgenic lines (4Alb, 21Alb, and 23Alb) expressing the transgene at different levels were obtained (Figure 6a, upper panel). Northern blot analysis revealed that the transgene was ex-pressed exclusively in the liver (Figure 6a, lower panel). As shown by Western blot analysis, hepatic expression of the Pref-1/hFc transgene produced a soluble form of the Pref-1/hFc fusion protein in circulation (Figure 6a, upper panel). Transgenic lines 4Alb and 21Alb, which express the highest levels of the transgene, were analyzed to investigate whether the soluble Pref-1/hFc secreted by the liver was able to affect adipose tissue development. The circulating levels of Pref-1/hFc fusion protein in Alb-Pref-1/hFc mice was similar to that of the 2AS aP2-Pref-1/hFc mice and was approximately 50% of that seen in the 3AS aP2-Pref-1/hFc line (Figures 6a and 1b). Compared with wild-type littermates, 21Alb transgenic mice did not show any change in body weight at birth, but did show a slight tendency to gain less weight during the study period, although the differences were not statistically significant (data not shown). The 21Alb Alb-Pref-1/hFc transgenic mice had smaller fat pad weight when compared with wild-type littermates, with 10- to 14-week-old mice showing significant 33.1, 21.2, and 24.1% reductions in renal, inguinal, and reproductive fat pad weight, respectively (Figure 6b). Moreover, a 12% reduction in brown adipose tissue was detected, although due to the variability it was not statistically significant. The lower fat mass was accompanied by decreased expression of adipocyte markers, including C/EBPα, FAS, SCD, ADSF/resistin, leptin, and adiponectin, indicating an impairment of adipogenesis in Alb-Pref-1/hFc transgenic mice (Figure 6c). The 4AlbPref-1/hFc transgenic line, which expressed comparable levels of transgene to 21Alb line, was unable to transmit the transgene to the F1 generation; therefore the fat pad weight of the founder was compared with the wild-type littermate. Similar to the 21Alb transgenic line, the 4AlbPref-1/hFc founder showed a 21% reduction in total fat pad weight as compared with wild-type littermates. Northern blot analysis also revealed a reduction in the expression of adipocyte markers (data not shown). A strong correlation between the expression level of Pref-1/hFc and the degree of decrease in fat pad weight was observed in the various transgenic lines of aP2-Pref-1/hFc and Alb-Pref-1/hFc, regardless of whether the transgene was expressed in adipose tissue or liver (Figure 7). The total fad pad weight showed a 45% reduction in adipose tissue mass in 3AS aP2-Pref-1/hFc transgenic line, the highest expresser of the transgene in adipose tissue. The decrease in total fat pad weight in 2AS aP2-Pref-1/hFc, 4Alb, and 21 Alb Alb-Pref-1/hFc lines was 27, 21, and 25%, respectively. These three lines of transgenic mice, whether the transgene is expressed in adipose tissue or in liver, had similar levels of circulating Pref-1/hFc protein corresponding to approximately 50% of that observed in the 3AS aP2-Pref-1/hFC transgenic mice. The ability of hepatic expression of Pref-1/hFc to affect fat pad weight to a similar degree suggests that the large soluble form of Pref-1 may act in an endocrine manner to inhibit adipocyte differentiation. It is also interesting to note that Delta, which was originally reported to function as a membrane form of a ligand, has been demonstrated to release a soluble extracellular domain capable of binding to and acting through Notch (30, 31). However, Pref-1 lacks the DSL domain that is conserved in all Notch ligands and mediates Notch receptor-ligand interaction (10). Thus, it is unlikely that Pref-1 acts as a Notch ligand. Identification of Pref-1–interacting proteins or its receptor will make possible to understand the molecular mechanisms underly-ing Pref-1 action.
Figure 6.
Reduced fat pad weight and decreased expression of adipocyte markers in Alb-Pref-1/hFc transgenic mice expressing Pref-1/hFc specifically in liver. (a) Circulating Pref-1/hFc was detected by Western blot analysis in three different lines of Alb-Pref-1/hFc using Pref-1 antibody. Serum Pref-1/hFc levels in Alb-Pref-1/hFc mice were compared with those in 3AS aP2-Pref-1/hFc mice (upper panel). The liver-specific expression of the transgene in Alb-Pref-1/hFc mice was assessed by Northern blot analysis (lower panel). H, heart; Li, liver; S, spleen; Lu, lung; Th, thymus; K, kidney; BAT, brown adipose tissue; WAT, white adipose tissue. (b) Weight of fat pads was reduced in 21Alb-Pref-1/hFc mice (black bars) compared with wild-type littermates (white bars). Results are mean ± SEM of fat pads from nine to ten mice, and statistical significance is indicated as *P < 0.05, **P < 0.01. Ren, renal fat pad; Ing, inguinal fat pad; Rep, reproductive fat pad. (c) Northern blot analysis of adipocyte marker expression in inguinal (Ing) and reproductive (Rep) depots of white adipose tissue from 21Alb-Pref-1/hFc mice. TG, transgenic.
Figure 7.
Reduction in fat pad mass was correlated with the expression levels of Pref-1/hFc in various transgenic lines. The high expresser line 3AS aP2-Pref-1/hFc showed a 45% reduction in total fad pad weight while lines 2AS aP2-Pref-1/hFc, 21Alb and 4Alb Alb-Pref-1/hFc, which express approximately 50% of that observed in 3AS, exhibited a 27, 21, and 25% reduction in total fat pad weight, respectively.
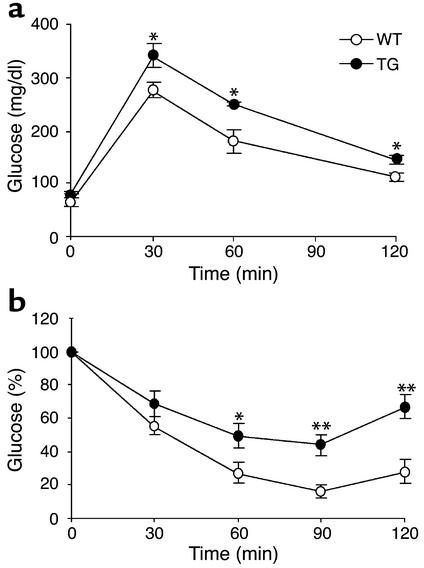
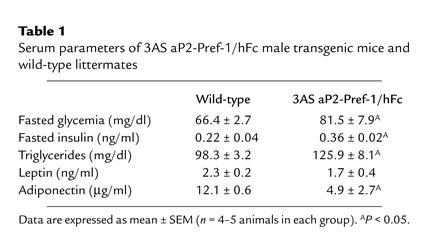
Changes in adipose tissue mass are frequently associated with alterations in glucose homeostasis. We therefore carried out insulin- and glucose-tolerance tests to determine if insulin and glucose homeostasis would be affected by ectopic expression of Pref-1. When glucose-tolerance tests were performed (Figure 8a) on the 3AS aP2-Pref-1/hFc transgenic line, mice expressing Pref-1/hFc showed higher levels of blood glucose (Table 1), and the glucose clearance rate was slower than in wild-type mice. Insulin-tolerance tests showed that the transgenic mice had overt insulin insensitivity (Figure 8b). Levels of insulin after overnight fasting were higher in transgenic mice than in their wild-type littermates (Table 1). We also measured serum triglyceride levels, since insulin resistance, hyperglycemia, and hyperinsulinemia have all been associated with dyslipidemia (32, 33). Plasma triglyceride levels were higher in transgenic mice than in wild-type mice (Table 1). These data suggest that expression of the Pref-1/hFc transgene and the resulting decrease in adipose tissue mass has promoted development of abnormalities in glucose homeostasis and insulin sensitivity. Increased fasting insulin levels in Pref-1 transgenic mice suggest that the capacity for insulin secretion by islet β cells is probably not impaired. Severe or complete loss of fat mass in the transgenic mice overexpressing dominant negative A-ZIP/F-1 or SREBP-1c in adipose tissue have been reported to accompany hypertriglyceridemia and syndromes similar to type II diabetes (34, 35). In addition to these animal models, studies in human patients with lipodystrophy also have revealed a strong correlation between decrease in adipose tissue mass and insulin resistance (36, 37). Insulin insensitivity in 3AS Pref-1/hFc transgenic mice accompanies the substantial loss of adipose tissue, although the loss is less complete than in lipoatrophic models. It is plausible that the reduced capacity for lipid storage in adipose tissue of these mice may contribute to these metabolic syndromes. Transgenic lines showing lower expression of the Pref-1/hFc fusion protein, 2AS aP2-Pref-1/hFc and 21Alb, 4Alb Alb-Pref-1/hFc, did not show significant differences in glucose clearance rates and insulin resistance (data not shown), indicating that the reduction in adipose tissue mass in those mice was not sufficient to bring about the insulin resistance and glucose intolerance associated with lipoatrophic models. The mechanisms, however, by which changes in adipose tissue mass cause insulin resistance still remain controversial (38). An increasing number of studies suggests an important role of fat-derived hormones in the etiology of insulin resistance associated with altered adipose tissue mass. Indeed, leptin and adiponectin have been reported to reverse insulin resistance associated with lipodystrophy (39, 40). On the other hand, ADSF/resistin, a recently discovered factor secreted by adipocytes (41, 42) has been suggested, although controversially, to contribute to insulin resistance (42). In this regard, it should be noted that the 3AS aP2-Pref-1/hFc transgenic mice exhibited a marked reduction in the expression and secretion of the insulin-sensitizing hormones leptin and adipo-nectin (Figure 5a and Table 1). The reduced levels of leptin and adiponectin in serum of 3AS aP2-Pref-1/hFc transgenic mice, which correlate with the expression levels in adipose tissue, may have contributed to the observed insulin resistance. The smaller reduction in leptin and adiponectin expression shown in adipose tissue from Alb-Pref-1/hFc transgenic mice may not have been sufficient to bring about the metabolic abnormalities observed in 3AS aP2-Pref-1/hFc transgenic mice. Further studies are needed to clarify the implications of the reduced expression of leptin and adiponectin in the insulin resistance and glucose intolerance observed in our Pref-1 transgenic mice.
Figure 8.
Glucose intolerance and insulin resistance in aP2-Pref-1/hFc transgenic mice that express high levels of Pref-1/hFc fusion protein. (a) Glucose-tolerance test. Overnight-fasted mice were given an intraperitoneal injection of glucose (2 mg/g body weight). Blood samples were collected from the tail at indicated time points and analyzed for glucose concentration. Results are means ± SEM from six animals in each group. (b) Insulin tolerance test. Insulin (0.5 U/kg body weight) was intraperitoneally injected after fasting for 5 hours. Data are presented as mean percentage of 0 min glucose value ± SEM from six mice in each group. Statistically significant differences between the groups are indicated as *P < 0.05 and **P < 0.01.
Table 1.
Serum parameters of 3AS aP2-Pref-1/hFc male transgenic mice and wild-type littermates
Since we observed lower body weight in transgenic mice at weaning, we examined embryonic expres-sion of the Pref-1/hFc transgene driven by the aP2 promoter in two transgenic lines. Pref-1/hFc transgene expression was detected at embryo day 12 and was further increased at later stages (Figure 9a). Interestingly, the 3AS aP2-Pref-1/hFc transgenic embryos showed growth retardation and skeletal abnormalities, primarily in the distal vertebra. The two other aP2-Pref-1/hFc transgenic lines also showed similar skeletal abnormalities, with the severity correlated with the levels of circulating Pref-1/hFc (Figure 9, b–d). Recently, pref-1/dlk1 has been reported to be a paternally expressed gene located in a chromosomal region containing six imprinted genes (18, 19, 43, 44). Paternal monoallelic expression of pref-1/dlk1 has been observed in the syntenic chromosomes 12, 18, and 14 in mice, sheep, and humans, respectively (18–20, 43, 44). Although the gene(s) responsible for the phenotype is not known, perturbation of imprinted gene expression in human paternal uniparental disomy (pUPD) 14 and mouse (pUPD) 12 causes growth retardation and bone malformation (45–55). The similar embryonic defects observed in the Pref-1/hFc transgenic mice suggest that Pref-1 expression may play a major role in these clinical features. Adipose and metabolic defects in these UPDs, however, have not yet been reported. This may be because the degree of overexpression of Pref-1 is probably lower than that in our transgenic mice, and the expression of the endogenous Pref-1 in preadipocytes controlled by its own promoter would be downregulated during adipose development. In contrast, in callipyge (clpg) sheep a regulatory mutation in this imprinted region of chromosome 18 results in overexpression of two genes, including pref-1/dlk1. This mutation produces lean animals with hypertrophic muscle (43, 56). Given our findings of decreased adipose tissue mass in Pref-1/hFc transgenic mice, it is possible that the lean phenotype of clpg sheep is due to the antiadipogenic action of Pref-1.
Figure 9.
Growth retardation and bone malformation in aP2-Pref-1/hFc transgenic mice. (a) Pref-1 expression in aP2-Pref-1/hFc embryos. Total protein extracts from embryo day 12 (E12) embryo from wild-type and 3AS line were subjected to Western blot analysis for Pref-1. Pref-1/hFc fusion protein was detected as a 75-kDa band only in transgenic embryo, while multiple forms of endogenous Pref-1 were detected at around 50-kDa in both wild-type and transgenic mice. (b) The appearances of wild-type and aP2-Pref-1/hFc transgenic littermates were compared at E18, showing the growth retardation of transgenic embryos. (c) The transgenic mice had kinky tails, the severity of which is correlated with levels of transgene expression. (d) Bone and cartilage staining. Embryos (E17) were stained with alizarin red for bone and alcian blue for cartilages. The Pref-1/hFc transgenic embryos had a smaller thoracic cavity with short ribs. Vertebrae were fused and disorganized, resulting in scoliosis.
In conclusion, we provide evidence that forced expression of Pref-1/hFc results in decrease in fat mass, fat cell size, and inhibition of adipogenesis. Moreover, we show that the large cleavage product of Pref-1 can act in an endocrine manner to inhibit adipocyte differentiation. We also show that substantial, although not complete, loss of fat mass and impairment of adipose function due to Pref-1 overexpression causes glucose intolerance and decreased insulin sensitivity, a metabolic syndrome observed previously in lipoatrophic models.
Acknowledgments
We thank David A. Bernlohr (University of Minnesota) for the aP2 promoter and Richard D. Palmiter (University of Washington) and Snorri S. Thorgeirsson (NIH) for the albumin promoter. We thank Eun June Yoon for histology. S. Lee was supported by Faculty Research Program 2000, Cheju National University. This work was supported by NIH grant DK-50828 to H.S. Sul.
Footnotes
J.A.Villena and K. Lee contributed equally to this work.
Kichoon Lee’s present address is: Louisiana State University AgCenter, Louisiana State University, Baton Rouge, Louisiana, USA.
Yang Soo Moon’s present address is: Department of Animal Sciences and Technology, School of Agricultural Biotechnology, Seoul National University, Seoul, Republic of Korea.
Sunjoo Lee’s present address is: Department of Chemistry, Cheju National University, Republic of Korea.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: preadipocyte factor-1 (Pref-1); adipocyte fatty acid–binding promoter (aP2); human immunoglobulin-γ constant (hFc); fatty acid synthase (FAS); stearoyl-CoA desaturase1 (SCD); uncoupling protein 1 (UCP-1); paternal uniparental disomy (pUPD); callipyge (clpg).
References
- 1.Faust IM, Johnson PR, Stern JS, Hirsch J. Diet-induced adipocyte number increase in adult rats: a new model of obesity. Am. J. Physiol. 1978;235:E279–E286. doi: 10.1152/ajpendo.1978.235.3.E279. [DOI] [PubMed] [Google Scholar]
- 2.Klyde BJ, Hirsch J. Increased cellular proliferation in adipose tissue of adult rats fed a high-fat diet. J. Lipid. Res. 1979;20:705–715. [PubMed] [Google Scholar]
- 3.Klyde BJ, Hirsch J. Isotopic labeling of DNA in rat adipose tissue: evidence for proliferating cells associated with mature adipocytes. J. Lipid. Res. 1979;20:691–704. [PubMed] [Google Scholar]
- 4.Darlington GJ, Ross SE, MacDougald OA. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 5.MacDougald OA, Mandrup S. Adipogenesis: forces that tip the scales. Trends Endocrinol. Metab. 2002;13:5–11. doi: 10.1016/s1043-2760(01)00517-3. [DOI] [PubMed] [Google Scholar]
- 6.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 7.Smas CM, Sul HS. Control of adipocyte differentiation. Biochem. J. 1995;309:697–710. doi: 10.1042/bj3090697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubin CS, Hirsch A, Fung C, Rosen OM. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J. Biol. Chem. 1978;253:7570–7578. [PubMed] [Google Scholar]
- 9.Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol. Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 10.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 11.Smas CM, Chen L, Sul HS. Cleavage of membrane-associated pref-1 generates a soluble inhibitor of adipocyte differentiation. Mol. Cell. Biol. 1997;17:977–988. doi: 10.1128/mcb.17.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smas CM, et al. Transcriptional control of the pref-1 gene in 3T3-L1 adipocyte differentiation. Sequence requirement for differentiation-dependent suppression. J. Biol. Chem. 1998;273:31751–31758. doi: 10.1074/jbc.273.48.31751. [DOI] [PubMed] [Google Scholar]
- 13.Zhou YT, Wang ZW, Higa M, Newgard CB, Unger RH. Reversing adipocyte differentiation: implications for treatment of obesity. Proc. Natl. Acad. Sci. USA. 1999;96:2391–2395. doi: 10.1073/pnas.96.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimomura I, et al. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smas CM, Chen L, Zhao L, Latasa MJ, Sul HS. Transcriptional repression of pref-1 by glucocorticoids promotes 3T3-L1 adipocyte differentiation. J. Biol. Chem. 1999;274:12632–12641. doi: 10.1074/jbc.274.18.12632. [DOI] [PubMed] [Google Scholar]
- 16.Floridon C, et al. Does fetal antigen 1 (FA1) identify cells with regenerative, endocrine and neuroendocrine potentials? A study of FA1 in embryonic, fetal, and placental tissue and in maternal circulation. Differentiation. 2000;66:49–59. doi: 10.1046/j.1432-0436.2000.066001049.x. [DOI] [PubMed] [Google Scholar]
- 17.Jensen CH, et al. Neurons in the monoaminergic nuclei of the rat and human central nervous system express FA1/dlk. Neuroreport. 2001;12:3959–3963. doi: 10.1097/00001756-200112210-00021. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 19.Takada S, et al. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr. Biol. 2000;10:1135–1138. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- 20.Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10:1711–1718. doi: 10.1101/gr.161600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen CH, et al. Studies on the isolation, structural analysis and tissue localization of fetal antigen 1 and its relation to a human adrenal-specific cDNA, pG2. Hum. Reprod. 1993;8:635–641. doi: 10.1093/oxfordjournals.humrep.a138110. [DOI] [PubMed] [Google Scholar]
- 22.Laborda J, Sausville EA, Hoffman T, Notario V. dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J. Biol. Chem. 1993;268:3817–3820. [PubMed] [Google Scholar]
- 23.Heckel JL, Sandgren EP, Degen JL, Palmiter RD, Brinster RL. Neonatal bleeding in transgenic mice expressing urokinase-type plasminogen activator. Cell. 1990;62:447–456. doi: 10.1016/0092-8674(90)90010-c. [DOI] [PubMed] [Google Scholar]
- 24.Sanderson N, et al. Hepatic expression of mature transforming growth factor beta 1 in transgenic mice results in multiple tissue lesions. Proc. Natl. Acad. Sci. USA. 1995;92:2572–2576. doi: 10.1073/pnas.92.7.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartsook EW, Hershberger TV, Nee JC. Effects of dietary protein content and ratio of fat to carbohydrate calories on energy metabolism and body composition of growing rats. J. Nutr. 1973;103:167–178. doi: 10.1093/jn/103.2.167. [DOI] [PubMed] [Google Scholar]
- 26.Smas CM, Green D, Sul HS. Structural characterization and alternate splicing of the gene encoding the preadipocyte EGF-like protein pref-1. Biochemistry. 1994;33:9257–9265. doi: 10.1021/bi00197a029. [DOI] [PubMed] [Google Scholar]
- 27.Kaneta M, et al. Role for pref-1 and HES-1 in thymocyte development. J. Immunol. 2000;164:256–264. doi: 10.4049/jimmunol.164.1.256. [DOI] [PubMed] [Google Scholar]
- 28.Ohno N, Izawa A, Hattori M, Kageyama R, Sudo T. dlk inhibits stem cell factor-induced colony formation of murine hematopoietic progenitors: Hes-1-independent effect. Stem Cells. 2001;19:71–79. doi: 10.1634/stemcells.19-1-71. [DOI] [PubMed] [Google Scholar]
- 29.Mei B, Zhao L, Chen L, Sul HS. Only the large soluble form of preadipocyte factor-1 (Pref-1), but not the small soluble and membrane forms, inhibits adipocyte differentiation: role of alternative splicing. Biochem J. 2002;364:137–144. doi: 10.1042/bj3640137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi H, et al. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 31.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 32.Cullen P, von Eckardstein A, Souris S, Schulte H, Assmann G. Dyslipidaemia and cardiovascular risk in diabetes. Diabetes Obes. Metab. 1999;4:189–198. doi: 10.1046/j.1463-1326.1999.00030.x. [DOI] [PubMed] [Google Scholar]
- 33.Betteridge DJ. Diabetic dyslipidaemia. Diabetes Obes. Metab. 2000;1(Suppl.):S31–S36. doi: 10.1046/j.1463-1326.2000.00021.x. [DOI] [PubMed] [Google Scholar]
- 34.Moitra J, et al. Life without white fat: a transgenic mouse. Genes Dev. 1998;12:3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebihara K, et al. Transgenic overexpression of leptin rescues insulin resistance and diabetes in a mouse model of lipoatrophic diabetes. Diabetes. 2001;50:1440–1448. doi: 10.2337/diabetes.50.6.1440. [DOI] [PubMed] [Google Scholar]
- 36.Robbins DC, et al. The effect of diet on thermogenesis in acquired lipodystrophy. Metabolism. 1979;28:908–916. doi: 10.1016/0026-0495(79)90090-8. [DOI] [PubMed] [Google Scholar]
- 37.Robbins DC, Horton ES, Tulp O, Sims EA. Familial partial lipodystrophy: complications of obesity in the non-obese? Metabolism. 1982;31:445–452. doi: 10.1016/0026-0495(82)90232-3. [DOI] [PubMed] [Google Scholar]
- 38.Ravussin E, Smith SR. Increased fat intake, impaired fat oxidation, and failure of fat cell proliferation result in ectopic fat storage, insulin resistance, and type 2 diabetes mellitus. Ann. NY Acad. Sci. 2002;967:363–378. doi: 10.1111/j.1749-6632.2002.tb04292.x. [DOI] [PubMed] [Google Scholar]
- 39.Shimomura I, Hammer RE, Ikemoto S, Brown MS, Goldstein JL. Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature. 1999;401:73–76. doi: 10.1038/43448. [DOI] [PubMed] [Google Scholar]
- 40.Yamauchi T, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 41.Kim KH, Lee K, Moon YS, Sul HS. A cysteine-rich adipose tissue-specific secretory factor inhibits adipocyte differentiation. J. Biol. Chem. 2001;276:11252–11256. doi: 10.1074/jbc.C100028200. [DOI] [PubMed] [Google Scholar]
- 42.Steppan CM, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 43.Charlier C, et al. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat. Genet. 2001;4:367–369. doi: 10.1038/86856. [DOI] [PubMed] [Google Scholar]
- 44.Charlier C, et al. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 2001;11:850–862. doi: 10.1101/gr.172701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fokstuen S, Ginsberg C, Zachmann M, Schinzel A. Maternal uniparental disomy 14 as a cause of intrauterine growth retardation and early onset of puberty. J. Pediatr. 1999;134:689–695. doi: 10.1016/s0022-3476(99)70282-9. [DOI] [PubMed] [Google Scholar]
- 46.Georgiades P, Watkins M, Surani A, Ferguson-Smith AC. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development. 2000;127:4719–4728. doi: 10.1242/dev.127.21.4719. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi S, et al. Mouse Peg9/Dlk1 and human PEG9/DLK1 are paternally expressed imprinted genes closely located to the maternally expressed imprinted genes: mouse Meg3/Gtl2 and human MEG3. Genes Cells. 2000;5:1029–1037. doi: 10.1046/j.1365-2443.2000.00390.x. [DOI] [PubMed] [Google Scholar]
- 48.Georgiades P, Chierakul C, Ferguson-Smith AC. Parental origin effects in human trisomy for chromosome 14q: implications for genomic imprinting. J. Med. Genet. 1998;35:821–824. doi: 10.1136/jmg.35.10.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takada S, et al. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Hum. Mol. Genet. 2002;11:77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]
- 50.Sutton VR, Shaffer LG. Search for imprinted regions on chromosome 14: comparison of maternal and paternal UPD cases with cases of chromosome 14 deletion. Am. J. Med. Genet. 2000;93:381–387. doi: 10.1002/1096-8628(20000828)93:5<381::aid-ajmg7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 51.Cotter PD, et al. Paternal uniparental disomy for chromosome 14: a case report and review. Am. J. Med. Genet. 1997;70:74–79. doi: 10.1002/(sici)1096-8628(19970502)70:1<74::aid-ajmg14>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 52.Coveler KJ, et al. A case of segmental paternal isodisomy of chromosome 14. Hum. Genet. 2002;110:251–256. doi: 10.1007/s00439-002-0688-4. [DOI] [PubMed] [Google Scholar]
- 53.Wang JC, Passage MB, Yen PH, Shapiro LJ, Mohandas TK. Uniparental heterodisomy for chromosome 14 in a phenotypically abnormal familial balanced 13/14 Robertsonian translocation carrier. Am. J. Hum. Genet. 1991;48:1069–1074. [PMC free article] [PubMed] [Google Scholar]
- 54.Walter CA, et al. Short-limb dwarfism and hypertrophic cardiomyopathy in a patient with paternal isodisomy 14: 45,XY,idic(14)(p11) Am. J. Med. Genet. 1996;65:259–265. doi: 10.1002/(SICI)1096-8628(19961111)65:4<259::AID-AJMG2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 55.Papenhausen PR, et al. Uniparental isodisomy of chromosome 14 in two cases: an abnormal child and a normal adult. Am. J. Med. Genet. 1996;59:271–275. doi: 10.1002/ajmg.1320590302. [DOI] [PubMed] [Google Scholar]
- 56.Freking BA, Keele JW, Nielsen MK, Leymaster KA. Evaluation of the ovine callipyge locus: II. Genotypic effects on growth, slaughter, and carcass traits. J. Anim. Sci. 1998;76:2549–2559. doi: 10.2527/1998.76102549x. [DOI] [PubMed] [Google Scholar]