Abstract
In prokaryotic genomes, some DNA methyltransferases form a restriction-modification gene complex, but some others are present by themselves. Dcm gene product, one of these orphan methyltransferases found in Escherichia coli and related bacteria, methylates DNA to generate 5′-CmCWGG just as some of its eukaryotic homologues do. Vsr mismatch repair function of an adjacent gene prevents C-to-T mutagenesis enhanced by this methylation but promotes other types of mutation and likely has affected genome evolution. The reason for the existence of the dcm-vsr gene pair has been unclear. Earlier we found that several restriction-modification gene complexes behave selfishly in that their loss from a cell leads to cell killing through restriction attack on the genome. There is also increasing evidence for their potential mobility. EcoRII restriction-modification gene complex recognizes the same sequence as Dcm, and its methyltransferase is phylogenetically related to Dcm. In the present work, we found that stabilization of maintenance of a plasmid by linkage of EcoRII gene complex, likely through postsegregational cell killing, is diminished by dcm function. Disturbance of EcoRII restriction-modification gene complex led to extensive chromosome degradation and severe loss of cell viability. This cell killing was partially suppressed by chromosomal dcm and completely abolished by dcm expressed from a plasmid. Dcm, therefore, can play the role of a “molecular vaccine” by defending the genome against parasitism by a restriction-modification gene complex.
Epigenetic methylation of DNA is a prominent phenomenon in many organisms. It occurs after passage of the replication fork and involves enzymatic transfer of a methyl group to specific bases on the DNA. The specific methylation of certain cytosine residues to 5-methylcytosine (5-mC) has been shown to affect gene expression in several organisms. In several eukaryotes, C-5 methylation is known to inactivate transposons and may have been maintained as a mechanism to defend the genome from genomic parasites (51). In the prokaryotes, many DNA methyltransferases are found as a component of a restriction-modification (RM) gene complex. Some methyltransferases are found paired with a restriction enzyme gene inactivated by a mutation (20, 25, 30), while others are not paired with a restriction enzyme homologue. Dcm in Escherichia coli and related bacteria is one of these “orphan” methyltransferases (39).
In the E. coli chromosome, approximately 1% of the cytosine residues are methylated by Dcm to generate the sequence 5′-CmCWGG (W = A or T) (39). 5-mC can deaminate to form thymine, which can lead to the production of C-to-T mutations. This mutation is prevented by the product of a contiguous gene, vsr, through the very-short-patch mismatch repair reaction (28, 46). However, Vsr can increase other types of mutation under some other conditions, and the actions of Dcm and Vsr most likely have affected genome evolution (6, 11, 13, 29, 31). The reason for the existence of the dcm-vsr gene pair in bacteria has remained unclear.
The only phenotype observed in dcm mutants is susceptibility of their DNA to restriction by EcoRII endonuclease (18, 45), which recognizes the same sequence as Dcm (according to the REBASE website [http://rebase.neb.com/rebase/rebase.html]). A type II restriction modification (RM) system, such as EcoRII, consists of a restriction endonuclease and a cognate methyltransferase that methylates the same recognition sequence to protect it from the cleavage (42). EcoRII methyltransferase (M · EcoRII) and Dcm share a large degree of sequence similarity (46) and are likely to be phylogenetically related to each other as detailed below. The EcoRII RM system was found in drug resistance transfer plasmids (48).
It is widely believed that the evolution and maintenance of RM genes was driven by the cell's need to protect itself from foreign DNA, such as viral or plasmid DNA. There are, however, several issues that are not satisfactorily explained only by this concept of cellular defense (23, 24). Previously we found that several type II RM gene complexes are not easily lost from their host cells when challenged by a competitor genetic element (16, 35, 36). The descendants of cells that fail to inherit an RM gene complex become unable to modify a sufficient number of restriction recognition sites on their chromosomes, which results in chromosome cleavage by the remaining pool of restriction endonuclease and cell lethality (14, 15, 35). This type of “postsegregational killing” has been recognized as a plasmid maintenance mechanism (9). However, we found that chromosomal RM gene complex also shows such host killing when challenged by an allelic gene (16). This cell killing would confer a competitive advantage on RM gene complexes. The capacity to act selfishly may have contributed to the spread and maintenance of RM systems (23, 24), although we think that RM elements would confer advantage to the host genome by fighting with invaders under many circumstances. There are increasing lines of observations in harmony with behavior of RM systems as selfish genetic elements (24). These include their mutual competition for recognition sequences (27), superinfection exclusion (37), restriction site avoidance in bacterial genomes (8, 43), and responses to DNA double-strand breaks by bacterial recombination repair systems (15). Furthermore, recent genome sequence comparisons strongly suggest that RM gene complexes are mobile genetic elements that have modeled bacterial genomes (1, 5, 24, 38). Indeed an attempt to replace a chromosomal RM gene complex led to genome rearrangements in the laboratory (16).
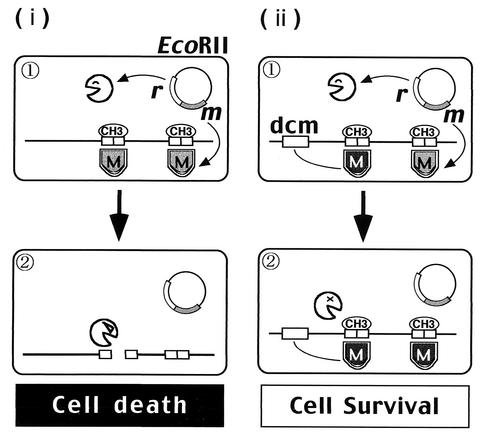
Dcm-proficient cells that have disturbed the EcoRII RM gene complex would be expected to survive if methylation of the EcoRII endonuclease (R · EcoRII) recognition site were assured by the Dcm methylase (Fig. 1, panel ii). In the present work, we tested this prediction. Our results suggest that Dcm can play the role of a live vaccine to defend the genome from parasitism by an RM gene complex.
FIG. 1.
Experimental design: postdisturbance killing by EcoRII and its prevention by Dcm methylase. (i) Postdisturbance killing by the EcoRII RM gene pair. (1) R · EcoRII and M · EcoRII are expressed from an EcoRII RM gene pair on a plasmid. M · EcoRII modifies its recognition sequence, 5′-CCWGG, on the chromosome. (2) Some disturbance of EcoRII RM occurs. When M · EcoRII cannot modify all the recognition sites along the newly replicated chromosome, the remaining level of R · EcoRII cleaves the chromosome at the unmodified sites and kills the cell. (ii) Prevention of the cell killing by Dcm methylation (prediction). (1) Dcm, in addition to M · EcoRII, modifies CCWGG. (2) When disturbance of EcoRII RM occurs, Dcm continues to modify the CCWGG sequence. R · EcoRII cannot cleave the chromosome, and the cell survives. Canvas software was used to produce this figure.
MATERIALS AND METHODS
Phylogenetic analysis.
Homologues of E. coli Dcm were sought with BLASTp (2) in the REBASE database and with tBLASTn in the GenBank Microbial Genome Databases (http://www.ncbi.nlm.nih.gov). Amino acid sequences of CCWGG-recognizing and related homologues were aligned with CLUSTALX (49). Their phylogenetic tree was inferred using a neighbor-joining algorithm in PAUP∗ (version 4; Sinauer Associates, Sunderland, Mass.). Bootstrap probabilities (in percentage) were obtained for 1,000 runs. The following sequences were used (with accession numbers in parentheses): M · EcoKDcm (gb/X13330), M · EcoRII (gb/X05050), M · HpaII (gb/L17342), M · SsoII (gb/M86545). M · StyD4I (gb/D73442), M · StyDcmIP (gb/AF076014), M · NmeAORF1500P (gb/AL162759), M · StyR27ORF154P (gb/AF250878), M · DsaV (gb/U10528), Salmonella enterica serovar Typhi (emb AL627272), and S. enterica serovar Enteritidis (gnl/UIUC-592), M · SpomI (gn/X82444), M · HsaIIP (gn/AJ223333), M · MmulIP (gn/AF045889), and M · DmeORFAP (gn/AE185647).
Bacterial strains.
The strains used were E. coli GM30 (F− λ− thr-1 araC14 leuB6 fhuA31 lacY1 tsx-78 glnV44 galK2 galT22 hisG4 rpsL136 xylA5 mtl-1 thi-1) (34) provided by M. Marinus; GM31 (F− λ− thr-1 araC14 leuB6 fhuA31 lacY1 tsx-78 glnV44 galK2 galT22 dcm-6 hisG4 rpsL136 xylA5 mtl-1 thi-1) (34) provided by M. Marinus; JC8679 [F− λ− thr-1 araC14 leuB6 · (gpt-proA)62 lacY1 tsx-33 glnV44 galK2 sbcA23 his-60 recC22 relA1 recB21 rpsL31 xylA5 mtl-1 argE3 thi-1] (10) from A. J. Clark; and BNY451, which is dcm-6 zed-508::Tn10 version of JC8679 made by transduction with P1vir phage.
Plasmids.
The plasmids used (along with RM genotype, replication unit, drug resistance, other properties, and source or reference) are as follows: N3 (EcoRII r+ m+, R factor, Tetr, S. Hattman [17]), pHSG415 (no RM, pSC101ts, Ampr Cmlr Kanr, temperature sensitive; J. Kato [14]),pKK-D (dcm+, ColE1, Ampr, dcm is overexpressed from the synthetic trc promoter; C. G. Cupples [31]), pKK-DV (dcm+ vsr+, ColE1, Ampr, dcm and vsr are overexpressed from the synthetic trc promoter; C. G. Cupples [31], and pKK233-2 (no RM, ColE1, Ampr, parental vector for pKK-D and pKK-DV; C. G. Cupples).
The drug resistance transfer factors N-3 and R15 (= RTF-2) control EcoRII RM specificity (44, 48). EcoRII restriction enzyme was isolated and purified from R15 (52). We used N-3 as a source of EcoRII restriction modification gene complex. A 3,026-bp fragment including the EcoRII RM gene pair was amplified with primers RII-1 (5′-GGCCCGGGCATAGTCGAGATTGGTGCAGA-3′) and RII-2 (5′-GGCCCGGGTCATCCATACCACGACCTCAA-3′) from plasmid N3. Each PCR primer has an artificial SmaI site in its 5′ end (underlined). The fragment was digested with SmaI before inserting it into the SmaI site of pUC19 to generate pNY30 (EcoRII r+ m+, ColE1, Ampr). Plasmid pNY35 (EcoRII r+ m+, pSC101ts, Cmlr) was constructed by replacing an FspI-SmaI fragment of pHSG415 with a SmaI fragment of pNY30. pNY45 (EcoRII r− m+, pSC101ts, Cmlr) was constructed by inserting a KpnI linker (TAKARA) into the BamHI site in the r gene of pNY35. These plasmids were transferred to JC8679 and BNY451.
Measurement of plasmid maintenance.
All the cultivation procedures were carried out at 30°C. A single colony on Luria-Bertani (LB) agar with chloramphenicol (CML) (20 μg/ml) was suspended in 5 ml of LB liquid medium with CML (20 μg/ml) and grown overnight with aeration. The saturated culture was diluted 1:100 and grown again in LB liquid medium with CML (20 μg/ml) until the the log phase was reached (1 × 108 to 2 × 108 cells/ml). Aliquots of the culture were spread onto LB agar to measure viable cells and onto LB agar with CML (20 μg/ml) to measure cells carrying the plasmid. The culture was diluted 10−5 to 10−6 to obtain approximately 103 cells/tube for growth without antibiotic selection in order to earn approximately 20 generations in one overnight period. The overnight culture was diluted 10−6 for growth. The fraction of cells carrying plasmid was calculated as number of Cmlr colony formers/the number of colony formers on LB agar without antibiotics. The generation number was calculated from the viable cell count.
Plate assay for detection of cell growth inhibition.
Strains carrying one of the above RM plasmids were grown at 30°C in LB broth containing appropriate selective antibiotics until the optical density at 660 nm (OD660) of the culture reached 0.3. Five microliters of the culture was streaked on two LB agar plates lacking the antibiotics selective for RM plasmids. One plate was incubated at 30°C, and another plate was incubated at 40°C. Plates were photographed after 20 h of incubation. Details of the procedure have been described previously (15).
Cell death following disturbance of the RM gene complex.
Strains carrying one of the RM plasmids were grown at 30°C in LB broth containing appropriate selective antibiotics until the OD660 of the culture reached 0.3. Then, the cells were centrifuged, suspended in LB broth lacking CML, and incubated at 40°C. The cells were diluted whenever the OD660 of the culture reached 0.3. Total cell numbers were counted with a microscope. The number of viable cells is defined as the number of colony formers on LB agar plates lacking CML. The number of cells carrying the RM plasmid is defined as the number of colony formers on LB agar plates containing CML and other appropriate selective antibiotics. The cells were harvested, fixed, and microscopically observed as described earlier (14, 15). Details of these procedures were described previously (14, 15).
Analysis of chromosomal DNA degradation.
Cultures of strains carrying an RM plasmid were shifted from 30 to 40°C. The cells were embedded in an agar plug and processed for pulsed-field gel electrophoresis. The DNA was electrophoresed by CHEF-DR III variable angle system (Bio-Rad, Hercules, Calif.) under the following conditions: (i) multistate mode, 20-h run time, 50-s switch time ramp, 120° included angle, 6 V/cm, 0.5× Tris-borate-EDTA at 14°C, 1.0% SeaKem GTG agarose (FMC, Rockland, Maine) or (ii) 20 h-run time, 4- to 50-s switch time ramp, 120° included angle, 6 V/cm, 0.5× Tris-borate-EDTA at 14°C, 1.0% SeaKem GTG agarose for XbaI-digested DNA. E. coli strain JC8679 and an isogenic dcm-6 strain BNY451 were used because it is difficult to detect chromosome degradation in recBC+ strains (14, 15). Cell killing was found to be attenuated by the chromosomal dcm gene and to be completely suppressed by the Dcm plasmid in this JC8679 background (data not shown).
RESULTS
Phylogenetic relationships between Dcm, M · EcoRII and other 5′-CCWGG-recognizing DNA C-5 methyltransferases.
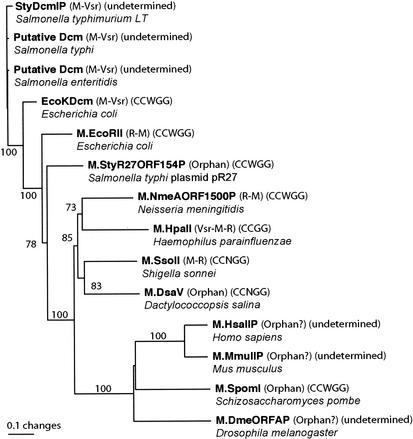
A phylogenetic tree for the homologues of E. coli Dcm is shown in Fig. 2. Some of the homologues have been shown to be a component of an RM system as EcoRII methylase, while some others do not appear so and might be orphan methylases as Dcm (REBASE). Close dcm homologues were present in Salmonella species, S. enterica serovar Typhi, S. enterica serovar Paratyphi (data not shown), S. enterica serovar Typhimurium, and S. enterica serovar Enteritidis. A dcm homologue in Klebsiella pneumoniae was truncated and excluded from analysis. Its distribution is in accord with the distribution of Dcm-type methylation at 5′-CCWGG sequences (12). Each of these Dcm homologues was followed by a sequence very similar to that of the E. coli vsr gene. E. coli Dcm and Salmonella Dcm homologues form a strongly supported monophyletic group. The dcm homologues, EcoRII methylase, and an orphan methylase found in a Salmonella plasmid form a distinct clade among these methylases as found in a preceding study (3).
FIG. 2.
Phylogenetic tree for Dcm and several CCWGG-recognizing DNA C-5 methyltransferases. A neighbor-joining tree from the amino acid sequences is shown with bootstrap values (in percentages) at the branches. Each methyltransferase name is followed by gene organization, recognition sequence, and organism name. Based on data taken from the REBASE and NCBI (http://www.ncbi.nlm.nih.gov) databases. Illustrator software was used to produce this figure. For current Salmonella nomenclature, see, for example, http://www.bacterio.cict.fr/salmonellanom.html.
EcoRII methylase recognizes the same sequence, 5′-CCWGG, as Dcm. RM systems recognizing this sequence are frequently encountered in diverse bacterial groups (REBASE). The seven methylases in the upper part of Fig. 2 that recognize (or likely recognize) this sequence shared homology in the target recognition domain as well as the motifs specific to C-5 methylases. Such sequence homology in the target recognition domain was also seen in M · SsoII and M · DsaV, which recognize CCNGG, but not in HpaII, which recognizes CCGG.
Among the eukaryotic C-5 methylases, which were distantly related to Dcm and M · EcoRII, one found in Schizosaccharomyces pombe recognizes the same sequence of CCWGG (40). This enzyme forms a clade with a Drosophila melanogaster methylase and Dnmt2 family of mammalian methylases (7) (Fig. 2). CmCWGG DNA methylation has been linked to gene silencing in lymphoma (32).
This phylogenetic tree is in harmony with the previous ones (3, 22, 33).
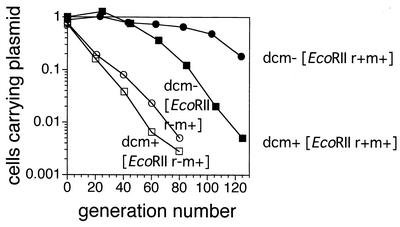
Effect of dcm on maintenance of an EcoRII plasmid.
Earlier we demonstrated that a plasmid carrying one of several RM gene complexes is stably maintained because its loss leads to cell death (postsegregational killing) (4, 14, 15, 35, 36, 37). We compared stability of maintenance of a plasmid carrying EcoRII RM gene complex in the dcm+ and dcm-6 backgrounds. We had cloned the EcoRII RM gene complex from plasmid N-3 and inserted it into a well-defined unstable plasmid pHSG415 (Materials and Methods). An E. coli strain, JC8679 (recBC sbcA), was used as a host at 30°C because of instability of plasmid maintenance. The large stabilization in the dcm-6 background was found reduced in the dcm+ background (Fig. 3). This result is in harmony with our hypothesis that genome methylation by Dcm defends genome from postsegregational attack by the EcoRII RM gene complex (Fig. 1).
FIG. 3.
Effect of dcm on plasmid stabilization by linkage of EcoRII RM gene complex. The cells were grown at 30°C in liquid medium in the absence of antibiotic selection. The fraction of cells carrying plasmid, i.e., the number of Cmlr colony formers/the number of colony formers on LB agar, were plotted as a function of the generation number. JC8679, its dcm-6 derivative carrying pNY35 (EcoRII r+ m+, Cmlr), or its restriction-negative derivative, pNY45, was examined.
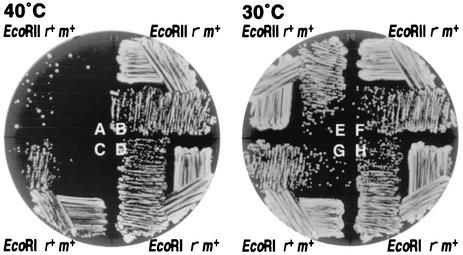
Strong cell death after disturbance of the EcoRII RM gene complex.
We also tried a plate assay for cell killing (15) with this vector (pHSG415) in a rec+ host strain (Fig. 4). The cells were grown at 30°C in liquid medium containing selective antibiotics for the plasmid and then streaked on agar medium without selective antibiotics. The plates were then incubated overnight at 30 or 40°C. Colony formation of cells was markedly inhibited at 40°C (Fig. 4), whereas colony formation of cells that had carried the control EcoRII restriction mutant plasmid was not inhibited (Fig. 4). At 30°C, the restriction-positive and restriction mutant cells showed comparable growth (Fig. 4). This growth inhibition can be explained by lethal attack of the restriction enzyme molecules on the chromosome of the cells. The growth inhibition was much stronger than that seen with EcoRI RM (15) (Fig. 4, lower parts of the circular plates) or with PaeR7I (A. Ichige and I. Kobayashi, unpublished data) using a similar plasmid construction.
FIG. 4.
Inhibition of colony formation after disturbance of the EcoRII RM gene complex. E. coli strain GM31 (rec+ dcm-6) carrying pNY35 (pSC101ts, EcoRII r+ m+, Cmlr), pNY45 (its r− version), pIK178 (pSC101ts, EcoRI r+ m+, Cmlr), or pIK179 (its r− version) was grown with antibiotics selective for the plasmid and then streaked on an agar medium without selective antibiotics as described in Materials and Methods. Photoshop software was used to produce this figure.
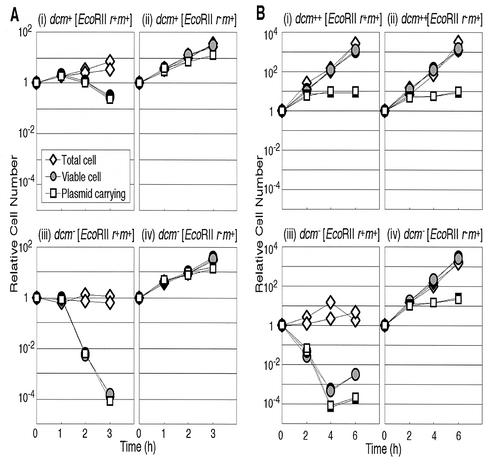
We then monitored the kinetics of cell growth inhibition after the temperature shift in liquid medium. As expected, the EcoRII restriction mutant cells continued to grow normally, while the increase in the number of plasmid-carrying viable cells gradually slowed down (Fig. 5A, panel iv). This is exactly as expected, because the pHSG415 replicon shows slower replication at 40°C. However, in cells with EcoRII RM, the temperature shift led to a decrease in the total number of viable cells and in the number of plasmid-carrying viable cells in the dcm-6 cells (Fig. 5A, panel iii). The decrease was by 4 orders of magnitude. This result is in sharp contrast to what we observed earlier with EcoRI RM and PaeR7 RM—the temperature shift leading to cessation or a slowing down of the increase of the viable cells and of plasmid-carrying viable cells (15)—as expected if only those cells that have lost a plasmid die. The viability loss observed in the present work is not expected if death occurs in a cell only after complete loss of the plasmid molecules. Rather, this suggests the possibility that the temperature shift somehow caused unbalance between R and M, reduced chromosome methylation by M · EcoRII, and led to chromosome breakage by EcoRII restriction at those unmodified EcoRII sites and cell killing. We might call this process presegregational killing or postdisturbance killing.
FIG. 5.
Dcm methylase suppresses cell killing after disturbance of EcoRII RM gene complex. (A) Partial suppression by chromosomal dcm. An E. coli strain GM30 (rec+ dcm+) or GM31 (its dcm-6 version) carrying pNY35 (pSC101ts, EcoRII r+ m+, Cmlr) or pNY45 (its r− version) was aerated at 30°C in LB broth with CML, which selects for plasmid-containing cells. Then CML was removed, and the culture was transferred to 40°C. Total cells, viable cells, and plasmid-carrying viable cells were estimated as described in Materials and Methods. Each value was divided by the value at time zero. (i) GM30 (dcm+) (pNY35 [EcoRII r+ m+]); (ii) GM30 (dcm+) (pNY45 [r− version]); (iii) GM31 (dcm-6) (pNY35 [EcoRII r+ m+]); (iv) GM31 (dcm-6) (pNY45 [r− version]). (B) Complete suppression by overexpressed Dcm. E. coli strain GM31 (rec+ dcm-6) carrying two plasmids was aerated at 30°C in LB broth with CML and ampicillin, which select for plasmid-containing cells. Then, CML was removed, and the culture was transferred to 40°C. Total cells, viable cells, and viable cells carrying the EcoRII plasmid were estimated. Each value was divided by the value at time zero. ++, Dcm was overexpressed from a plasmid. (i) GM 31 (pKK-D [dcm overexpressed, Ampr], pNY35 [pSC101ts, EcoRII r+ m+, Cmlr]); (ii) GM31 (pKK-D, pNY45 [r− version of pNY35]); (iii) GM31 (pKK233-2 [vector for pKK-D], pNY35); (iv) GM31 (pKK233-2, pNY45). DeltaGraph and Canvas softwares were used to produce this figure.
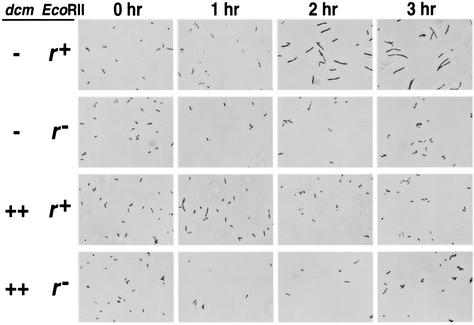
The total cell number measured under a microscope showed no marked increase during the 3-h observation period (Fig. 5A, panel iii). Most of the cells became filamentous (Fig. 6, first row) as in the case with EcoRI (15). This filamentation was not observed in the restriction mutant control (Fig. 6, second row).
FIG. 6.
Dcm methylase suppresses cell filamentation after disturbance of EcoRII RM gene complex. The E. coli strain GM31 (dcm-6) (pKK233-2 [vector], pNY35 [EcoRII r+ m+]), GM31 (pKK233-2, pNY45 [r− version]), GM31 (pKK-D [dcm overexpressed], pNY35 [EcoRII r+ m+]), or GM31 (pKK-D, pNY45 [r− version]) was grown, harvested, fixed, and microscopically observed as described earlier (14, 15). Photoshop software was used to produce this figure.
Suppression of cell death by Dcm.
Dcm, which recognizes the same DNA sequence (5′-CCWGG) as EcoRII, would be expected to inhibit cell killing induced by the disturbance of the EcoRII RM gene complex (Fig. 1). We compared the kinetics of cell death of isogenic dcm+ strain and dcm-6 strain in liquid culture. The drop in the total viable cell counts and in the plasmid-carrying viable cell counts was not as dramatic in the dcm+ culture compared to the dcm mutant (Fig. 5A, panel i). In the experiment shown, at 3 h after the temperature shift, the cell viability (number of viable cell/total number of viable cells) was 6.4 × 10−2. This was approximately 400-fold higher than that of the dcm strain. The number of plasmid-carrying viable cells was close to the number of total viable cells in the dcm+ strain and dcm strain. Most of the plasmid molecules appeared to have been lost during the cell killing process. The increase in the total cell count was larger than that in the dcm strain (Fig. 5A, panel iii) but not as large as in the restriction mutant control (Fig. 5A, panel ii). Most of the cells remained filamentous (data not shown).
The dcm-6 mutation has a polar effect on vsr. When we used a plasmid (pKK-D) in which dcm was expressed from a strong, constitutive promoter (trc) (31) in a dcm-6 strain, cell killing after temperature shift of the cells carrying EcoRII genes was completely suppressed (Fig. 5B, panel i). There was no decrease in the total viable cell count or in the total cell count. The number of plasmid-carrying viable cells stopped increasing and remained constant. These curves were indistinguishable from those of the restriction-minus control (Fig. 5B, panel ii). In a control experiment, the growth inhibition elicited by temperature shift of the cells carrying the EcoRI RM gene complex (as opposed to EcoRII RM) was not affected by dcm on the chromosome or by dcm on the plasmid (data not shown). The suppression effect of Dcm was, therefore, specific for EcoRII. The cell filamentation was also suppressed by pKK-D (Fig. 6, third row). Suppression was also observed with pKK-DV (31) in which dcm and vsr were overexpressed (data not shown).
Suppression of chromosomal DNA degradation.
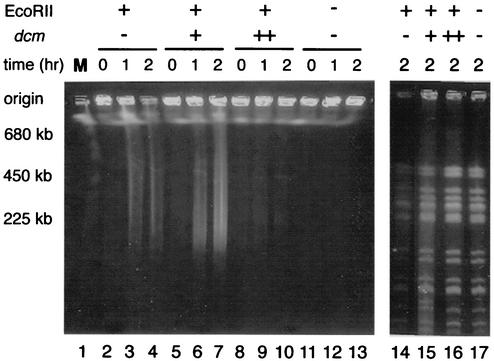
To confirm that degradation causes cell death after cleavage by EcoRII in vivo and that this process is modulated by dcm, whole cellular DNA was analyzed by pulsed-field gel electrophoresis. The cells were harvested at various time intervals after the temperature shift, embedded in agarose plugs, and lysed in situ to liberate the DNA for electrophoresis. In order to monitor the amount of undegraded DNA, DNA was also treated with the rare-cutting restriction enzyme XbaI in vitro before electrophoresis in a control experiment.
In dcm cells, the temperature shift of the EcoRII r+ m+ gene complex resulted in the appearance of a smear composed of DNA fragments of 200 to 700 kb (Fig. 7, lanes 3 and 4). Two hours after the temperature shift, the DNA in the well also started to disappear (lane 4). The decrease in the amount of DNA (that is detectable within this gel) was clearly seen after XbaI digestion in vitro (lane 14). No degradation of DNA was detected in the restriction mutant control (lanes 11 to 13) or in cells overexpressing dcm (lanes 8 to 10). In dcm+ cells after temperature shift of the EcoRII RM gene complex (lanes 5 to 7), DNA loss from the well was not as pronounced as in the dcm cells, and the smear indicative of relatively large DNA fragments (200 to 700 kb) was more abundant. The amount of DNA found after XbaI digestion in vitro (lane 15) was again between that of the dcm cells (lane 14) and that of the Dcm-overproducing cells (lane 16). We conclude that the extent of DNA degradation paralleled the extent in cell killing seen with different combinations of dcm and EcoRII (Fig. 7).
FIG. 7.
Dcm methylase suppresses chromosomal DNA degradation after disturbance of the EcoRII RM gene complex. DNA was not treated (left panel) or treated (right panel) with XbaI in vitro. A culture of E. coli strain BNY451 (pKK233-2, pNY35) (recBC sbcA, dcm, EcoRII r+ m+), JC8679 (pKK233-2, pNY35) (recBC sbcA dcm+, EcoRII r+ m+), BNY451 (pKK-D, pNY35) (recBC sbcA dcm++ [dcm overexpressed], EcoRII r+ m+), or BNY451 (pKK233-2, pNY45) (recBC sbcA dcm EcoRII r− m+) was grown at 30°C and transferred to 40°C. At the indicated time intervals after transfer, chromosomal DNA was prepared and electrophoresed through a pulsed-field gel without or after digestion with XbaI as described in Materials and Methods. Lane 1 contains Saccharomyces cerevisiae chromosomes as markers. Photoshop software was used to produce this figure.
DISCUSSION
Strong cell killing after disturbance of EcoRII RM gene complex.
The temperature shift of the cells carrying EcoRII gene complex connected with pHSG415 led to a severe decrease in the numbers of viable cells and of plasmid-carrying viable cells. This is in contrast to the results obtained with EcoRI, PaeR7I, and EcoRV, which were carried by a similar plasmid construct and tested by the same experimental procedures (14, 15, 27, 35, 37). It appears that the cell killing took place before the cell had completely lost its RM gene complex. This is not postsegregational cell killing in the strict sense of the word and may be called presegregational cell killing. Similar inhibition of colony formation at 40°C was observed with pBR322 and with pUC19 vectors (N. Takahashi and I. Kobayashi, unpublished observation). What we observed may have been due to the lowered concentration and/or activity of the EcoRII methylase after the temperature shift, which had resulted in the exposure of unmethylated sites on the DNA to lethal attack by the EcoRII restriction enzyme. Whatever the mechanisms, the concept of postsegregational cell killing needs to be enlarged to cover the concept of postdisturbance cell killing. The latter term implies that disturbance of the RM gene complexes can lead to attack on the genome and cell killing.
The number of EcoRII recognition sites (5′-CCWGG) on the E. coli genome is much larger than the number of EcoRI or PaeR7I recognition sites (5′-GAATTC or 5′-CTCGAG). However, the number of recognition sites cannot be the sole determinant of the extent of cell killing, because SsoII, which recognizes the sequence CCNGG and, therefore, has more recognition sites on the E. coli genome than EcoRII, showed less cell killing than EcoRII (4).
The sequence 5′-CCWGG represents one of the most common recognition sequences for RM systems identified so far. For example, out of the 3392 RM systems in the REBASE database (42), 225 recognize CCWGG. Indeed, the sequence 5′-CCWGG is significantly less frequent than expected in the genome of Neisseria meningitidis and Vibrio cholerae (J. Posfai and T. Vincze, REBASE).
Action of Dcm as a molecular vaccine against a parasitic RM gene complex.
E. coli Dcm methyltransferase, which shares the same recognition site (CCWGG) as EcoRII, suppressed the plasmid stabilization by linkage of EcoRII RM gene complex and the cell death induced by the disturbance of the EcoRII RM gene complex. In other words, the E. coli genome can protect itself from the virulence of a parasitic RM gene complex by utilizing a methyltransferase gene that recognizes the same site as the RM complex. We do not know yet whether Dcm is maintained because of this live vaccine effect, which would defend the genome against parasitism by EcoRII RM complexes and/or its isoschizomers.
The homology between M · EcoRII and Dcm suggests that they may have a common ancestor (Fig. 2). From the phylogenetic analysis, one may speculate that some ancient bacterium, threatened by parasitism with the EcoRII family, took up an M · EcoRII gene, which later evolved into the dcm gene in a group of bacteria. This type of “orphan” DNA methylases turned out to be abundant, especially in the sequenced genomes (REBASE). Figure 2 shows an example of a pair of orphan methylase and a methylase of an RM system recognizing CCNGG. A modification enzyme homologue is often found next to a mutationally inactivated restriction homologue (for example, see references 20, 25, and 30), and it would play the same role and could be an intermediate form in evolution.
The natural level of expression of dcm on the chromosome was effective against cutting by the EcoRII restriction endonuclease, but was insufficient to completely prevent cell killing following the EcoRII RM replication block. Consistent with this observation, some specific dcm sites on the E. coli chromosome were found to be unmethylated in vivo (41). On the other hand, overexpression of dcm completely protected against cell death. If Dcm functions as a live vaccine, why was the level of dcm expression insufficient to prevent cell death? One possible explanation is the cost of cytosine methylation. One kind of cost is gene expression, although there is no report of effects of dcm on gene expression.
Another kind of cost of Dcm action is in its mutagenic potential (see the introduction). The vsr gene, which can prevent the mutation at 5-methylcytosine, can also introduce other types of mutation and likely affects genome composition. Thus, Vsr may be regarded as a sort of secondary drug to attenuate the adverse side effects of the first drug but causes other undesirable effects. This might explain why this very-short-patch repair system is present in the first place. Indeed, several RM gene complexes have been found linked to a vsr homologue (Fig. 2) (26; REBASE). Vsr has structural similarity with restriction enzymes and may well be evolutionarily related to them (50).
Some bacteriophages carry an orphan methylase that would protect itself from attack by certain RM gene complexes (REBASE). If they become lysogenized as a prophage, they may protect bacterial genomes against RM systems. On the other hand, a bacteriophage can move an orphan methylase by transduction. The same argument is possible with plasmids and other mobile genetic elements. The defense of genome of bacteriophage and other mobile elements against RMs by carriage of orphan methylase may be related with the defense of bacterial genomes against RMs.
The hypothesis of spreading and maintenance of an orphan methylase as a live vaccine is not so unnatural when we consider antibiotics resistance genes. Some of them are likely derived from the self-tolerance genes in a gene cluster for antibiotics biosynthesis (for example, see reference 47). We can postulate a relationship of “rock, scissors, and paper” among bacteria carrying orphan M, RM, and neither. Such a relationship allows stable maintenance of the three groups of bacteria under certain conditions (21).
If orphan methylases serve as a live vaccine against RM gene complexes, there would be a specific relationship between their distribution patterns among diverse bacterial groups as found with other types of selfish genetic elements (19). Unfortunately, our present knowledge of their distribution is too little to identify such a relationship (REBASE).
In several eukaryotic organisms, including fungi, plants and mammals, DNA methylation at C-5 inactivates parasitic genetic elements such as retroviruses and transposons (see the introduction). This role could be related to the role of Dcm in counteracting an RM complex, as demonstrated in the present work. Some eukaryotic C-5 methylases seem to be phylogenetically related to Dcm, and at least two of them share the recognition sequence with Dcm (Fig. 2) (32, 40). Further experiments are necessary to fully understand mechanisms underlying the interaction between Dcm and RM. These include the fates of restriction enzyme, modification enzyme, and Dcm after the temperature shift.
Acknowledgments
We thank S. Hattman, J. Kato, C. G. Cupples, M. Marinus, and A. J. Clark for gifts of biological materials and the Genome Sequencing Center, Washington University, St. Louis (http://genome.wustl.edu/gsc/), the Department of Microbiology at the University of Illinois (http://www.salmonella.org), and the Sanger Center (http://www.sanger.ac.uk/) for making available DNA sequence data prior to publication. We are grateful to F. Taddei, C. Cupples, S. Hattman, M. Marinus, and S. Turner for comments. We acknowledge the Human Genome Center, whose facilities were used for sequence analysis.
This work was supported by grants to I.K. from MEXT, NEDO, and the Uehara Memorial Foundation and by grants to N.H. from the Japan Science Society. N.H. is supported by a JSPS fellowship.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. DeJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bujnicki, J. M., and M. Radlinska. 1999. Molecular phylogenetics of DNA 5mC-methyltransferases. Acta Microbiol. Pol. 48:19-30. [PubMed] [Google Scholar]
- 4.Chinen, A., Y. Naito, N. Handa, and I. Kobayashi. 2000. Evolution of sequence recognition by restriction-modification enzymes: selective pressure for specificity decrease. Mol. Biol. Evol. 17:1610-1619. [DOI] [PubMed] [Google Scholar]
- 5.Chinen, A., I. Uchiyama, and I. Kobayashi. 2000. Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction-modification genes with large genome polymorphisms. Gene 259:109-121. [DOI] [PubMed] [Google Scholar]
- 6.Doiron, K. M., S. Viau, M. Koutroumanis, and C. G. Cupples. 1996. Overexpression of vsr in Escherichia coli is mutagenic. J. Bacteriol. 178:4294-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, A., J. A. Yoder, X. Zhang, L. Zhou, T. H. Bestor, and X. Cheng. 2001. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 29:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelfand, M. S., and E. V. Koonin. 1997. Avoidance of palindromic words in bacterial and archaeal genomes: a close connection with restriction enzymes. Nucleic Acids Res. 25:2430-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes, K., S. Ayora, I. Canosa, P. Ceglowski, R. Diaz, T. Franch, A. P. Gultyaev, R. B. Jensen, I. Kobayashi, C. Macpherson, D. Summers, C. Thomas, and U. Zielenkiewicz. 2000. Plasmid maintenance systems, p. 49-86. In M. Couturier, K. Gerdes, and C. M. Thomas (ed.),The horizontal gene pool. Harwood Academic Publishers GmbH, Amsterdam, The Netherlands.
- 10.Gillen, J. R., D. K. Willis, and A. J. Clark. 1981. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K12. J. Bacteriol. 145:521-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glasner, W., R. Merkl, V. Schellenberger, and H. J. Fritz. 1995. Substrate preferences of Vsr DNA mismatch endonuclease and their consequences for the evolution of the Escherichia coli K-12 genome. J. Mol. Biol. 245:1-7. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Eichelmann, M. C., A. Levy-Mustri, and J. Ramirez-Santo. 1991. Presence of 5-methylcytosine in CC(A/T)GG sequences (Dcm methylation) in DNAs from different bacteria. J. Bacteriol. 173:7692-7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez, G., J. Casadesus, J. L. Oliver, and A. Marin. 1994. Compositional heterogeneity of the Escherichia coli genome: a role for VSP repair? J. Mol. Evol. 39:340-346. [DOI] [PubMed] [Google Scholar]
- 14.Handa, N., and I. Kobayashi. 1999. Post-segregational killing by restriction modification gene complexes: observations of individual cell deaths. Biochimie 81:931-938. [DOI] [PubMed] [Google Scholar]
- 15.Handa, N., A. Ichige, K. Kusano, and I. Kobayashi. 2000. Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol. 182:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handa, N., Y. Nakayama, M. Sadykov, and I. Kobayashi. 2001. Experimental genome evolution: large-scale genome rearrangements associated with resistance to replacement of a chromosomal restriction-modification gene complex. Mol. Microbiol. 40:932-940. [DOI] [PubMed] [Google Scholar]
- 17.Hattman, S., and L. Cousens. 1972. Location of the region controlling host specificity (hsII) with respect to drug resistance markers on the fi− R factor, N-3. J. Bacteriol. 112:1428-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattman, S., S. Schlagman, and L. Cousens. 1973. Isolation of a mutant of Escherichia coli defective in cytosine-specific deoxyribonucleic acid methylase activity and in partial protection of bacteriophage lambda against restriction by cells containing the N-3 drug-resistance factor. J. Bacteriol. 115:1103-1110. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst, L. D., A. Atlan, and B. O. Bengtsson. 1996. Genetic conflicts. Q. Rev. Biol. 71:317-364. [DOI] [PubMed] [Google Scholar]
- 20.Ibanez, M., I. Alvarez, J. M. Rodriguez-Pena, and R. Rotger. 1997. A ColE1-type plasmid from Salmonella enteritidis encodes a DNA cytosine methyltransferase. Gene 196:145-158. [DOI] [PubMed] [Google Scholar]
- 21.Kerr, B., M. A. Riley, M. W. Feldman, and B. J. Bohannan. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171-174. [DOI] [PubMed] [Google Scholar]
- 22.Klimasauskas, S., J. L. Nelson, and R. J. Roberts. 1991. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 19:6183-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, I. 1996. DNA modification and restriction: selfish behavior of an epigenetic system. p. 155-172. In V. Russo, R. Martienssen, and A. Riggs (ed.), Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong, H., L. F. Lin, N. Porter, S. Stickel, D. Byrd, J. Posfai, and R. J. Roberts. 2000. Functional analysis of putative restriction-modification system genes in the Helicobacter J99 genome. Nucleic Acids Res. 28:3216-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulakauskas, S., J. M. Barsomian, A. Lubys, R. J. Roberts, and G. G. Wilson. 1994. Organization and sequence of the HpaII restriction-modification system and adjacent genes. Gene 142:9-15. [DOI] [PubMed] [Google Scholar]
- 27.Kusano, K., T. Naito, N. Handa, and I. Kobayashi. 1995. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl. Acad. Sci. USA 92:11095-11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieb, M. 1991. Spontaneous mutation at a 5-methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genetics 128:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieb, M., and S. Rehmat. 1995. Very short patch repair of T:G mismatches in vivo: importance of context and accessory proteins. J. Bacteriol. 177:660-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, L. F., J. Posfai, R. J. Roberts, and H. Kong. 2001. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 98:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macintyre, G., K. M. Doiron, and C. G. Cupples. 1997. The Vsr endonuclease of Escherichia coli: an efficient DNA repair enzyme and a potent mutagen. J. Bacteriol. 179:6048-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malone, C. S., M. D. Miner, J. R. Doerr, J. P. Jackson, S. E. Jacobsen, R. Wal, and M. Teitell. 2001. CmC(A/T)GG DNA methylation in mature B cell lymphoma gene silencing. Proc. Natl. Acad. Sci. USA 98:10404-10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 34.Marinus, M. G., and N. R. Morris. 1973. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J. Bacteriol. 114:1143-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naito, T., K. Kusano, and I. Kobayashi. 1995. Selfish behavior of restriction-modification systems. Science 267:897-899. [DOI] [PubMed] [Google Scholar]
- 36.Naito, Y., T. Naito, and I. Kobayashi. 1998. Selfish restriction modification genes: resistance of a resident R/M plasmid to displacement by an incompatible plasmid mediated by host killing. Biol. Chem. 379:429-436. [DOI] [PubMed] [Google Scholar]
- 37.Nakayama, Y., and I. Kobayashi. 1998. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl. Acad. Sci. USA 95:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobusato, A., I. Uchiyama, S. Ohashi, and I. Kobayashi. 2000. Insertion with long target duplications: a mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene 259:99-108. [DOI] [PubMed] [Google Scholar]
- 39.Palmer, B. R., and M. G. Marinus. 1994. The dam and dcm strains of Escherichia coli—a review. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 40.Pinarbasi, E., J. Elliott, and D. P. Hornby. 1996. Activation of a yeast pseudo DNA methyltransferase by deletion of a single amino acid. J. Mol. Biol. 12:804-813. [DOI] [PubMed] [Google Scholar]
- 41.Ringquist, S., and C. L. Smith. 1992. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc. Natl. Acad. Sci. USA 89:4539-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts, R. J., and D. Macelis. 2001. REBASE—restriction enzymes and methylases. Nucleic Acids Res. 29:268-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rocha, E. P., A. Danchin, and A. Viari. 2001. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 11:946-958. [DOI] [PubMed] [Google Scholar]
- 44.Schlagman, S., and S. Hattman. 1974. Mutants of the N-3 R-factor conditionally defective in hspII modification and deoxyribonucleic acid-cytosine methylase activity. J. Bacteriol. 120:234-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlagman, S., S. Hattman, M. S. May, and L. Berger. 1976. In vivo methylation by Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase protects against in vitro cleavage by the RII restriction endonuclease (R·Eco RII). J. Bacteriol. 126:990-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sohail, A., M. Lieb, M. Dar, and A. S. Bhagwat. 1990. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J. Bacteriol. 172:4214-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiyama, M., C. J. Thompson, T. Kumagai, K. Suzuki, R. Deblaere, R. Villarroel, and J. Davies. 1994. Characterisation by molecular cloning of two genes from Streptomyces verticillus encoding resistance to bleomycin. Gene 151:11-16. [DOI] [PubMed] [Google Scholar]
- 48.Takano, T., Watanabe, T., and T. Fukasawa. 1968. Mechanism of host-controlled restriction of bacteriophage lambda by R factors in Escherichia coli K12. Virology 34:290-302. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsutakawa, S. E., T. Muto, T. Kawate, H. Jingami, N. Kunishima, M. Ariyoshi, D. Kohda, M. Nakagawa, and K. Morikawa. 1999. Crystallographic and functional studies of very short patch repair endonuclease. Mol. Cell 3:621-628. [DOI] [PubMed] [Google Scholar]
- 51.Yoder, J. A., Walsh, C. P., and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 52.Yoshimori, R. N. 1971. A genetic and biochemical analysis of the restriction and modification of DNA by resistance transfer factors, p. 1-73. Ph.D. thesis. University of California, San Francisco.