Abstract
Tolerance of environmental stress, especially low pH, by Streptococcus mutans is central to the virulence of this organism. The Clp ATPases are implicated in the tolerance of, and regulation of the response to, stresses by virtue of their protein reactivation and remodeling activities and their capacity to target misfolded proteins for degradation by the ClpP peptidase. The purpose of this study was to dissect the role of selected clp genes in the stress responses of S. mutans, with a particular focus on acid tolerance and adaptation. Homologues of the clpB, clpC, clpE, clpL, clpX, and clpP genes were identified in the S. mutans genome. The expression of clpC and clpP, which were chosen as the focus of this study, was induced at low pH and at growth above 40°C. Inactivation of ctsR, the first of two genes in the clpC operon, demonstrated that CtsR acts as a repressor of clp and groES-EL gene expression. Strains lacking ClpP, but not strains lacking ClpC, were impaired in their ability to grow under stress-inducing conditions, formed long chains, aggregated in culture, had reduced genetic transformation efficiencies, and had a reduced capacity to form biofilms. Comparison of two-dimensional protein gels from wild-type cells and the ctsR and clpP mutants revealed many changes in the protein expression patterns. In particular, in the clpP mutant, there was an increased production of GroESL and DnaK, suggesting that cells were stressed, probably due to the accumulation of denatured proteins.
In order to survive adverse environmental conditions, bacteria undergo a complex program of differential gene expression that leads to the transient induction of a subset of proteins that protect the cell from damage caused by the accumulation of unfolded and misfolded proteins (22). Most of these polypeptides, also known as heat shock proteins (HSPs), are molecular chaperones or proteases that play crucial roles in promoting protein folding, assembly, and degradation during normal growth and, in particular, under stress-inducing conditions (5, 13). The HSP100/Clp ATPases constitute a large family of closely related proteins that perform important housekeeping functions, including protein reactivation and remodeling activities typical of molecular chaperones, and target specific proteins for degradation by the ClpP peptidase (35, 40). The number of identified members of the Clp family has significantly increased in the past few years (33). These highly conserved and ubiquitously distributed proteins are classified on the basis of the presence of one or two ATP-binding domains and the occurrence of specific signature sequences (33). The class 1 Clp proteins, such as ClpA, ClpB, and ClpC, have two ATP-binding domains and are distinguished by the length of the spacer regions separating the ATP-binding domains, whereas the class 2 Clp proteins (such as ClpX and ClpY) display a single domain. Although designated a Clp protein, for caseinolytic protease, ClpP itself is unrelated to the Clp ATPase family and has only peptidase activity. However, when ClpP associates with members of the Clp ATPase family, it acts as a serine protease and prevents the accumulation of altered proteins that might be toxic for the bacteria under stress-inducing conditions (25).
Despite their ubiquity and high degree of conservation, mechanisms for the regulation of heat shock genes in bacteria are fairly diverse. In Escherichia coli, transcription of the heat shock regulon requires either the σ32 subunit of RNA polymerase or the minor sigma factor σE (4). It was demonstrated that the DnaK, DnaJ, and GrpE chaperones play a central role in the control of the general stress response by targeting σ32 for degradation by the FtsH protease. In Bacillus subtilis, at least four classes of stress-regulated genes have been identified (10, 16). Genes coding for the ClpP protease and for the ClpC and ClpE ATPases of B. subtilis are designated as part of the class 3 stress regulon (10, 11, 20). Class 3 stress genes are negatively controlled by CtsR, which is the product of the first gene of the clpC operon and which binds to a directly repeated heptad that usually overlaps with the transcriptional initiation site or the −35 and −10 sequences of the ctsR, clpE, and clpP genes. This target sequence and the CtsR repressor were also shown to be present in the regulatory region of clp genes from several gram-positive bacteria, including some Streptococcus species (10).
Streptococcus mutans is recognized as one of the major etiological agents of human dental caries (23). In the oral cavity, S. mutans colonizes the teeth and is able to withstand substantial environmental insults, including pH fluctuations, severe carbohydrate restriction, and oxidative stresses. Among the environmental stresses encountered in dental biofilms, rapid and significant acidification may be the most extreme challenge. S. mutans is intrinsically more acid resistant than many oral bacteria and can mount an acid tolerance response (ATR), which allows the organisms to adapt to growth at a low pH. The ATR of S. mutans is characterized thus far by enhanced glycolytic capacities, increased activity of the proton-translocating F-ATPases, enhanced DNA repair capacity, and acquisition of cross-protection against other stresses (2, 14, 15, 31). The scope of the ATR appears significant as assessed by two-dimensional (2-D) gel electrophoresis (36), but few of the gene products that appear to be necessary for acid tolerance in S. mutans have been identified and essentially nothing is known about genetic control of the expression of the ATR.
Our laboratory is primarily interested in dissecting the role of stress genes in acid tolerance and the regulation of adaptation to acidic conditions. Previously, it was demonstrated that the major molecular chaperones, DnaK and GroEL, are controlled by the HrcA repressor and are intimately involved in responses to environmental acidification (18, 21). In addition, we have shown that DnaK is essential for cell survival and that reduction in the levels of this chaperone leads to an acid-sensitive phenotype (21). Here, we identified and analyzed the transcriptional regulation of the genes coding for the Clp protease machinery in S. mutans. The expression and functional significance of the S. mutans ClpC and ClpP proteins in stress tolerance, competence development, and biofilm formation were also investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain DH10B was grown in Luria broth, and S. mutans strain UA159 was grown in brain heart infusion (BHI) broth. When needed, kanamycin (40 μg ml−1 for E. coli or 500 μg ml−1 for S. mutans) or ampicillin (100 μg ml−1) was added to the media. For heat shock of S. mutans, cells were grown in BHI at 37°C to mid-exponential phase (optical density at 600 nm [OD600] ≅ 0.5) and aliquots of the cultures were then transferred to a water bath at 42°C for various periods of time. For studies involving acid shock and acid adaptation, S. mutans was grown in continuous chemostat culture at pH 7.0 or 5.0 as previously described (18). To evaluate the impact of the loss of the clpP, clpC, or ctsR gene on stress resistance, the wild-type and mutant strains were grown overnight in BHI, diluted 50-fold into BHI, and incubated at 37°C (control) or at high temperatures (40, 42, or 43°C), at a low pH (pH 5.0, 37°C), or at 37°C in the presence of streptomycin (25 μg ml−1). The ability to form stable biofilms was assessed by growing the cells in a 96-well (flat-bottom) cell culture cluster (Costar 3595; Corning, Inc., Corning, N.Y.) containing biofilm medium as originally described by Loo et al. (24), with minor modifications for S. mutans developed by Bhagwat et al. (3).
DNA methods.
Chromosomal DNA was prepared from S. mutans as previously described (6). Plasmid DNA was isolated from E. coli by using QIAgen columns (Qiagen, Chatsworth, Calif.), and restriction and DNA-modifying enzymes were obtained from Life Technologies, Inc. (Gaithersburg, Md.), New England Biolabs (Beverly, Mass.), or MBI Fermentas (Amherst, N.Y.). PCRs were carried out with 100 ng of S. mutans chromosomal DNA by using Taq DNA polymerase, and PCR products were purified using the QIAquick kit (Qiagen). DNA was introduced into S. mutans by natural transformation (30) and into E. coli by electroporation (32). Southern blot analyses were carried out under high-stringency conditions as detailed by Sambrook et al. (32).
Strain construction.
The strains used in this study are listed in Table 1. To construct insertion mutations in ctsR, clpC, and clpP, DNA fragments, each approximately 1.7 kb in length, containing the genes of interest were amplified by PCR and cloned onto pGEM5 (Promega, Madison, Wis.) as SphI-SpeI fragments. The resulting plasmids were used to insert a kanamycin resistance gene (Kanr) that is flanked by strong transcription and translation terminators (ΩKan) (29) into the ctsR, clpC, and clpP genes. A second clpP mutant was generated by the insertion of a nonpolar Kanr marker (19) to exclude the possibility that the phenotype observed for the clpP polar insertion was due to polar effects on downstream genes. To inactivate ctsR in a way that would allow for the expression of clpC, which is located immediately downstream of ctsR, a nonpolar construct with the ΩKan element followed by the Streptococcus salivarius urease promoter (PureI) (8) was used. This approach was successfully used to generate a nonpolar insertion in the S. mutans hrcA gene that allowed for expression of the genes downstream of hrcA in the dnaK operon (21). Plasmids containing the Kanr gene marker were then isolated and used to transform S. mutans. Transformants were selected on BHI agar with kanamycin, and Southern blot analysis was used to confirm that the gene inactivation desired had occurred as planned.
TABLE 1.
S. mutans strains used in this study
| Strains | Relevant genotype | Source or reference |
|---|---|---|
| UA159 | Wild-type | Laboratory stock |
| JL-ctsR | ctsR::ΩKan | This study |
| JL-NPctsR | NP-ctsR::ΩKan with S. salivarius urease promoter | This study |
| JL-clpC | clpC::ΩKan | This study |
| JL-NPclpP | clpP::nonpolar Kan | This study |
| JL-clpP | clpP::ΩKan | This study |
Assay of genetic competence.
Transformation efficiencies of S. mutans UA159 and its derivatives were determined by preparing competent cells as previously described and transforming them with plasmid-borne copies of the luxS or brpA (for biofilm regulatory protein A) genes of S. mutans into which erythromycin antibiotic resistance (Ermr) markers had been inserted (41). Transformants were enumerated by plating cells on BHI agar containing erythromycin (5 μg ml−1).
2-D polyacrylamide gel electrophoresis.
2-D electrophoresis was performed at Kendrick Labs, Inc. (Madison, Wis.) by following the protocol described by O'Farrell (28) but using conditions described elsewhere (21). The protein concentration of the samples was determined by a bicinchoninic acid assay (Sigma, St. Louis, Mo.). Gels containing equal amounts of protein were silver stained and dried between sheets of cellophane.
RNA methods.
Total RNA was extracted from chemostat-grown cultures of S. mutans or from mid-exponential phase batch cultures by following a protocol described by Chen et al. (8). Total RNA was separated on a 1.0% formaldehyde gel as described elsewhere (1). RNAs were UV cross-linked to the membranes, and filter membranes were probed with internal fragments of the S. mutans clpC, clpE, clpP, ctsR, or groEL gene labeled with [α-32P]dATP (NEN Life Science, Boston, Mass.). All hybridizations and washes were carried out under high-stringency conditions. Signals obtained on autoradiographs were quantified using an IS1000 digital imaging system (Alpha Innotech Co., San Leandro, Calif.). Primer extensions were carried out with the oligonucleotide designated EXTCTSR (5′-CTGACTGGGAACAACAC-3′) by a protocol described previously (1), with primer annealing and reverse transcription performed at 42°C.
RESULTS
Identification of clp genes in S. mutans.
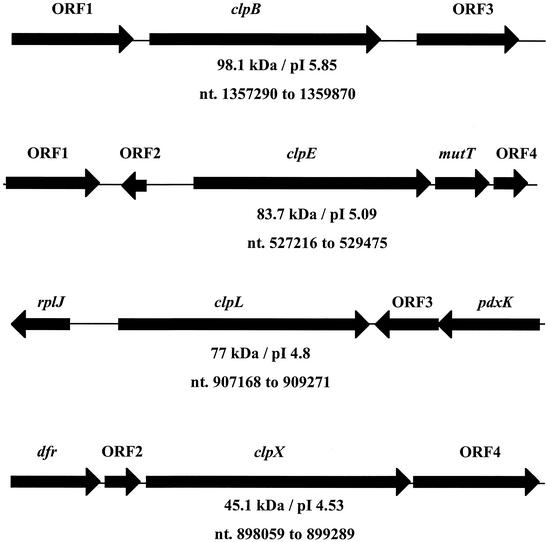
The highly conserved amino acid sequence of the Clp proteins of E. coli, B. subtilis, Streptococcus pneumoniae, and Lactococcus lactis were used to identify putative clp genes in the S. mutans genome by using resources available at the University of Oklahoma's Advanced Center for Genome Technology. A total of five open reading frames (ORFs) were identified within the S. mutans genome. BLAST search analysis of the deduced amino acid sequences of the five ORFs indicated high degrees of similarity to ClpB, ClpC, ClpE, ClpL, and ClpX from other organisms and, therefore, the genes were designated clpB, clpC, clpE, clpL, and clpX. The structural organization and location of the clpB, clpE, clpL, and clpX genes in the chromosome are shown in Fig. 1, and the clpC operon organization is shown in Fig. 2. We also took advantage of the complete sequence of the S. mutans genome to identify the clpP gene and the gene coding for the putative repressor of clp gene expression in gram-positive bacteria, ctsR. In order to begin investigating the CtsR regulon in S. mutans, we began a more detailed analysis of the ctsR, clpC, and clpP genes. The clpC gene (2,373 bp in length), which is located at nucleotides 1899196 to 1901634 in the S. mutans genome, encoded a polypeptide with a predicted molecular mass of 87.8 kDa that shared the highest levels of similarity with a putative S. pyogenes ClpC (73% identity). Immediately upstream, and overlapping by 1 nucleotide with the clpC start codon, was another ORF of 463 bp. The 17.5-kDa product encoded by this ORF showed 47% identity (65% similarity) with the class 3 heat shock gene repressor CtsR from B. subtilis. Finally, elsewhere in the chromosome (positions 1587548 to 1588136), a clpP homologue coding for a predicted 21.6-kDa protein that shared 86% identity with a putative S. pyogenes ClpP and high levels of homology to other known ClpP proteins was identified.
FIG. 1.
Schematic diagram of the clpB, clpE, clpL, and clpX loci and flanking regions. ORF1 and ORF3 in the clpB locus encode putative phosphosugar mutase and dihydrolipoamide dehydrogenase proteins, respectively. ORF1 in the clpE locus encodes a putative transposase, and mutT codes for a MutT/nudix family protein. The rplJ and pdxK genes in the clpL locus encode putative 50S ribosomal L10 and pyridoxal kinase proteins, respectively. The dfr gene in the clpX locus is predicted to encode a dihydrofolate reductase protein, and ORF4 encodes a putative GTP-binding protein. ORF2 and ORF4 in the clpE locus, ORF3 in the clpL locus, and ORF2 in the clpX locus code for hypothetical proteins. See the text for more details. Arrows represent the orientation of the genes. The molecular mass, isoelectric points, and nucleotide positions (nt.) in the chromosome of the putative Clp proteins are shown underneath the genes.
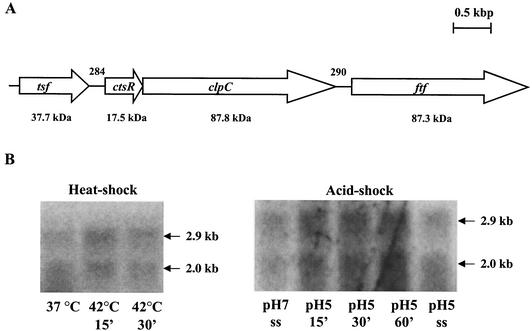
FIG. 2.
(A) Genetic organization of the S. mutans ctsR/clpC operon and flanking regions. The tsf gene is predicted to encode an elongation factor, and ftf encodes a levansucrase precursor. Arrows indicate the direction of transcription. The estimated molecular masses from deduced amino acid sequences are shown below the genes, and the numbers above the schematic indicate the size of the intergenic regions in base pairs. (B) Northern blot analysis of S. mutans UA159 clpC. RNA was isolated from batch- or chemostat-grown cells that were subjected to heat shock (42°C) or acid shock (pH 5.0) (ss [steady state]). Total RNA (10 μg per lane) was separated in a 1.0% gel, transferred to a nylon membrane, and hybridized to a clpC-specific probe.
Transcriptional analysis of the clpC operon.
The clpC and ctsR genes were found to be part of a bicistronic operon. An ORF coding for a protein highly similar to a putative elongation factor (Tsf) from S. pyogenes was found 5′ to ctsR, and a levansucrase precursor from S. mutans GS-5 (ftf [34]) was identified 3′ to clpC (Fig. 2A). A promoter was mapped by primer extension (data not shown) 42 bases upstream of the ctsR start codon with a −10 sequence identical to σA-type promoters and a partially conserved −35 sequence (TTCCAA versus TTGACA of σA-type promoters) located 16 bp upstream of the −10 sequence.
Northern analyses with RNA isolated from the wild-type strain by using ctsR or clpC internal fragments as probes revealed a transcript of approximately 2.9 kb, indicating that ctsR and clpC are cotranscribed from the σA-type promoter located upstream of ctsR (Fig. 2B). Curiously, a smaller transcript of approximately 2 kb in size was detected with the ctsR and clpC probes, but the size of this transcript does not correlate well with the known sizes of ctsR and clpC. However, both transcripts were equally induced after heat and acid stresses (twofold when cells were subjected to heat shock at 42°C for 15 min and fourfold after 30 min following acid shock at pH 5.0). No significant differences in the clpC mRNA levels in steady-state cells grown at a pH of either 7.0 or 5.0 were observed (Fig. 2B).
Transcriptional analysis and stress induction profile of clpP.
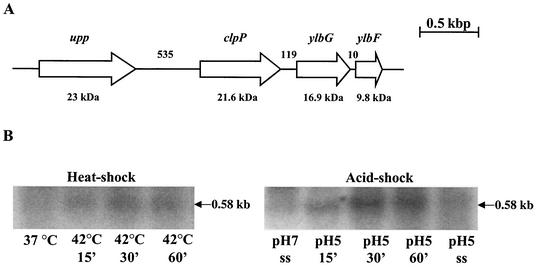
Recently, a σA-type promoter was identified upstream of the clpP gene in S. pneumoniae (7). Analysis of the sequence of the region upstream of the S. mutans clpP gene revealed a putative σA-type promoter identical in sequence to the one that was mapped in S. pneumoniae (TTGACC-N17-TATAAT). Upstream from clpP and separated by 535 bp, an ORF that coded for a protein that shares 93% identity with an S. salivarius uracil phosphoribosyltransferase (upp) was identified (Fig. 3A). Analysis of the region 3′ to the clpP stop codon revealed two ORFs with the highest degree of similarity to those coding for the B. subtilis ylbF and ylbG gene products (35 and 43% identities, respectively). Recently, it was demonstrated that, in B. subtilis, YlbF and perhaps YlbG are involved in sporulation and in competence development by increasing the stability or synthesis of the competence factor ComK at the posttranscriptional level (37).
FIG. 3.
(A) Schematic representation of the S. mutans clpP gene and flanking regions. The upp gene is predicted to encode a uracil phosphoribosyltransferase, and ylbF and ylbG have been implicated in competence development and sporulation in B. subtilis. Arrows indicate the direction of transcription. The estimated molecular masses from deduced amino acid sequences are shown below the genes, and the numbers above the schematic indicate the size of the intergenic regions in base pairs. (B) Northern blot analysis of S. mutans UA159 clpP. RNA was isolated from batch- or chemostat-grown cells that were subjected to heat shock (42°C) or acid shock (pH 5.0) (ss [steady state]). Total RNA (10 μg per lane) was separated in a 1.0% gel, transferred to a nylon membrane, and hybridized to a clpP-specific probe.
Northern blot analysis with RNA isolated from the wild-type strain was used to demonstrate a stress-inducible transcript of 0.58 kb, which corresponds to the length of clpP, as well as two larger transcripts that were not stress inducible but nevertheless corresponded in size to the 16S and 23S ribosomal RNAs (Fig. 3B). It has been demonstrated in other organisms that ClpP is an HSP whose synthesis can be induced by a variety of stresses. Densitometric analysis of Northern blots of RNAs from batch- or chemostat-grown cells revealed that the levels of clpP mRNA were increased threefold when cells were subjected to heat shock at 42°C for 30 min or fourfold after 30 min following acid shock at pH 5.0 (Fig. 3B). Comparisons of the clpP mRNA levels from steady-state cultures at pH 7.0 or 5.0 showed a 1.5-fold induction at low pH, similar to what has been observed for the dnaK gene (18).
Effects of the inactivation of ctsR, clpC, and clpP on growth under stress-inducing conditions.
The clpC, clpP, and ctsR genes were inactivated by inserting the ΩKan cassette in the 5′ portion of the genes to give the strains listed in Table 1. Although there was strong evidence from Northern analyses that clpP is a monocistronic transcript, a second clpP mutant strain (JL-NPclpP) was constructed using a nonpolar kanamycin cassette to eliminate the possibility that the phenotype observed in the clpP mutant strain could be due to polar effects on the downstream genes. Similarly, a second CtsR-deficient strain (JL-NPctsR) was constructed with a nonpolar element that allowed for clpC transcription. Cell morphology, competence development, and stress sensitivities of the polar and nonpolar ctsR and clpP mutants were examined, but no differences were observed. Therefore, because we had already conducted a detailed characterization of the nonpolar ctsR mutant (JL-NPctsR) and the polar clpP mutant strain (JL-clpP), these two strains were used throughout the studies.
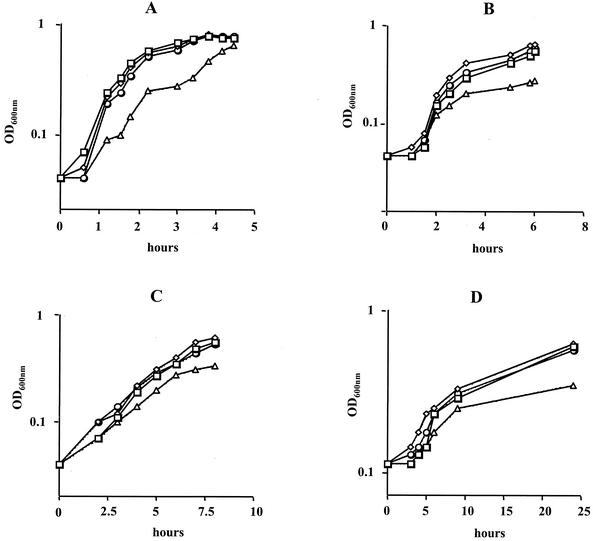
Under standard growth conditions, i.e., 37°C in a 5% CO2 atmosphere, the JL-NPctsR and JL-clpC mutant strains grew as well as the wild-type strain (Fig. 4A). At temperatures above 37°C, the wild-type, JL-NPctsR, and JL-clpC strains were able to grow at temperatures of up to 43°C. However, at temperatures above 40°C, growth rates of the strains were reduced and the cells tended to clump and adhere to the sides of the glass culture tubes. We also tested the ability of the strains to grow at pH 5.0 or in the presence of streptomycin (25 μg ml−1), an antibiotic known to increase mistranslation. Under all those conditions, the JL-NPctsR and JL-clpC strains displayed growth rates comparable to those for the wild type (Fig. 4B, C, and D).
FIG. 4.
Growth curves of S. mutans wild-type (□), JL-clpC (◊), JL-NPctsR (○), and JL-clpP (▵) strains grown at 37°C (A), at 42°C (B), in the presence of streptomycin (25 μg ml−1) (C), and at pH 5.0 (D). A typical result representative of three independent experiments is shown.
Under standard growth conditions, the clpP mutant grew more slowly than the wild-type strain but was able to reach a similar OD600 before entering stationary phase (Fig. 4A). Although the ClpP-deficient strain was able to grow at temperatures of up to 40°C, growth of JL-clpP was impaired at 42°C (Fig. 4B) and completely inhibited at 43°C (data not shown). When grown at 42°C at pH 5.0 or in the presence of streptomycin (25 μg ml−1), the lag phase and early log phase of the clpP mutant were comparable to those of the wild-type strain. However, under every stress-inducing condition tested, the cell yields achieved by the clpP mutant strain were significantly lower than those achieved by the wild-type strain (Fig. 4B, C, and D). These results clearly indicated a stress-sensitive phenotype for the clpP mutant strain.
Inactivation of CtsR leads to high-level expression of clpC, clpE, clpP, and groEL.
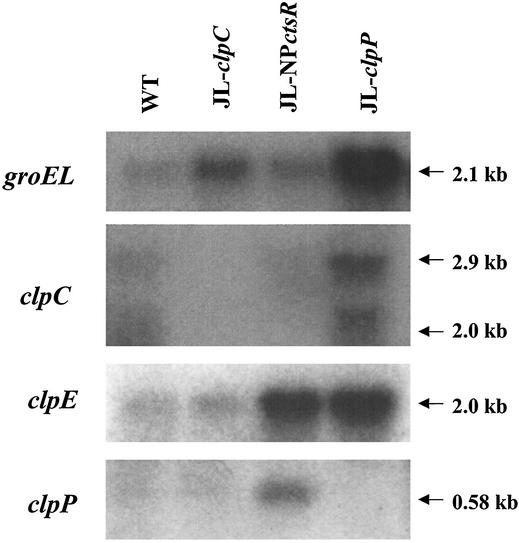
Northern blot analysis with RNA isolated from mid-exponential-phase cultures of CtsR-deficient (JL-NPctsR) and wild-type strains of S. mutans demonstrated that groEL, clpE, and clpP mRNAs were present in greater amounts in the mutant strain (two-, six-, and sixfold increases, respectively) (Fig. 5). It was not possible to determine the effect of ctsR inactivation on clpC expression, since the transcription from the cognate promoter of clpC in the ctsR mutant was disrupted by a polar Kanr marker and clpC was driven by the S. salivarius urease promoter. The levels of expression of groEL, clpC, clpE, and clpP mRNA were also compared in the wild-type strain and the clpC and clpP mutants. In the JL-clpC strain, the levels of clpE and clpP mRNA were comparable to what was found for the wild-type strain whereas the level of groEL mRNA was increased about fourfold (Fig. 5). This finding suggests that ClpC may have a regulatory effect on groEL expression or that the levels of groEL mRNA were elevated due to the accumulation of damaged proteins that were once targeted by ClpC to the protease machinery. Northern analyses with RNA isolated from the clpP mutant strain grown to mid-log phase under normal conditions revealed that levels of groEL, clpC, and clpE mRNA were induced nine-, three-, and sixfold, respectively (Fig. 5). Thus, lack of ClpP also causes an effective induction of these stress genes.
FIG. 5.
Northern blot analysis of UA159 (WT) and its derivatives. Cells were grown in BHI at 37°C to mid-exponential phase. RNA (10 μg per lane) was isolated from cells and separated in a 1.0% gel, transferred to a nylon membrane, and hybridized to groEL-, clpC-, clpE-, and clpP-specific probes.
Protein expression patterns of clp and ctsR mutants.
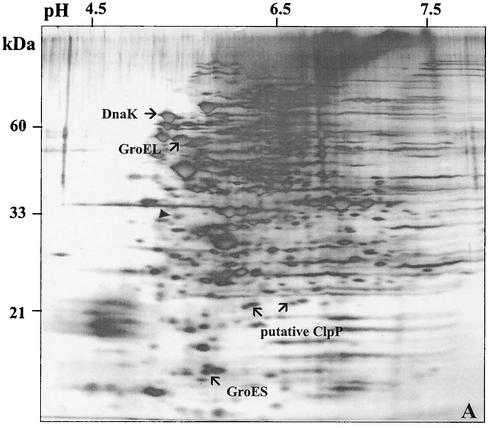
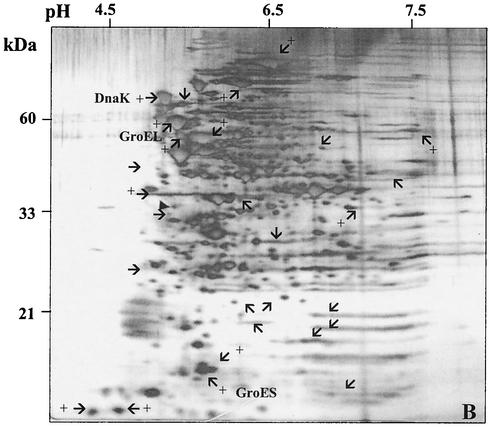
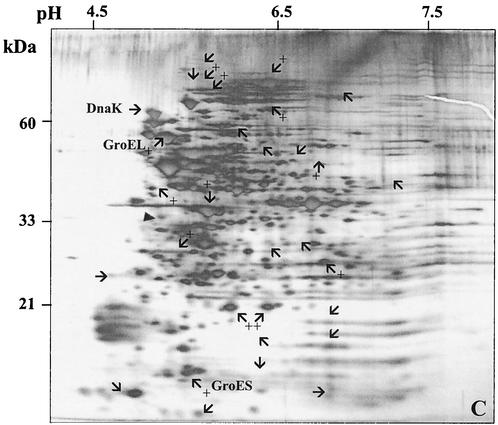
A 2-D gel approach was used to determine whether there were detectable changes in the synthesis of the proteome of S. mutans due to the loss of ClpP (JL-clpP) or CtsR (JL-NPctsR). In the clpP mutant strain, the levels of the molecular chaperones DnaK and GroEL-ES were elevated at least twofold, indicating that cells were perceiving a stress-inducing condition, probably due to the accumulation of denatured proteins that are normally degraded by ClpP. Although the twofold increase of GroEL levels in the clpP mutant strain does not correlate very well with the mRNA levels (ninefold increase), it has been observed that large increases in the amount of groEL mRNA are needed to effect significant changes in the absolute quantity of this protein in the cells (21). Additionally, the levels of at least 25 other proteins were altered (10 up-regulated, 15 down-regulated) in the clpP mutant compared to those in the wild-type strain growing under the same conditions (Fig. 6B). When compared to the wild-type 2-D pattern, at least 13 proteins were up-regulated in the JL-NPctsR strain whereas 17 proteins showed reduced synthesis (Fig. 6C). Two protein spots, approximately 22 kDa each, were strongly expressed in the JL-NPctsR strain and absent in the JL-clpP strain. The predicted molecular mass of the S. mutans ClpP protein is 21 kDa, so perhaps one or both of the two spots is the ClpP protein.
FIG. 6.
2-D protein gels of S. mutans strains UA159 (A), JL-clpP (B), and JL-NPctsR (C). Cells were grown in BHI to an OD600 of 0.5. The silver-stained proteins that exhibited more obvious differences in the clpP and ctsR mutants in comparison to the wild type are indicated. Proteins with enhanced expression are indicated with arrows and a plus symbol, and proteins with reduced synthesis are indicated by arrows only. The DnaK, GroEL, GroES, and putative ClpP proteins are indicated as such. The filled triangle indicates tropomyosin protein (33 kDa, pI 5.2) loaded as an internal control.
Role of ClpC and ClpP in competence development.
To assess the role of the S. mutans ClpC and ClpP proteins in competence, the efficiency of transformation of the ctsR, clpC, and clpP mutants was evaluated by transforming the strains with constructs containing an Ermr marker as detailed in Materials and Methods. Similar results were obtained regardless of the transforming DNA. As shown in Table 2, the JL-clpC strain showed a modest although not statistically significant increase in transformation efficiency whereas the inactivation of ctsR had no effect. When compared to the parental strain, the clpP mutant showed a fourfold reduction in the number of transformants (Table 2).
TABLE 2.
Transformation frequency of S. mutans strains
| Strain | Competence frequency (%)a |
|---|---|
| UA159 | 100 |
| JL-clpC | 140 (± 52.9) |
| JL-NPctsR | 71 (± 23.8) |
| JL-clpP | 26 (± 13.1) |
The numbers presented here are the means (and standard deviations) from three independent experiments.
Role of clp genes in biofilm formation.
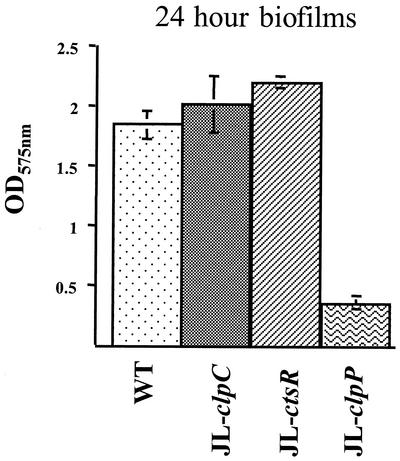
The capacity of the JL-NPctsR, JL-clpC, and JL-clpP strains to form biofilms in the wells of 96-well microtiter plates was evaluated. The data obtained after 24 h of growth indicated that the inactivation of clpC and ctsR led to a slight, albeit consistent, increase in biofilm formation (>9 and >19%, respectively) compared to that in the wild-type strain. Inactivation of clpP resulted in a fivefold reduction in biofilm formation (Fig. 7). To exclude the possibility that the differences observed were due to variations in the growth capacity of the strains, the final OD600 of the cultures in microtiter plates was assessed. All the strains grew equally well over a 24-h period (data not shown), but the clpP strain showed a consistent four- to fivefold reduction in stable biofilm formation after 24, 48, and 72 h.
FIG. 7.
Biofilm formation by S. mutans UA159 (WT) and its derivatives. Cultures were grown in a 96-well microtiter plate containing biofilm medium at 37°C in 5% CO2 for 24 h. Absorbance at 575 nm of the crystal violet extracted from the 24-h biofilms with ethanol is indicated. The graph shows the averages and standard deviations from three independent experiments.
DISCUSSION
It is recognized that protein degradation is a critical process that ensures the optimal functioning of the cell by preventing the accumulation of denatured proteins or by regulating the stability of regulatory proteins. The Clp protein complex, which possesses both proteolytic and chaperone activities, has been implicated in stress survival in many organisms. In B. subtilis, mutations in the clpC, clpP, and clpX genes were shown to affect competence development, motility, growth at high temperatures, degradative enzyme synthesis, and sporulation (26, 38, 39). In L. lactis, clpE mutants formed smaller colonies on agar media containing puromycin although growth of this mutant as well as of the ΔclpB and ΔclpC strains did not seem to be affected by heat or high concentrations of salt (17). It was also shown that inactivation of the L. lactis clpP gene results in a stress-sensitive phenotype (12). Finally, in S. pneumoniae, it was shown that ClpP and ClpE are required for thermotolerance but that ClpC is dispensable for growth at a high temperature (7).
In this study, we have identified and characterized the clpC and clpP genes of S. mutans. Additional members of the Clp ATPase family, namely, clpB, clpE, clpL, and clpX, were also identified in the S. mutans genome, but the roles of these genes in homeostasis and stress tolerance remain to be elucidated. Similar to what has been observed in other gram-positive organisms, loss of ClpP in S. mutans resulted in a stress-sensitive phenotype. The clpP mutant grew more slowly than the wild-type strain, formed long chains, aggregated, and had an 80% reduction in biofilm formation. Moreover, the levels of the chaperones DnaK, GroEL, ClpC, and ClpE were elevated in the ClpP-deficient strain, indicating that ClpP deficiency induced a stress response in the cells, probably as a result of the accumulation of denatured proteins that are normally targeted to the ClpP protease. It has been observed that clp genes play a role in cell division. In B. subtilis, ClpC- and ClpP-deficient strains produce elongated cells and form long chains, especially under stress-inducing conditions. The longer chains observed in the S. mutans clpP mutant clearly suggest an impairment in cell septation and/or autolysis. However, the precise role of ClpP in cell division and autolysis remains to be investigated. In contrast to the ClpP-deficient strains, the ClpC-deficient strain was able to grow as well as the wild-type strain and, under certain stress-inducing conditions, the clpC mutant grew slightly faster and reached a higher yield than the parental strain. Additionally, the clpC mutant did not show alterations in cell morphology, chain length, or biofilm-forming capabilities. Therefore, unlike what was observed in B. subtilis, it appears that ClpC does not play a major role in stress tolerance in S. mutans and possibly in other lactic acid bacteria.
In B. subtilis, the cellular concentration of the competence factor comK has been shown to be controlled by the ClpP and ClpC proteolytic machinery, along with the MecA protein, which acts as an adapter, connecting ComK to ClpC (38, 39). There has been a report of a MecA-like gene in S. mutans, but it is not known whether this gene product has a function similar to that of B. subtilis MecA (9). By searching the S. mutans databases, we found no evidence of a comK homologue. In S. pneumoniae, it appears that ClpP is responsible for controlling the levels of the competence genes comC, comD, and comE whereas ClpC does not appear to be involved in competence (7). The efficiency of transformation of the S. mutans clpP, clpC, and ctsR mutants was determined by the acquisition of an Ermr marker via homologous recombination. Compared to the parental strain, the clpP mutant displayed a fourfold reduction in the number of transformants. In contrast, there was a relatively small increase in the frequency of transformation for the JL-clpC and JL-NPctsR strains. The reduced competence in the S. mutans clpP mutant strain differs from the complete absence of transformation in a B. subtilis clpP mutant (26). Notably, the clpP gene is located upstream of two ORFs, with homologies to the products encoded by ylbF and ylbG of B. subtilis. In B. subtilis, disruption of ylbF leads to a dramatic decrease in the expression of comK (37). There was no evidence of a comK homologue in the S. mutans databases, and a nonpolar clpP mutant strain also showed reduced competence (data not shown). However, we did not analyze the expression of ylbF and ylbG in the clpP mutant strains and it is possible that the observed effects on competence in the clpP mutant are due, at least in part, to the altered expression of downstream genes. In S. pneumoniae, it was shown that ClpP acts as a negative regulator of comCDE expression under conditions that do not support competence development (7). In order to definitively address whether ClpP has a similar regulatory function in competence development by S. mutans, further studies to investigate expression of the S. mutans com genes in the clpP mutant strain are under way.
In gram-positive bacteria, there are at least two different transcriptional repressors, HrcA and CtsR, that regulate expression of heat shock genes (27). In many gram-positive organisms, the levels of ClpC, ClpE, and ClpP are controlled by the CtsR repressor that binds to a consensus heptad direct-repeat sequence (A/GGTCAAA-NAN-A/GGTCAAA) located in the regulatory regions of these genes (10). More recently, it was demonstrated in S. pneumoniae that the CtsR repressor binds to DNA elements in the clpC, clpE, clpP, and groESL promoter regions that differ in sequence from the consensus sequence originally suggested for CtsR binding (7). Comparisons of the regulatory regions of the clpC, clpE, clpP, and groESL genes from S. pneumoniae and S. mutans disclosed almost identical target sequences for CtsR binding, strongly supporting the idea that CtsR negatively controls the expression of those genes in S. mutans. The role of CtsR in the regulation of clpP and groES-EL was confirmed in the ctsR mutant in this study by Northern blotting and 2-D gel electrophoresis. Previously, it was shown that HrcA is a major regulator of groESL expression (21), and thus this is the second report (7) to demonstrate that the groESL operon is under dual heat shock control by the HrcA and CtsR repressors. In addition to GroES-EL and the Clp proteins, which have been shown to be repressed by CtsR, proteomic analysis revealed that levels of an additional 13 proteins were elevated in the ctsR mutant strain. It is reasonable to speculate that the up-regulated proteins in the strain bearing the ctsR mutation may constitute new members of the CtsR regulon.
It is also interesting to note that the clpP mutant strain showed a reduced capacity to form biofilms. Although the basis for this phenotype is unknown, we can speculate that ClpP may act by controlling the stability or activity of transcriptional regulators of biofilm maturation. Another possibility, which is not mutually exclusive, may be that tolerance of nutrient limitation or accumulation of growth-inhibitory metabolites resulting from mass transport limitations in biofilms may require a functional ClpP for protein turnover. The linkage of stress tolerance with biofilm formation is currently under investigation in our laboratory.
In conclusion, this study describes the genetic and transcriptional organization, and begins to shed light on the physiological significance, of the clp genes of S. mutans. The data obtained demonstrate that ClpP plays a critical role in the survival of common environmental stresses, most likely by preventing the accumulation of denatured proteins. In contrast, ClpC does not appear to be essential, although proteomic analysis reveals that there is a significant impact of ClpC deficiency on the proteome of S. mutans (data not shown). The disparity between the changes in expression profiles and the lack of apparent stress sensitivity may be explained by compensation for ClpC deficiency by other chaperones. In addition, CtsR and ClpP may be part of complex regulatory networks, either through the direct repression of expression or by the modulation of the stability of other proteins. Work is in progress to identify the proteins with altered expression in the ctsR and clpP mutants, with the goals of revealing members of the CtsR regulon and substrates of the Clp protease and of shedding light on how these gene products interact to achieve physiologic homeostasis and stress resistance.
Acknowledgments
We thank Jacqueline Abranches and Margaret Chen for critical reading of the manuscript.
This study was supported by grants DE12236 and DE13239 from the National Institute for Dental and Craniofacial Research.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Belli, W. A., and R. E. Marquis. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57:1134-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 4.Bukau, B. 1993. Regulation of the Escherichia coli heat shock response. Mol. Microbiol. 9:671-680. [DOI] [PubMed] [Google Scholar]
- 5.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 6.Burne, R. A., K. Schilling, W. H. Bowen, and R. E. Yasbin. 1987. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J. Bacteriol. 169:4507-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y. M., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.1 urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cvitkovitch, D. G. 2001. Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:217-243. [DOI] [PubMed] [Google Scholar]
- 10.Derré, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 11.Derré, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 12.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815-823. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, K., R. C. Faustoferri, and R. G. J. Quivey. 1999. Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol. Microbiol. 31:1489-1498. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton, I. R., and N. D. Buckley. 1991. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol. Immun. 6:65-71. [DOI] [PubMed] [Google Scholar]
- 16.Hecker, M., W. Schumann, and U. Volker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 17.Ingmer, H., F. K. Vogensen, K. Hammer, and M. Kilstrup. 1999. Disruption and analysis of the clpB, clpC, and clpE genes in Lactococcus lactis: ClpE, a new Clp family in gram-positive bacteria. J. Bacteriol. 181:2075-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 19.Kremer, B. H. A., M. Kraan, P. J. Crowley, I. R. Hamilton, L. J. Brady, and A. S. Bleiweis. 2001. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J. Bacteriol. 183:2543-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruger, E., and M. Hecker. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 180:6681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemos, J. A. C., Y. Y. M. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and the role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindquist, S., and E. A. Craig. 1988. The heat shock proteins. Annu. Rev. Genet. 22:631-677. [DOI] [PubMed] [Google Scholar]
- 23.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurizi, M. R., W. Clark, S. H. Kim, and S. Gottesman. 1990. ClpP represents a unique family of serine proteases. J. Biol. Chem. 265:12546-12552. [PubMed] [Google Scholar]
- 26.Msadek, T., V. Dartois, F. Kunst, M. L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 27.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 3:1-8. [DOI] [PubMed] [Google Scholar]
- 28.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor protein of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry, D., and H. K. Kuramitsu. 1981. Genetic transformation of Streptococcus mutans. Infect. Immun. 32:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quivey, R. G., Jr., R. C. Faustoferri, K. A. Clancy, and R. E. Marquis. 1995. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol. Lett. 126:257-262. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Schirmer, E. C., J. R. Glover, M. A. Singer, and S. Lindquist. 1996. Hsp100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21:289-296. [PubMed] [Google Scholar]
- 34.Shiroza, T., and H. K. Kuramitsu. 1988. Sequence analysis of the Streptococcus mutans fructosyltransferase gene and flanking regions. J. Bacteriol. 170:810-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Squires, C., and C. L. Squires. 1992. The Clp proteins: proteolysis regulators or molecular chaperones? J. Bacteriol. 174:1081-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svensäter, G., B. Sjogreen, and I. R. Hamilton. 2000. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology 146:107-117. [DOI] [PubMed] [Google Scholar]
- 37.Tortosa, P., M. Albano, and D. Dubnau. 2000. Characterization of ylbF, a new gene involved in competence development and sporulation in Bacillus subtilis. Mol. Microbiol. 35:1110-1119. [DOI] [PubMed] [Google Scholar]
- 38.Turgay, K., L. W. Hamoen, G. Venema, and D. Dubnau. 1997. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, that controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11:3097-3104. [DOI] [PubMed] [Google Scholar]
- 39.Turgay, K., J. Hahn, J. Burghhorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wawrzynow, A., B. Banecki, and M. Zylicz. 1996. The Clp ATPases define a novel class of molecular chaperones. Mol. Microbiol. 21:895-899. [DOI] [PubMed] [Google Scholar]
- 41.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]