Abstract
Proline-rich regions have been identified in many surface proteins of pathogenic streptococci and staphylococci. These regions have been suggested to be located in cell wall-spanning domains and/or to be required for surface expression of the protein. Because little is known about these regions, which are found in extensively studied and biologically important surface proteins, we characterized the proline-rich region in one such protein, the β protein of group B streptococci. The proline-rich region in β, designated the XPZ region, has a proline at every third position, and the sequence is highly periodic in other respects. Immunochemical analysis showed that the XPZ region was not associated with the cell wall but was exposed on the bacterial surface. Moreover, characterization of a β mutant lacking the XPZ region demonstrated that this region was not required for surface expression of the β protein. Comparison of the XPZ region in different β proteins showed that it varied in size but always retained the typical sequence periodicity. Circular dichroism spectroscopy indicated that the XPZ region had the structure of a polyproline II helix, an extended and solvent-exposed structure with exactly three residues per turn. Because of the three-residue sequence periodicity in the XPZ region, it is expected to be amphipathic and to have distinct nonpolar and polar surfaces. This study identified a proline-rich structure with unique properties that is exposed on the surface of an important human pathogen.
Proline-rich regions play important roles in many protein-protein interactions, such as signaling events involving SH3 domains in eukaryotic cells (27, 59, 61). In other cases, proline-rich regions are important structural elements, e.g., in the hinge region of immunoglobulin A1 (IgA1) (11). However, for many proline-rich regions, the cellular localization and function are unclear.
Many surface proteins in pathogenic streptococci and staphylococci have been shown to include a proline-rich region. For example, such regions have been identified in the M6, SclA, and SclB proteins of Streptococcus pyogenes (20, 37, 38, 49, 50, 58), in protein A of Staphylococcus aureus (18), in protein G of group G streptococci (15), in PspA and PspC of Streptococcus pneumoniae (6, 13), in the P1 adhesin of Streptococcus mutans (12), in protein L of Peptostreptococcus magnus (26), and in FnBA of Streptococcus dysgalactiae (36). In all of these cases, the proline-rich region is located in the C-terminal, wall-proximal half of the protein. It has therefore been proposed that such proline-rich regions are associated with the bacterial cell wall (6, 16, 18, 45) or are required for cell surface expression (12), but their role remains unclear. Indeed, it is not known whether the proline-rich regions referred to above have similar or different functions. Because of the prevalence of proline-rich regions with unknown structure and function in extensively studied and biologically important bacterial surface proteins, we have characterized the proline-rich region in one such protein, the streptococcal β protein.
The ∼125-kDa β protein (also known as β C and Bac) is expressed by many strains of group B streptococci (GBS), human pathogens that are the most common cause of life-threatening bacterial infections in the neonatal period (5, 52). The β protein elicits protective immunity (9) and has therefore been evaluated as a possible component in a vaccine against GBS disease (40). Immunochemical analysis has demonstrated that β has separate binding sites for human IgA-Fc (19, 24, 25, 47, 51) and the complement regulator factor H (3) (Fig. 1), properties that may allow β to interfere with IgA- and complement-mediated opsonization (3, 47).
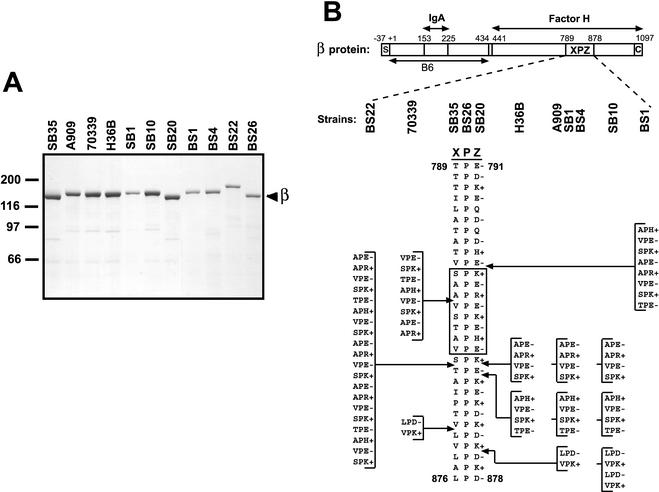
FIG. 1.
Size and sequence variability in the XPZ region of the β protein. (A) Analysis by SDS-PAGE of β proteins expressed by 11 different GBS strains. These β proteins were isolated by incubating washed bacteria at elevated pH, a procedure that allows recovery of almost pure β protein (35). The major band at ∼125 kDa represents the β protein (see text). (B) Schematic representation of the streptococcal β protein and of the proline-rich XPZ region. Binding sites for human IgA-Fc and factor H are located in the N- and C-terminal halves of β, respectively, as indicated. The XPZ region, which is located in the C-terminal part of the protein, is composed of tandemly arranged three-residue XPZ motifs, in which the first residue (X) is uncharged, the second residue (P) is a proline, and the third residue (Z) is almost invariably charged. Moreover, the third residue is alternately of positive and negative charge, as indicated. The XPZ region is not required for binding of factor H but may enhance binding of this ligand (3; T. Areschoug, unpublished data). The region designated B6 represents an N-terminal fragment of β used to make antiserum (see text). S, signal peptide; C, C-terminal membrane anchor. The numbers refer to amino acid positions in the processed protein, with numbering according to Hedén et al. (19). The lower part of the figure shows the sequence of the XPZ region in the 11 different β proteins analyzed. Strain designations are indicated at the top. The different XPZ sequences may all be derived from the shortest sequence by the addition of blocks containing multiples of six amino acid residues, as indicated. For example, the amino acid sequence in strain SB10 can be derived from that in strain SB35 by the addition of three 12-residue blocks; the first two of these blocks are the same as those indicated for strain H36B. Due to the periodic sequence of the XPZ region at the protein and DNA levels, other sites of insertion are also possible. The boxed region corresponds to a synthetic peptide used for CD analysis and for raising antiserum to the XPZ region.
Unlike most surface proteins of GBS and other gram-positive bacteria (28, 44, 57), the β protein does not contain any long repeats, but its C-terminal part includes a proline-rich sequence, the XPZ region, which exhibits a unique sequence periodicity (19, 24). This region is composed of tandemly arranged three-residue XPZ motifs, in which the first residue (X) is invariably uncharged, the second residue (P) is proline, and the third residue (Z) is almost invariably charged. Moreover, the third residue alternates between a positive and a negative charge (Fig. 1). Thus, the distribution of amino acid residues in the XPZ region is highly periodic. The function and cellular location of this region have remained unknown, but several investigators have proposed that it is located within the cell wall-spanning part of the β protein (8, 24, 32).
The gene encoding β, the bac gene, has been sequenced in two unrelated GBS strains (19, 24). Comparison of the two deduced β sequences shows that they have XPZ regions containing 30 and 40 XPZ motifs, respectively, but apart from that difference the amino acid sequences are identical except for a single residue. These limited data suggest that the β protein varies in size due to size variability in the XPZ region.
We show here that the XPZ region is exposed on the bacterial surface and is not required for cell wall anchoring of the β protein. Moreover, we present spectroscopic evidence that the XPZ region adopts the conformation of a left-handed polyproline II (PPII) helix, an extended and solvent-exposed structure (55, 59). The length and sequence of the XPZ region varied in different β proteins, but the sequence periodicity was strictly conserved. Together, these data indicate that the proline-rich XPZ region of the β protein represents a novel type of surface structure in bacteria.
MATERIALS AND METHODS
Bacterial strains and media.
The GBS strains used were of capsular serotypes Ia, Ib, and II. Strain SB35 was used to sequence the bac gene, which encodes β (19). Strains A909 and H36B (51) were obtained from C. Schalén (Lund University, Lund, Sweden), and strain 70339 (9) was from L. Bevanger (University of Trondheim, Trondheim, Norway). The other β-expressing strains used in this study were identified in a collection of GBS strains obtained from the Clinical Microbiology Laboratory at Lund University Hospital. Strains were identified as β-expressing on the basis of their ability to bind IgA-Fc. A β-negative mutant, designated the Δbac strain, in which the bac gene of strain A909 was replaced with a kanamycin resistance cassette, has been described (3). In the transcomplemented Δbac/pLZbac strain, the bac gene is expressed from plasmid pLZ12Spec (3). Bacteria were grown at 37°C in Todd-Hewitt broth (Oxoid, Basingstoke, Hampshire, United Kingdom). GBS strains containing derivatives of plasmid pLZ12Spec were grown in the presence of spectinomycin (70 μg/ml).
Construction of a β protein lacking the XPZ region.
The regions upstream and downstream of the sequence in the bac gene that encodes the XPZ region were amplified by PCR, with plasmid pBAC601 (19) as the template. The upstream region was amplified with the synthetic oligonucleotides 5′-AAATTTGAATTCTGCAGGAAGTTATTATTCCGAATG-3′ and 5′-AAATTTGGATCCATTTGTCTCTAGACCTTTTTGAAATT-3′. The resulting PCR fragment was digested with EcoRI and BamHI, recognition sequences for which had been introduced through the primers. This fragment was ligated into plasmid pLZ12Spec (21) which had been digested with EcoRI and BamHI, resulting in plasmid pLZ-UP.
The region in bac downstream of the XPZ coding region was amplified with synthetic oligonucleotides 5′-AAATTTGGATCCGGGTTAAATAAAGTTGGACAAGCA-3′ and 5′-AAATTTAAGCTTGTATTTTCATTGCCCTCAACATCA-3′. This PCR fragment was digested with BamHI and HindIII, recognition sequences for which had been introduced through the primers. This fragment was ligated into pLZ-UP which had been digested with BamHI and HindIII. The resulting plasmid, designated pLZΔxpz, contains a bac gene in which the entire region encoding the XPZ region has been deleted and replaced with a BamHI cleavage site. The pLZΔxpz plasmid was transformed into the β-negative GBS Δbac mutant, as described before (17), generating the Δbac/pLZΔxpz strain.
Synthetic peptides.
A 24-residue linear peptide, designated (XPZ)8, with the sequence SPKAPEAPRVPESPKTPEAPHVPE was purchased from Neosystem Laboratoire, Strasbourg, France. The (XPZ)8 peptide corresponds to amino acid residues 819 to 842 in the XPZ region of the β protein expressed by strain SB35 (19). The N terminus of the peptide was acetylated, and the C terminus was amidated. A second peptide, (XPZ)8-Cys, with the sequence SPKAPEAPRVPESPKTPEAPHVPEC, was used in raising specific antibodies.
Proteins and antisera.
The β protein expressed by strain SB35 was purified as described previously (54). Briefly, a suspension of washed bacteria was incubated at an elevated pH, which causes selective release of almost pure β protein (35), and the protein was purified to homogeneity by a combination of ion-exchange and molecular sieve chromatographies. β proteins expressed by other GBS strains were recovered after incubation of the bacteria at an elevated pH (35) and used without further purification. Human polyclonal IgA was from Cappel-Organon Teknika (Turnhout, Belgium). Protein G was from Amersham Pharmacia Biotech (Uppsala, Sweden). Antibodies against the XPZ region were raised by immunizing a rabbit with the (XPZ)8-Cys peptide conjugated to the carrier bovine serum albumin via the C-terminal cysteine (Neosystem Laboratoire, Strasbourg, France). The antiserum was raised by immunizing a rabbit subcutaneously on the back with 200 μg of conjugate, with complete Freund's adjuvant for the first immunization and incomplete Freund's adjuvant for three boosters, which were given at monthly intervals. An antiserum directed against the N-terminal B6 fragment of β, corresponding to the first 434 amino acid residues of the mature protein, has been described (19).
PCR and DNA sequencing.
Chromosomal DNA from different GBS isolates was used as the template in PCR analysis with the primers 5′-2555CAAAAATGAATGCTACTGTTGC2576-3′ (forward primer) and 5′-2934CGTAACCTTAGGAATTCCATCAGTTG2909-3′ (reverse primer). The figures indicate the position in the bac sequence described previously by Hedén et al. (19). The reverse primer was constructed so that an EcoRI site was created. The PCR-generated fragments were digested with XbaI, cutting at a site located just downstream of the forward primer, and EcoRI and analyzed by electrophoresis in 3% agarose and Tris-acetate buffer at 100 V. The digested fragments were further cloned into M13mp18 and M13mp19 (60), and the sequences were determined by standard procedures.
CD spectroscopy.
Circular dichroism (CD) spectra were recorded at 5, 25, 45, 65, and 85°C with a Jasco J-720 spectropolarimeter supplied with a Peltier thermostate and quartz cuvettes with a 0.1-mm path length. The spectra were recorded between 250 and 185 nm with a scan speed of 5 nm/min, a response time of 8 s, and a step resolution of 1 nm. The (XPZ)8 peptide was dissolved in 5 mM potassium phosphate with 0.15 M NaF, and the pH was adjusted to 1.5, 7.0, or 12.0 with orthophosphoric acid or NaOH. For salt effect studies, aliquots of concentrated peptide stock at pH 7 were diluted in 5 mM potassium phosphate and/or 5 mM potassium phosphate with 0.7 M NaF. The concentration of the peptide was determined by amino acid analysis after acid hydrolysis (Biomedical Center, Uppsala, Sweden).
Other methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as described previously (54). Analysis of surface expression of β and of the XPZ region was performed by a two-step binding assay, with rabbit antiserum and radiolabeled protein G, as described previously (54); controls with preimmune rabbit serum were included in all tests and were negative. Analysis of the ability of bacteria to bind IgA-Fc or factor H was performed as described previously (3, 34). Reactivity of antibodies with β protein immobilized in microtiter wells was performed essentially as described previously (53), with wells coated with pure β protein (1 μg/ml; 50 μl/well). Proteins were labeled with carrier-free 125INa (Amersham International, Amersham, United Kingdom), with the chloramine-T method.
Nucleotide sequence accession numbers.
The nucleotide sequences corresponding to the XPZ regions reported here are as follows: BS22, AF522266; 70339, AF522264, BS26, AF522267; SB20, AF522272; H36B, AF522269; A909, AY126439; SB1, AF522270; BS4, AF522268; SB10, AF522271; and BS1, AF522265. For strain SB35, the sequence of the entire bac gene, including the part encoding the XPZ region, has been reported previously (19).
RESULTS
Size variation and sequence periodicity in the XPZ region.
The two published sequences of the β protein include 30 and 40 XPZ motifs (19, 24). However, the β protein encoded by one of these strains (24) is expressed at a very low level (31), which made it unclear whether size variation in the XPZ region is compatible with normal surface expression of the β protein. The first part of this study was therefore aimed at analyzing whether the XPZ region is present in all surface-expressed β proteins, whether it varies in length in these β proteins, and whether the characteristic sequence periodicity is present in all XPZ regions. For this purpose, we studied 11 GBS strains that expressed β on the surface, as judged by their ability to bind IgA-Fc (data not shown). These strains included SB35, which was used for sequencing of the structural gene for β, the bac gene (19).
The β proteins expressed by the 11 GBS strains were isolated in almost pure form by incubating bacterial suspensions at elevated pH, a procedure that causes selective release of β protein (35). Analysis of these β proteins by SDS-PAGE demonstrated a limited size variability (Fig. 1A). Western blot analysis showed that all proteins bound IgA-Fc and were recognized by anti-β antibodies, confirming that they were β proteins. Moreover, all of these β proteins were recognized by an antiserum specific for the XPZ region, implying that they all contained an XPZ region (data not shown).
To analyze whether the size variation between different β proteins could be explained by a change in the length of the XPZ region, the corresponding region of the gene was amplified by PCR. The size of the PCR product correlated with the size of the purified β protein, suggesting that the limited size variability was indeed due to variation in the length of the XPZ region. In contrast, the PCR product corresponding to a region located upstream of the XPZ region did not show any variation in size (data not shown).
Sequence analysis of the XPZ region in the 11 β-expressing strains (Fig. 1B) showed that the size of this region varied from 30 three-residue XPZ motifs (in strains SB35, BS26, and SB20) to 50 such motifs (in strain BS22). The amino acid sequence of the XPZ region was identical in some strains, and a total of seven different sequences were identified in the 11 β proteins. The characteristic sequence periodicity was strictly maintained in all XPZ regions. As indicated in Fig. 1B, the different protein sequences may all be derived from the shortest sequence by the addition of blocks containing multiples of six amino acid residues, i.e., by the addition of multiples of six codons at the DNA level.
To analyze whether the XPZ region also varies in size within a bacterial strain, strains A909 and H36B were grown in 10 ml of broth and diluted 100-fold for 10 consecutive days, corresponding to ∼66 generations of growth. For each of the final cultures, 15 colonies were picked, and the length of the XPZ region was analyzed by PCR. No size variation was seen in this analysis, which would have detected a change in length corresponding to six codons. Thus, the XPZ region appears to be stable within a strain under laboratory conditions.
The XPZ region is not required for surface expression of β protein.
For some surface proteins of streptococci and staphylococci, it has been suggested that the proline-rich region threads through the peptidoglycan layer (16, 18, 45), suggesting that this region is important for surface localization of the protein. Moreover, it has been reported that a proline-rich region in the P1 adhesin of Streptococcus mutans is required for surface expression (12). This situation made it of interest to analyze whether the XPZ region was needed for surface expression of β. For this purpose, we constructed a bac gene with an in-frame deletion covering the entire XPZ region, and a plasmid carrying this construct (plasmid pLZΔxpz) was used to transform a GBS strain (the Δbac strain) in which the bac gene had been deleted, generating the Δbac/pLZΔxpz strain. Surface expression of β was analyzed in this strain and in a strain expressing the wild-type β protein (the Δbac/pLZbac strain), with the Δbac strain as the negative control (Fig. 2A).
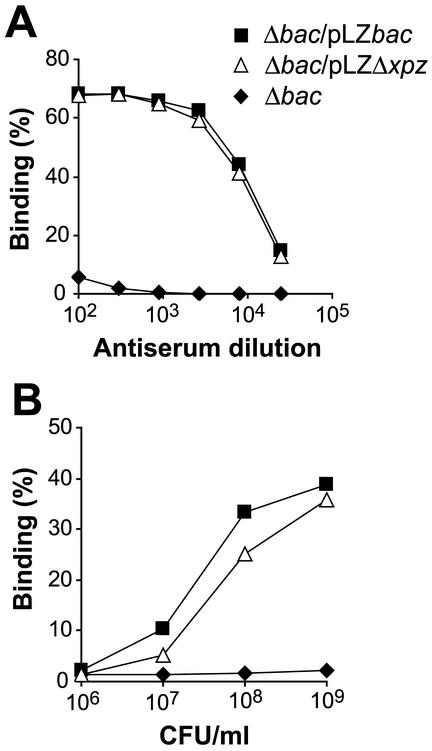
FIG. 2.
A mutant of the β protein lacking the XPZ region is expressed on the bacterial surface. (A) Surface expression of β was analyzed in the Δbac strain, in which the chromosomal bac gene (encoding β) has been deleted, in the Δbac strain transcomplemented with a plasmid carrying the intact bac gene (Δbac/pLZbac), and in the Δbac strain transcomplemented with a plasmid encoding a β protein (βΔXPZ) lacking the XPZ region (Δbac/pLZΔxpz). The analysis was performed with an antiserum directed against the N-terminal B6 fragment of β, which does not include the XPZ region (19) (Fig. 1). This antiserum did not react with the β-negative strain but reacted equally well with the strain expressing the wild-type β protein and that expressing βΔXPZ, implying that the XPZ region is not required for surface expression of the β protein. (B) Binding of radiolabeled IgA to the three strains described for panel A. The Δbac/pLZΔxpz strain bound IgA almost as well as the control Δbac/pLZbac strain, implying that the XPZ region is not required for the ability of surface-exposed β to bind IgA-Fc. The experiments were performed at least three times, with similar results.
For this analysis, we used an antiserum directed against the N-terminal B6 fragment of β, which does not include the XPZ region (Fig. 1B). This antiserum reacted equally well with the strain expressing the wild-type β protein and that expressing the XPZ deletion variant of β (designated βΔXPZ) but did not react with the β-negative Δbac strain (Fig. 2A). Moreover, the binding of IgA-Fc was only slightly reduced for the strain expressing βΔXPZ compared to the strain expressing the wild-type protein (Fig. 2B). These data indicate that the XPZ region is not required for surface expression of the β protein and that it is not required for IgA-Fc binding to surface-expressed β.
The XPZ region is exposed on the bacterial cell surface.
An antiserum directed against a synthetic peptide derived from the XPZ region (Fig. 1B) was used to analyze whether this region is exposed on the bacterial surface. The specificity of this antiserum was first studied by Western blot analysis of purified preparations of the β protein and the β variant lacking the XPZ region, the βΔXPZ protein (Fig. 3A). Both proteins reacted with antibodies to the N-terminal B6 fragment of β, but the anti-XPZ serum reacted only with the wild-type β protein. Thus, the anti-XPZ serum could be used to specifically detect the XPZ region.
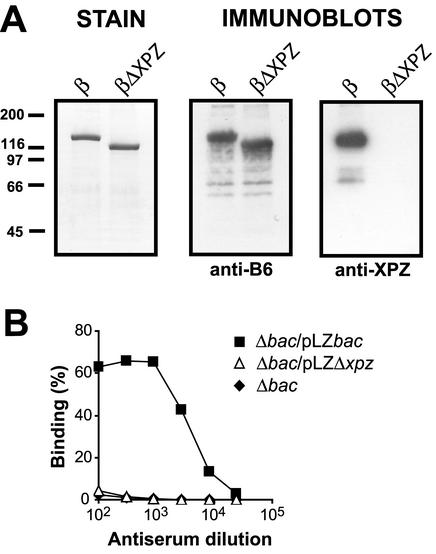
FIG. 3.
Use of specific antibodies to demonstrate that the XPZ region is exposed on the surface of GBS. (A) Analysis of the specificity of antibodies directed against the XPZ region. Similar amounts of purified wild-type β protein or the βΔXPZ mutant protein, which lacks the XPZ region, were subjected to Western blot analysis. A Coomassie-stained gel is shown on the left. Two identical blotting membranes were probed with antibodies to the N-terminal B6 fragment of β (anti-B6) or antibodies directed against a 24-residue synthetic peptide derived from the XPZ region (anti-XPZ), as indicated. Bound antibodies were detected by incubation with radiolabeled protein G, followed by autoradiography. In a control blot with preimmune serum, no signals were obtained. (B) Use of specific anti-XPZ antibodies to analyze whether the XPZ region is exposed on the bacterial surface. The anti-XPZ serum described for panel A was analyzed for reactivity with the β-negative Δbac mutant, with the Δbac strain transcomplemented with the wild-type bac gene (Δbac/pLZbac), and with the Δbac/pLZΔxpz strain, which expresses the β mutant lacking the XPZ region. Bound antibodies were detected with radiolabeled protein G. The strain expressing the wild-type protein but not the other two strains reacted with anti-XPZ antibodies, indicating that the XPZ region is exposed on the surface of GBS. The experiments were performed three times, with similar results.
The anti-XPZ serum reacted with bacteria expressing the β protein but not with the isogenic β-negative Δbac mutant (Fig. 3B), indicating that the XPZ region is at least partially exposed on the bacterial surface and is not buried in the cell wall. As expected, the anti-XPZ serum did not react with a GBS strain expressing the βΔXPZ protein. Although these data indicated that the XPZ region is expressed on the surface of GBS, it seemed possible that the result was due to β protein that had been released from the bacteria, followed by (nonspecific) readsorption to the surface. A control experiment was therefore performed in which a suspension of the β-negative Δbac mutant (109 CFU/ml) was incubated for 30 min at 23°C in the presence of pure wild-type β protein (100 μg/ml). The bacteria were then washed and analyzed for reactivity with anti-XPZ serum. In this experiment, the anti-XPZ antibodies did not bind to the bacteria (data not shown), implying that the reactivity of these antibodies with the wild-type strain was not due to β that had been released and readsorbed.
Although the analysis with anti-XPZ antibodies indicated that the XPZ region is exposed on the bacterial surface, the data did not exclude the possibility that this region is surface exposed in only some β proteins or only on some bacteria. To analyze this problem, we compared the reactivity of antiserum directed against the N-terminal surface-exposed B6 region of β with that of antiserum against the XPZ region. These two antisera were first analyzed for reactivity with pure β immobilized in microtiter wells, i.e., under conditions in which all parts of the β protein should be equally accessible to antibodies. Under these conditions, the titer of the anti-XPZ serum was ∼2-fold lower than the titer of the anti-B6 serum (Fig. 4A). When the same two antisera were analyzed for reactivity with β present on the bacterial surface, the titer of the anti-XPZ serum was also ∼2-fold lower (Fig. 4B). This result shows that the N-terminal region and the XPZ region are equally accessible to antibodies whether β is present in pure form or is exposed on the surface of GBS. Finally, electron microscopic analysis with immunogold techniques showed that β is surface exposed on virtually all bacteria (data not shown). Together, these data indicate that the XPZ region is fully surface exposed on a GBS strain expressing the β protein.
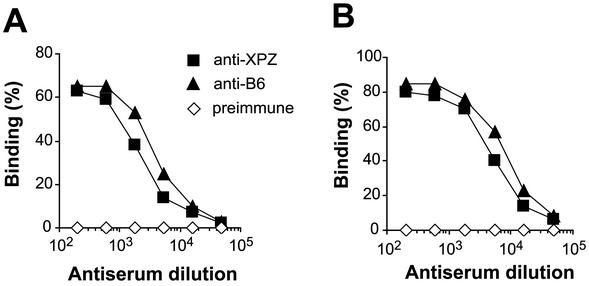
FIG. 4.
The XPZ region and the N-terminal B6 region of β are equally accessible to antibodies on the surface of GBS. (A) Reactivity of antibodies directed against the XPZ region or the B6 region with pure β immobilized in microtiter wells. The titer of the anti-XPZ serum was ∼2-fold lower than that of the anti-B6 serum. As expected, a preimmune control serum did not react with β. (B) Reactivity of the same three sera with β expressed on the surface of GBS strain A909. The data are similar to those shown in panel A, implying that the XPZ region and the B6 region are equally accessible to antibodies on the bacterial surface. The experiments were performed three times, with similar results.
CD spectroscopy indicates that the XPZ region has PPII helix structure.
A sequence of four or more prolines in a row adopts the conformation of a left-handed PPII helix, an extended structure with exactly three residues per turn (29, 55, 59). This conformation is also known to be adopted by many regions that are rich in proline but contain other amino acids, including some regions in which every third residue is proline (59).
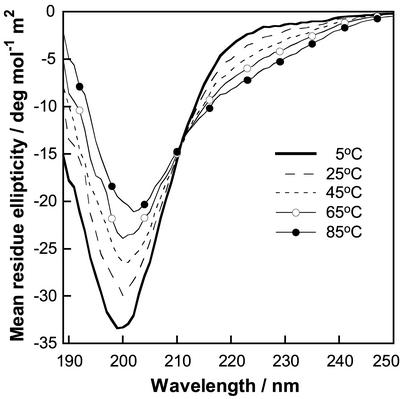
To analyze whether the XPZ region adopts a PPII structure, we characterized a 24-residue synthetic peptide derived from the XPZ region. This peptide, designated (XPZ)8, was studied by circular dichroism (CD) spectroscopy (Fig. 5). At 5°C, the CD spectrum of (XPZ)8 showed a deep negative peak at 200 nm (−33 deg mol−1 m2). The CD spectrum of polyproline was characterized by a large negative band (−59 deg mol−1 m2) at 205 nm and a small positive band (+4.6 deg mol−1 m2) at 228 nm (23). However, many other peptides with the PPII helical structure display spectra very similar to the one we observed here for (XPZ)8, with a minimum at around 200 and the positive peak at 228 nm missing (2, 39, 48).
FIG. 5.
CD spectra of a 24-residue synthetic peptide derived from the XPZ region. The peptide studied, designated (XPZ)8, includes eight XPZ motifs, and its sequence is shown in the box in Fig 1B. This figure shows CD spectra for 54 μM (XPZ)8 in 0.15 M NaF and 5 mM potassium phosphate buffer, pH 7.0. Each spectrum is an average of eight scans in a 1-mm quartz cuvette at 5°C, 25°C, 45°C, 65°C, and 85°C.
The spectral shifts relative to polyproline are often attributed to slight deviation from the ideal PPII helical structure, the presence of cis-proline residues, or formation of higher-order helical forms (4, 43). Raising the temperature to 25, 45, 65, and 85°C causes a gradual decrease in intensity of the negative band, a shift towards a higher wavelength, and the appearance of a negative shoulder at 220 to 230 nm. Similar temperature effects are seen for many other PPII structures and are due to unfolding to an unordered state. In summary, we may conclude that (XPZ)8 adopts a left-handed PPII helix that is disrupted at elevated temperature.
Because of the intriguing sequence of alternating positive and negative charges in the Z position, CD spectra were recorded as a function of salt and pH to evaluate the electrostatic contribution to the structure. Spectra recorded at 0.005, 0.15, and 0.7 M ionic strength superimposed completely (data not shown), indicating that screening of the electrostatic interactions does not affect the structure of (XPZ)8. Another way to perturb potentially stabilizing contributions from attractive electrostatic interactions between opposite charges is to go to extreme pHs. One may expect that the Z position carries alternating positive and uncharged residues at pH 1.5 and alternating negative and uncharged side chains at pH 12. The CD spectra obtained at pH 1.5 and pH 12 were, however, very similar to that at pH 7, but with a small decrease in the intensity at 200 nm (by 8% at pH 1.5 and by 12% at pH 12) (data not shown). The lack of substantial salt and pH effects shows that ionic interactions are only minor determinants of the PPII structure of (XPZ)8, which is hence governed by other forces.
DISCUSSION
The data reported here indicate that the proline-rich XPZ region of the streptococcal β protein is exposed on the bacterial surface and adopts the conformation of a left-handed PPII helix. Although the exact function of the XPZ region is not yet known, these findings shed new light on the properties of proline-rich regions in surface proteins of streptococci and staphylococci. In particular, our data indicate that the XPZ region is not required for cell wall anchoring and/or surface expression of the β protein. Rather, the surface exposure of this region and the knowledge that proline-rich regions commonly participate in protein-protein interactions (59) suggest that the XPZ region could promote interactions with host proteins or with other bacterial surface proteins. The XPZ region could also have a structural role, e.g., it might act as a scaffold that allows surface exposure of other regions of the β protein. However, our data do not support this hypothesis, because deletion of the XPZ region had little or no effect on surface exposure of the N-terminal IgA-binding part of β.
The conclusion that the XPZ region adopts the conformation of a PPII helix was based on CD analysis of the (XPZ)8 peptide, which exhibits the characteristic sequence periodicity. Because this periodicity is conserved throughout the XPZ region, it seems likely that the intact XPZ region is a PPII helix. This extended and solvent-exposed conformation, which has been studied extensively due to its importance for protein-protein interactions (1, 29), may even occur in regions lacking proline (1). The importance of this conformation is underlined by its presence in each of the three strands of collagen (7, 59) and by the finding that class II major histocompatibility complex molecules bind peptides in a PPII-like conformation (22, 42).
Because there are exactly three residues per turn in a PPII helix (29, 59) and because the XPZ region exhibits three-residue periodicity, the helix corresponding to the XPZ region is expected to be amphipathic and to have three surfaces with very different properties, corresponding to the X, P, and Z residues. The X surface is mainly hydrophobic, although with a fair number of hydroxyl groups. The proline rings will line up to form an extended hydrophobic surface, while the Z residues will form a hydrophilic stretch of alternating charges. This separation of surface properties may allow the XPZ region to interact simultaneously with molecules having binding sites of very different characters. Possibly, the alternating charges of the third residue could also contribute to dimerization of two staggered XPZ regions by a zipper-like mechanism (46).
The XPZ region varies in size between different β proteins and is present in all β proteins analyzed, and the sequence periodicity is strictly maintained in XPZ regions of different lengths. These data suggest that the XPZ region is important for the function of the β protein, and they are in good agreement with recent studies that employed PCR to demonstrate size variation in the XPZ region (8, 30). The mechanisms that cause size variation in the XPZ region are not known, and it is not clear whether the mechanisms commonly used to explain size variability in repetitive sequences (10, 14) can also explain size variation in the XPZ region, because this region has a periodic rather than a repetitive sequence. Possibly, the size variability arises through an error-prone process (41), causing duplications and deletions of multiples of six codons, followed by a selection that eliminates mutants that do not retain the typical periodicity.
In this context, it is of interest that three of the four proline codons are used for the central residue in the XPZ motif, suggesting that the XPZ region is indeed subject to a high mutation rate and to a selection process responsible for conservation of the periodicity (33). The selection of XPZ variants that retain the sequence periodicity may reflect a structural requirement, while the sequence variability could reflect immunological selection of antigenically different variants, a hypothesis implying that sequence variability in the XPZ region allows GBS to escape host immunity.
In summary, we have identified a PPII helix with a highly periodic sequence that is exposed on the surface of GBS. To our knowledge, a surface structure of this type has not previously been identified in bacteria. Finally, we note that “natural” antibodies directed against self-antigens preferentially recognize epitopes rich in proline and were suggested to provide a first line of defense against pathogens expressing proline-rich surface proteins (56). Conversely, it seems possible that surface-exposed proline-rich regions in pathogens may contribute to the emergence of natural antibodies or even to the development of autoimmunity. This argument is particularly relevant with regard to the XPZ region studied here, because the PPII helix structure of this region may favor presentation by major histocompatibility complex class II molecules (22, 42).
Acknowledgments
We are grateful to Lars Stenberg and Charlotta Bergquist for contributions in the early stages of this study and to Erik Lindahl for advice. We also thank the students in the Prokaryotic Molecular Genetics course at Lund University for help with sequence analysis.
This work was supported by grants from the Swedish Research Council (grant 09490 to G.L. and grant 11552 to S.L.), the Medical Faculty of Lund University, the Royal Physiographic Society in Lund, the Swedish Society for Medical Research, and the Golje, Groschinsky, Hedberg, Hierta, Jerring, Kock, Lundström, and Österlund foundations.
REFERENCES
- 1.Adzhubei, A. A., and M. J. E. Sternberg. 1993. Left-handed polyproline II helices commonly occur in globular proteins. J. Mol. Biol. 229:472-493. [DOI] [PubMed] [Google Scholar]
- 2.Anthonyraj, K. J., T. Karunakaran, and P. A. Raj. 1998. Bactericidal activity and poly-L-proline II conformation of the tandem repeat sequence of human salivary mucin glycoprotein (MG2). Arch. Biochem. Biophys. 356:197-206. [DOI] [PubMed] [Google Scholar]
- 3.Areschoug, T., M. Stålhammar-Carlemalm, I. Karlsson, and G. Lindahl. 2002. Streptococcal β protein has separate binding sites for human factor H and IgA-Fc. J. Biol. Chem. 277:12642-12648. [DOI] [PubMed] [Google Scholar]
- 4.Arnott, S., and S. D. Dover. 1968. The structure of poly-L-proline II. Acta Crystallogr. B 24:599-601. [DOI] [PubMed] [Google Scholar]
- 5.Baker, C. J., and M. S. Edwards. 1995. Group B streptococcal infection, p. 980-1054. In J. S. Remington and J. O. Klein (ed.), Infectious diseases of the fetus and the newborn infant. W. B. Saunders Company, Philadelphia, Pa.
- 6.Beall, B., G. Gherardi, R. R. Facklam, and S. K. Hollingshead. 2000. Pneumococcal pspA sequence types of prevalent multiresistant pneumococcal strains in the United States and of internationally disseminated clones. J. Clin. Microbiol. 38:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bella, J., M. Eaton, B. Brodsky, and H. M. Berman. 1994. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science 266:75-81. [DOI] [PubMed] [Google Scholar]
- 8.Berner, R., M. Ruess, S. Bereswill, and M. Brandis. 2002. Polymorphisms in the cell wall-spanning domain of the C protein β-antigen in clinical Streptococcus agalactiae isolates are caused by genetic instability of repeating DNA sequences. Pediatr. Res. 51:106-111. [DOI] [PubMed] [Google Scholar]
- 9.Bevanger, L., and A. I. Naess. 1985. Mouse-protective antibodies against the Ibc proteins of group B streptococci. Acta Pathol. Microbiol. Immunol. Scand. B 93:121-124. [DOI] [PubMed] [Google Scholar]
- 10.Bi, X., and L. F. Liu. 1996. recA-independent DNA recombination between repetitive sequences: mechanisms and implications. Prog. Nucleic Acid Res. Mol. Biol. 54:253-292. [DOI] [PubMed] [Google Scholar]
- 11.Boehm, M. K., J. M. Woof, M. A. Kerr, and S. J. Perkins. 1999. The Fab and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: a study by X-ray and neutron solution scattering and homology modelling. J. Mol. Biol. 286:1421-1447. [DOI] [PubMed] [Google Scholar]
- 12.Brady, L. J., D. G. Cvitkovitch, C. M. Geric, M. N. Addison, J. C. Joyce, P. J. Crowley, and A. S. Bleiweis. 1998. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect. Immun. 66:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bzymek, M., and S. T. Lovett. 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. USA 98:8319-8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahnestock, S. R., P. Alexander, J. Nagle, and D. Filpula. 1986. Gene for an immunoglobulin-binding protein from a group G streptococcus. J. Bacteriol. 167:870-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischetti, V. A. 2000. Surface proteins of gram-positive bacteria, p. 11-24. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 17.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guss, B., M. Uhlén, B. Nilsson, M. Lindberg, J. Sjöquist, and J. Sjödahl. 1984. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur. J. Biochem. 138:413-420. (Correction, Eur. J. Biochem. 143:685.) [DOI] [PubMed] [Google Scholar]
- 19.Hedén, L.-O., E. Frithz, and G. Lindahl. 1991. Molecular characterization of an IgA receptor from group B streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. Eur. J. Immunol. 21:1481-1490. [DOI] [PubMed] [Google Scholar]
- 20.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1986. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus. Repetitive structure and membrane anchor. J. Biol. Chem. 261:1677-1686. [PubMed] [Google Scholar]
- 21.Husmann, L. K., J. R. Scott, G. Lindahl, and L. Stenberg. 1995. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect. Immun. 63:345-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jardetzky, T. S., J. H. Brown, J. C. Gorga, L. J. Stern, R. G. Urban, J. L. Strominger, and D. C. Wiley. 1996. Crystallographic analysis of endogenous peptides associated with HLA-DR1 suggests a common, polyproline II-like conformation for bound peptides. Proc. Natl. Acad. Sci. USA 93:734-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenness, D. D., C. Sprecher, and W. C. Johnson, Jr. 1976. Circular dichroism of collagen, gelatin, and poly(proline) II in the vacuum ultraviolet. Biopolymers 15:513-521. [DOI] [PubMed] [Google Scholar]
- 24.Jerlström, P. G., G. S. Chhatwal, and K. N. Timmis. 1991. The IgA-binding β antigen of the c protein complex of group B streptococci: sequence determination of its gene and detection of two binding regions. Mol. Microbiol. 5:843-849. [DOI] [PubMed] [Google Scholar]
- 25.Jerlström, P. G., S. R. Talay, P. Valentin-Weigand, K. N. Timmis, and G. S. Chhatwal. 1996. Identification of an immunoglobulin A binding motif located in the β-antigen of the c protein complex of group B streptococci. Infect. Immun. 64:2787-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kastern, W., U. Sjöbring, and L. Björck. 1992. Structure of peptostreptococcal protein L and identification of a repeated immunoglobulin light chain-binding domain. J. Biol. Chem. 267:12820-12825. [PubMed] [Google Scholar]
- 27.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 28.Kehoe, M. A. 1994. Cell-wall-associated proteins in Gram-positive bacteria. New Comp. Biochem. 27:217-261. [Google Scholar]
- 29.Kelly, M. A., B. W. Chellgren, A. L. Rucker, J. M. Troutman, M. G. Fried, A.-F. Miller, and T. P. Creamer. 2001. Host-guest study of left-handed polyproline II helix formation. Biochemistry 40:14376-14383. [DOI] [PubMed] [Google Scholar]
- 30.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J. Clin. Microbiol. 40:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreikemeyer, B., and P. G. Jerlström. 1999. An Escherichia coli-Enterococcus faecalis shuttle vector as a tool for the construction of a group B Streptococcus heterologous mutant expressing the β antigen (Bac) of the C protein complex. FEMS Microbiol. Lett. 180:255-262. [DOI] [PubMed] [Google Scholar]
- 32.Lachenauer, C. S., C. J. Baker, M. J. Baron, D. L. Kasper, C. Gravekamp, and L. C. Madoff. 2002. Quantitative determination of immunoglobulin G specific for group B streptococcal β C protein in human maternal serum. J. Infect. Dis. 185:368-374. [DOI] [PubMed] [Google Scholar]
- 33.Lévi-Strauss, M., M. C. Carroll, M. Steinmetz, and T. Meo. 1988. A previously undetected MHC gene with an unusual periodic structure. Science 240:201-204. [DOI] [PubMed] [Google Scholar]
- 34.Lindahl, G., and B. Åkerström. 1989. Receptor for IgA in group A streptococci: cloning of the gene and characterization of the protein expressed in Escherichia coli. Mol. Microbiol. 3:239-247. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl, G., B. Åkerström, J.-P. Vaerman, and L. Stenberg. 1990. Characterization of an IgA receptor from group B streptococci: specificity for serum IgA. Eur. J. Immunol. 20:2241-2247. [DOI] [PubMed] [Google Scholar]
- 36.Lindgren, P.-E., M. J. McGavin, C. Signäs, B. Guss, S. Gurusiddappa, M. Höök, and M. Lindberg. 1993. Two different genes coding for fibronectin-binding proteins from Streptococcus dysgalactiae. The complete nucleotide sequences and characterization of the binding domains. Eur. J. Biochem. 214:819-827. [DOI] [PubMed] [Google Scholar]
- 37.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, R. M. Ireland, S. D. Reid, G. G. Adams, and J. M. Musser. 2000. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun. 68:6542-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, B. J. Shelvin, E. A. Graviss, and J. M. Musser. 2001. Identification and characterization of a second extracellular collagen-like protein made by group A Streptococcus: control of production at the level of translation. Infect. Immun. 69:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma, K., L.-S. Kan, and K. Wang. 2001. Polyproline II helix is a key structural motif of the elastic PEVK segment of titin. Biochemistry 40:3427-3438. [DOI] [PubMed] [Google Scholar]
- 40.Madoff, L. C., L. C. Paoletti, J. Y. Tai, and D. L. Kasper. 1994. Maternal immunization of mice with group B streptococcal type III polysaccharide-beta C protein conjugate elicits protective antibody to multiple serotypes. J. Clin. Investig. 94:286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcotte, E. M., M. Pellegrini, T. O. Yeates, and D. Eisenberg. 1998. A census of protein repeats. J. Mol. Biol. 293:151-160. [DOI] [PubMed] [Google Scholar]
- 42.Murthy, V. L., and L. J. Stern. 1997. The class II MHC protein HLA-DR1 in complex with an endogenous peptide: implications for the structural basis of the specificity of peptide binding. Structure 5:1385-1396. [DOI] [PubMed] [Google Scholar]
- 43.Naganagowda, G. A., T. L. Gurunaja, and M. J. Levine. 1998. Delineation of conformational preferences in human salivary statherin by 1H, 31P NMR and CD studies: sequential assignment and structure-function correlations. J. Biomol. Struct. Dyn. 16:91-107. [DOI] [PubMed] [Google Scholar]
- 44.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pancholi, V., and V. A. Fischetti. 1988. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. J. Bacteriol. 170:2618-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perutz, M. 1994. Polar zippers: their role in human disease. Protein Sci. 3:1629-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pleass, R. J., T. Areschoug, G. Lindahl, and J. M. Woof. 2001. Streptococcal IgA-binding proteins bind in the Cα2-Cα3 interdomain region and inhibit binding of IgA to human CD89. J. Biol. Chem. 276:8197-8204. [DOI] [PubMed] [Google Scholar]
- 48.Raj, P. A., and M. Edgerton. 1995. Functional domain and poly-L-proline II conformation for candidacidal activity of bactenecin 5. FEBS Lett. 368:526-530. [DOI] [PubMed] [Google Scholar]
- 49.Rasmussen, M., and L. Björck. 2001. Unique regulation of SclB—a novel collagen-like surface protein of Streptococcus pyogenes. Mol. Microbiol. 40:1427-1438. [DOI] [PubMed] [Google Scholar]
- 50.Rasmussen, M., A. Edén, and L. Björck. 2000. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 68:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell-Jones, G. J., E. C. Gotschlich, and M. S. Blake. 1984. A surface receptor specific for human IgA on group B streptococci possessing the Ibc protein antigen. J. Exp. Med. 160:1467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuchat, A. 1999. Group B streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 53.Stålhammar-Carlemalm, M., T. Areschoug, C. Larsson, and G. Lindahl. 2000. Cross-protection between group A and group B streptococci due to cross-reacting surface proteins. J. Infect. Dis. 182:142-149. [DOI] [PubMed] [Google Scholar]
- 54.Stålhammar-Carlemalm, M., L. Stenberg, and G. Lindahl. 1993. Protein Rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177:1593-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stapley, B. J., and T. P. Creamer. 1999. A survey of left-handed polyproline II helices. Protein Sci. 8:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tchernychev, B., S. Cabilly, and M. Wilchek. 1997. The epitopes for natural polyreactive antibodies are rich in proline. Proc. Natl. Acad. Sci. USA 94:6335-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wästfelt, M., M. Stålhammar-Carlemalm, A.-M. Delisse, T. Cabezon, and G. Lindahl. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J. Biol. Chem. 271:18892-18897. [DOI] [PubMed] [Google Scholar]
- 58.Whatmore, A. M. 2001. Streptococcus pyogenes sclB encodes a putative hypervariable surface protein with a collagen-like repetitive structure. Microbiology 147:419-429. [DOI] [PubMed] [Google Scholar]
- 59.Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 61.Yu, H., J. K. Chen, S. Feng, D. C. Dalgarno, A. W. Brauer, and S. L. Schreiber. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76:933-945. [DOI] [PubMed] [Google Scholar]