Abstract
During the production of fermented dairy products, virulent bacteriophages infecting Lactococcus lactis can delay or stop the milk acidification process. A solution to this biological problem consists of introducing natural phage barriers into the strains used by the dairy industry. One such hurdle is called abortive infection (Abi) and causes premature cell death with no or little phage progeny. Here, we describe the isolation and characterization of a novel Abi mechanism encoded by plasmid pED1 from L. lactis. The system is composed of two constitutively cotranscribed genes encoding putative proteins of 127 and 213 amino acids, named AbiTi and AbiTii, respectively. Site-directed mutagenesis indicated that a hydrophobic region at the C-terminal extremity of AbiTi is essential to the antiphage phenotype. The AbiT system is effective against phages of the 936 and P335 species (efficiency of plaquing between 10−5 and 10−7) and causes a 20-fold reduction in the efficiency to form centers of infection as well as a 10- to 12-fold reduction in the burst size. Its efficacy could be improved by raising the plasmid copy number, but changing the intrinsic ratio of AbiTi and AbiTii did not greatly affect the antiphage activity. The monitoring of the intracellular phage infection process by DNA replication, gene expression, and electron microscopy as well as the study of phage mutants by genome mapping indicated that AbiT is likely to act at a later stage of the phage lytic cycle.
Bacteria have several molecular weapons to defend themselves against invading bacteriophages. Conversely, phages have simple and sophisticated means of avoiding cellular gatekeepers. The dairy fermentation environment provides an enticing and dynamic model for the study of these phage-host interactions, as it comprises a limited number of host strains that are constantly exposed to new phages. Lactococcus lactis is a gram-positive bacterium extensively used for its lactic acid production capability during the manufacture of fermented dairy products. Although phage concentration is normally low in raw and pasteurized milk (36), a specific population can increase very rapidly if phage-sensitive cells are present in the lactococcal starter culture. The consequent lysis of a large number of sensitive cells will delay or halt the fermentation process (2, 38).
In the last two decades, several studies have confirmed the biodiversity of the lactococcal phage population within the dairy plant. These viruses are currently classified into 12 genetically distinct species (32). Recently, it was proposed to reduce the number of lactococcal phage species to 11 (35). Nonetheless, only 936-, c2-, and P335-like phages are repeatedly isolated in the dairy industry (38). Another clear recognition was that several L. lactis strains have plasmids that code for defense mechanisms against bacteriophages. These antiphage systems are classified based on the moment of their action in the phage lytic cycle (17). Some of them prevent the adsorption of the virion to the cell, others block the ejection of the phage DNA (26). Restriction-modification systems act by cutting the foreign DNA that has entered the cell. Finally, abortive infection (Abi) systems can act at any moment between DNA ejection and cell lysis. Contrary to other types of cell defenses, the Abi+ host does not survive the phage infection. However, a low proportion of cells release virions, and the burst size is reduced. Nineteen Abi systems have been characterized at the molecular level in L. lactis (12, 21, 25, 52). Although the number of known Abi systems is rather impressive in this microorganism, very little is known about the specific mechanisms they use to halt phage infection.
Phage-insensitive L. lactis strains have been constructed by introducing natural plasmids coding for phage resistance into phage-sensitive strains. The extensive use of these phage-resistant cultures is plagued by the emergence of phage mutants capable of overcoming the introduced antiviral systems. The mutants characterized to date acquired DNA from plasmids or from the host chromosome (1, 7, 22, 29, 42) and could also overcome Abi systems by point mutations (5, 18). Since the viral populations can adapt to these selective pressures, the search for distinct antiphage barriers is still warranted. This paper describes the molecular characterization of a novel Abi system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown at 37°C in Luria-Bertani medium, and L. lactis was grown at 30°C in M17 (Quélab) supplemented with 0.5% glucose (GM17) or, in the case of W51, with 0.5% lactose (LM17). When needed, antibiotics were added to the media at the following concentrations: 50 μg of ampicillin per ml or 20 μg of chloramphenicol per ml for E. coli, and 5 μg of chloramphenicol or erythromycin per ml for L. lactis.
TABLE 1.
Strains, plasmids, and bacteriophages used in this study
| Strain, plasmid, or phage | Relevant characteristic(s)a | Source |
|---|---|---|
| L. lactis | ||
| W51 | Multiple plasmids, including pED1 | This study |
| LM0230 | Plasmid-free host for 936- and c2-like phages | 37 |
| MG1363 | Plasmid-free host for 936- and c2-like phages | 27 |
| SMQ481 | Host for P335-like phages | 7 |
| E. coli | ||
| DH5alpha | Cloning host | Invitrogen |
| GM48 | dam−/dcm− host | |
| Plasmids | ||
| pBS | Cloning vector for DNA sequencing, Apr, 2.9 kb | Stratagene |
| pSA3 | E. coli-L. lactis shuttle vector, Cmr Tcr Emr, 10.2 kb | 13 |
| pMIG3 | E. coli-L. lactis shuttle vector, Cmr, 5.5 kb | 55 |
| pNZ123 | High-copy vector for L. lactis, Cmr, 2.8 kb | 16 |
| pMG36c | Expression vector for L. lactis, Cmr, 3.4 kb | 53 |
| pED1 | Resident plasmid of W51, Abi+, 12.2 kb | This study |
| Bacteriophages | ||
| p2 | 936 species, small isometric headed, cos type | 43 |
| p2k | 936 species, small isometric headed | 8 |
| sk1 | 936 species, small isometric headed | 10 |
| jj50 | 936 species, small isometric headed | 33 |
| ul36 | P335 species, small isometric headed, pac type | 41 |
| Q30 | P335 species, small isometric headed | 39 |
| Q33 | P335 species, small isometric headed | 39 |
| φ31 | P335 species, small isometric headed | 1 |
| φ50 | P335 species, small isometric headed | 1 |
| c2 | c2 species, prolate headed | 49 |
| eb1 | c2 species, prolate headed | 43 |
| ml3 | c2 species, prolate headed | 43 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Emr, erythromycin resistance; Abi+, active Abi mechanism.
Bacteriophage propagation and microbiological assays.
Bacteriophages used in this study are listed in Table 1. They were propagated as described previously (41). The efficiency of plaquing (EOP) was calculated by dividing the phage titer on the tested strain by the titer on the sensitive control strain. Adsorption assays were conducted as described by Sanders and Klaenhammer (49), and cell survival was assayed by the method of Behnke and Malke (4). One-step growth curves were performed as described previously (40). The efficiency to form centers of infection (ECOI) was calculated by dividing the phage titer, obtained with resistant infected cells, by the titer obtained with the sensitive strain immediately after the adsorption step. The burst size was determined by dividing the average titer after the exponential phase by the average titer before cells began to release virions. The phage lytic cycle was monitored at time intervals by electron microscopy in AbiT− and AbiT+ strains as described previously (8).
DNA isolation and manipulations.
E. coli plasmid DNA was isolated by the method of Birnboim (6). L. lactis plasmid DNA was isolated as described by O'Sullivan and Klaenhammer (46). Phage DNA was prepared as described previously (42), and large amounts were obtained with the Qiagen Lambda Maxi Kit (Qiagen). Total intracellular DNA was isolated by the method of Hill et al. (28). In brief, strains were grown in 10 ml of GM17 until the optical density (600 nm) reached 0.5. Phages were added at a final concentration of 5 × 107 PFU/ml (multiplicity of infection, <1). Isolated DNA was dissolved in 100 μl, and 10 μl was digested and loaded per well. All DNA manipulations were carried out as described by Sambrook and Russell (48). E. coli was transformed with the Gene Pulser II apparatus as indicated by the manufacturer and L. lactis electrotransformation was done as specified by Holo and Nes (31).
DNA sequencing and sequence analysis.
Fragments were cloned in pBS and sequenced on both strands with an ABI Prism 3100 apparatus using the universal forward and reverse primers as well as synthetic primers (Invitrogen). DNA and deduced protein sequences were analyzed with the Wisconsin Package version 9.0 (Genetics Computer Group) (15). The open reading frames (ORFs) were compared with the available databases using BLAST version 2.1.1 (3).
Plasmid construction.
DNA fragments from pED1 cloned into the cloning vectors pMIG3, pNZ123, and pSA3 were designated by the prefixes pED1XX, pED2XX, and pED3XX, respectively. The XX corresponds to the DNA fragments presented in Fig. 1 or below in Table 2. When needed, pBS was first used to clone restriction fragments and to introduce deletions or frameshifts, as it contains more convenient restriction sites. The abiT genes were amplified using primers containing restriction sites (in bold) to generate pED419 and pED420 (primers PED1.1, 5′-GAATTCGAATTCGATTATAAGAAACGGAGGGAC-3′ and PED1.2, 5′-TCTAGATCTAGACCGTTTTCGTAGTATCACGT-3′) as well as pED107 and pED307 (primers PED1.2 and PED1.3, 5′-GTCGACGTCGACGGTGCTTTTTGTCTTTCTAGTT-3′). The PCR products were digested and purified as described elsewhere (20). Site-directed mutagenesis was done by the megaprimer PCR method (48). The mutagenic primers (mutation in bold) were as follows: R108T, 5′-GCTACTTGTAACTGTTAATGATATG-3′; A115E, 5′-TTCCTGTTTCCACTAAGCTACT-3′; A115V, 5′-TTCCTGTTACCACTAAGCTACT-3′; L122R, 5′-ACTTGACTACGTAGACCAAGTA-3′; L112P, 5′-ACTTGACTAGGTAGACCAAGTA-3′.
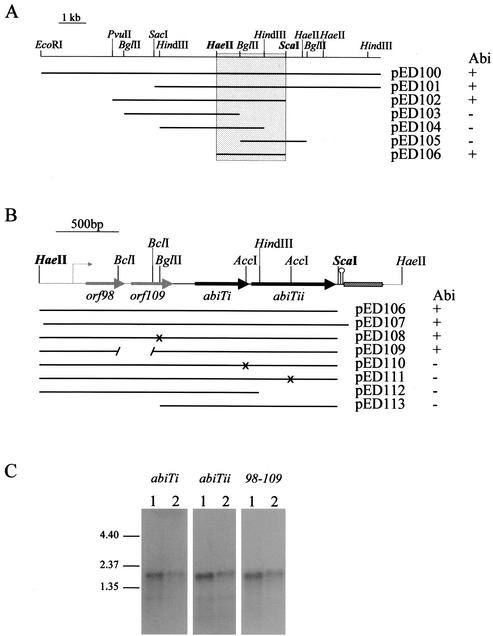
FIG. 1.
Identification and expression of the genetic determinants of AbiT encoded on pED1. (A) Restriction map and pertinent subclones of pED1 with their respective Abi phenotype. The smallest Abi+ region obtained is delimited by a gray box. (B) DNA sequence analysis of the 2,711-bp HaeII fragment and inactivation of the ORFs. The putative promoter is indicated by a thin arrow, and predicted genes are indicated by large arrows. Regions of homology with lactococcal plasmids are shown in gray; the hatched box is a region of homology with E. faecalis and S. pneumoniae transposons. X, frameshift. (C) Hybridization of RNA isolated from AbiT-containing L. lactis strains with a labeled DNA fragment of abiTi, abiTii, and orf98-orf109. Lane 1, MG1363(pED100); lane 2, MG1363(pED107). The size of the transcripts was evaluated with a 0.24- to 9.5-kb RNA ladder (Invitrogen).
TABLE 2.
Site-directed mutagenesis of the C-terminal region of AbiTi
| Plasmid | La 107 | R 108 | V 109 | T 110 | S 111 | S 112 | L 113 | V 114 | A 115 | T 116 | G 117 | I 118 | L 119 | G 120 | L 121 | L 122 | S 123 | O 124 | V 125 | G 126 | I 127 | EOP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pED107 | 6.9 × 10−6 | |||||||||||||||||||||
| pED114 | T | 1.1 × 10−2 | ||||||||||||||||||||
| pED115 | E | 1.0 | ||||||||||||||||||||
| pED116 | V | 8.7 × 10−6 | ||||||||||||||||||||
| pED117 | R | 1.0 | ||||||||||||||||||||
| pED118 | P | 1.0 |
Hydrophobic residues are in bold.
RNA isolation and manipulations.
Cells were grown in 100 ml of GM17 to an optical density of 0.2 and pelleted. For phage RNA isolation, the following steps were added: the pellet was resuspended in 10 ml of GM17 with CaCl2 at 10 mM and phage was added at a multiplicity of infection of 1. Aliquots of 1 ml were taken at different time points, mixed with 100 μl of rifampin at 10 mg/ml, and centrifuged, and the pellet was immediately frozen at −80°C. Total RNA was isolated with the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. After a treatment with RNase-free DNase I (Roche Diagnostic), 4 μg of each RNA sample was migrated on a formaldehyde gel and transferred to a nylon membrane as described by Sambrook and Russell (48). The following DNA fragments were labeled with [α-32P]dATP (Amersham Pharmacia Biotech) by random priming and used as probes: the 256-bp BclI fragment of pED1 for orf98 and orf109, PCR products corresponding to abiTi and abiTii (coordinates 1233 to 1534 and 1599 to 2202), to the orf35 homologue and the mcp gene of p2 (coordinates 20478 to 21179 and 6837 to 7259 in sk1; accession no. AF011378).
Nucleotide sequence accession number.
The DNA sequence of AbiT was deposited in the GenBank database (accession no. AF483000).
RESULTS
Plasmid isolation and classification of the antiphage mechanism.
L. lactis W51 was previously isolated from raw milk and was shown to be resistant to a cocktail of phages (data not shown). However, this strain produces off-flavors when cultivated in milk, thereby preventing its use in commercial applications. We set forth to determine the mechanisms responsible for the phage resistance phenotype in L. lactis W51. The cryptic plasmids of W51 were coelectroporated with the plasmid vector pSA3 (containing an erythromycin resistance gene) into the phage-sensitive, plasmid-free strain LM0230 (Table 1). Colonies isolated on erythromycin-containing media were then tested for phage resistance with phage p2 of the 936 species. All phage-resistant isolates contained, in addition to pSA3, a 12.2-kb plasmid named pED1. Curing of pSA3 produced an erythromycin-sensitive phage-resistant strain, which confirmed the involvement of pED1 in the antiphage activity.
The EOP of the small isometric-headed phage p2 (936 species) on LM0230(pSA3, pED1) was 10−6 at 20, 30, and 39°C. Plaques isolated on the strain remained insensitive to the antiphage mechanism after propagation through a sensitive host, which demonstrates that it is not a restriction-modification system. Plasmid pED1 did not affect the first step of the phage lytic cycle, since the adsorption of phage p2 was 93% ± 3% on LM0230(pSA3) and 96% ± 2% on LM0230(pSA3, pED1). Finally, cell survival was 11% ± 4% for the sensitive strain and 16% ± 6% for the pED1-containing strain. The fact that resistant cells do not survive phage infection proves that the antiphage system carried by pED1 is an Abi mechanism. It was therefore named AbiT.
Cloning and identification of the genetic determinants of AbiT.
Plasmid pED1 was subcloned into the shuttle vector pMIG3 after restriction mapping. The different clones were then introduced into plasmid-free phage-sensitive L. lactis MG1363 and their Abi phenotype was tested with phage p2. The 2.2-kb HaeII-ScaI fragment was the smallest region that conferred phage resistance (Fig. 1A). This restriction fragment was cloned in pBS for DNA sequencing. Bioinformatic analysis revealed that it comprises four ORFs (Fig. 1B) preceded by a putative promoter (coordinates 234 to 262, TTGTAG-17 nucleotides-AATAAT). Since no transcription terminator could be identified on the 2.2-kb HaeII-ScaI fragment, the 2.7-kb HaeII fragment was entirely sequenced (Fig. 1). Two inverted repeats (coordinates 2225 to 2252 and 2264 to 2288) and a stretch of 7 T nucleotides (2289 to 2295) that could act as a rho-independent terminator (ΔG = −22.2 kcal/mol) were localized immediately after the ScaI site. The four putative genes are therefore organized as an operon.
The sequenced region of 2,711 bp had a G+C content of 32.3%. Its 5′ and 3′ regions (coordinates 1 to 960 and 2620 to 2711) had more than 90% identity at the DNA level with the lactococcal plasmids pNZ4000 and pAH82 (Fig. 1B). Another segment (coordinates 2302 to 2562) had 89% identity with Tn916 of Enterococcus faecalis (accession no. U09422) and Tn5251 of Streptococcus pneumoniae (accession no. X90941). The different ORFs were inactivated by the introduction of deletions or frameshifts, and only orf127 and orf213 were necessary to obtain an Abi+ phenotype as long as they were preceded by the putative promoter (Fig. 1B). They were renamed abiTi and abiTii. These genes are encoded on a DNA segment that does not share similarity with any other lactococcal plasmids, but it has a typical G+C content for this bacterium (33.3%). The predicted gene product of abiTi has a molecular mass of 11.7 kDa, a pI of 5.67, and does not share homology with proteins in the databases. AbiTii has a predicted size of 24.4 kDa, a pI of 5.00, and has 28% identity with a putative protein of Xylella fastidiosa (accession no. AE004052).
Expression of AbiT.
To confirm that the first 585 bp of the HaeII fragment included the promoter required for phage resistance, abiTi and abiTii were cloned as a PCR product (coordinates 1151 to 2313) in both orientations in the lactococcal expression vector pMG36c. The two constructs (pED419 and pED420) were introduced into MG1363, and their effect on phage p2 was tested. When cloned in the sense orientation (pED419) the two genes affected phage proliferation, whereas no antiphage activity was observed in the antisense orientation (pED420). Thus, the 5′ region of the fragment can be replaced by a constitutive promoter of L. lactis.
To determine whether abiTi and abiTii are constitutively expressed from their own promoter and are located on the same transcript, total RNA of MG1363 containing pertinent subclones of pED1 was isolated. Following separation on gels and Northern blotting, RNA was hybridized with DNA fragments internal of each of the abiT genes and with a fragment covering orf98 and orf109. The hybridization patterns were identical for all probes (Fig. 1C), which demonstrates that abiTi and abiTii are coexpressed. A unique transcript of about 2 kb was observed with pED100 and pED107 (Fig. 1A and C). Transcripts containing the genetic determinants of AbiT, but not the putative terminator, were of various lengths (data not shown). This confirms the location and function of the predicted terminator.
Site-directed mutagenesis of AbiTi.
The putative AbiTii protein does not contain a particular motif. However, the putative AbiTi protein has a hydrophobic region at its C-terminal extremity (residues 107 to 127) (Table 2). To determine whether this region is essential to the antiphage phenotype, two of the hydrophobic residues were replaced by a proline, a charged residue, or a different hydrophobic residue by site-directed mutagenesis. Clones containing wild-type or mutant abiTi and wild-type abiTii were confirmed by DNA sequencing and tested for their ability to abort phage p2 infection. Replacement of A115 and L122 by a proline or a charged residue resulted in the loss of the antiphage activity, while mutant A115V was as phage resistant as the wild-type strain (Table 2). Replacement of R108, the first charged residue upstream of the hydrophobic region, with a threonine reduced partially the efficacy of AbiT. DAS, TMpred, and Toppred2 programs (11, 30, 54) predicted this segment of AbiTi as a transmembrane helix but could not predict its orientation. Translational fusions between AbiTi and LacZ failed to prove that this region was located in the cell membrane, as all plasmid constructions conferred beta-galactosidase activity to E. coli and L. lactis (data not shown).
Effect of modification of the AbiTi/AbiTii ratio.
Since both putative proteins are involved in the phage resistance phenotype, we investigated further to determine whether one of them was in limiting concentration. The AbiT system was therefore first cloned into the low-copy plasmid pSA3 (Emr) to generate pED307. Then, DNA fragments encoding one functional and one nonfunctional AbiT protein were cloned into the high-copy plasmid pNZ123 (Cmr) to generate pED209, pED211, pED217, and pED218 (see plasmid construction description in Materials and Methods). Combinations of these clones were then introduced into L. lactis MG1363 to create a condition in which the wild-type AbiTi/AbiTii ratio was modified.
It was first concluded that the efficacy of AbiT on phage p2 depended on plasmid copy number, since the EOP was much lower (10−8) when the wild-type abiT genes were cloned on the high-copy vector pNZ123 (pED209) (Table 3). An increase in the level of AbiTi had no effect on the EOP values (10−2), whereas an increase of the AbiTii/AbiTi ratio slightly reduced the EOP (10−3) (Table 3). These data suggest that the arrangement in the wild-type plasmid pED1 guarantees a balanced stoichiometry between AbiTi and AbiTii for maximum antiphage activity.
TABLE 3.
Efficiency of plaquing and plaque size of phage p2 with different concentrations of AbiTi and AbiTii
| Plasmids | AbiTi | AbiTii | EOPa | Plaque size |
|---|---|---|---|---|
| pED307, pNZ123 | + | + | (5.4 ± 2.7) × 10−2 | 1 mm |
| pED307, pED209 | +++ | +++ | ≤1 × 10−8 | —b |
| pED307, pED211 | +++ | + | (3.0 ± 2.6) × 10−2 | 1 mm |
| pED307, pED217 | + | +++ | (6.4 ± 0.8) × 10−3 | Pinpoint |
| pED307, pED218 | + | +++ | (5.6 ± 3.4) × 10−3 | Pinpoint |
| pSA3, pED211 | +++ | − | 1.0 | 2-3 mm |
| pSA3, pED217 | − | +++ | 1.0 | 2-3 mm |
| pSA3, pED218 | − | +++ | 1.0 | 2-3 mm |
n = 3.
—, no plaque.
Efficacy of AbiT against different phage species.
As recently proposed (38), the EOP of the phage resistance mechanism AbiT was determined against members of the three main lactococcal phage species (936, c2, and P335). The four members of the 936 species (p2, p2k, sk1, and jj50) were sensitive to the antiphage system, with EOP values ranging from 2.1 × 10−5 to 1.4 × 10−6. Proliferation of four members of the P335 species (ul36, Q30, φ31, and φ50) was also severely affected by AbiT (EOPs from 4.5 × 10−6 to 1.6 × 10−7) with the notable exception of phage Q33 (EOP, 1.0). DNA restriction and Southern blot analyses indicated that a segment of the ul36 genome corresponding to the lysogeny module (coordinates 678 to 3270) and another segment covering the major part of the late genes (20651 to 35403) have no similarity with the genome of Q33 (data not shown). None of the three c2-like phages (c2, eb1, and ml3) was AbiT sensitive.
The ECOI and the burst size of phage p2 (936 species) and phage ul36 (P335 species) were then evaluated with one-step growth curves (Table 4). Only 4 to 7% of AbiT+-infected cells released phage, and the number of virions released was reduced 10- to 12-fold. Severe reductions in the ECOI and the burst size are typical of Abi systems. The fact that the microbiological impacts of pED1, pED100, and pED109 (Fig. 1A and B) were identical confirms that only abiTi, abiTii, and the promoter are required for a complete phage resistance phenotype.
TABLE 4.
Microbiological effects of AbiT on phage p2 of the 936 species and phage u136 of the P335 species
| Phage | Host | EOPa | ECOIa (%) | Burst sizea |
|---|---|---|---|---|
| p2 | LM0231(pSA3) | 1.0 | 100 | 58 ± 14 |
| LM0231 (pSA3, pED1) | (9.5 ± 3.9) × 10−6 | 6.5 ± 3.5 | 6 ± 1 | |
| MG1363(pMIG3) | 1.0 | 100 | 87 ± 3 | |
| MG1363(pED100) | (2.0 ± 1.2) × 10−6 | 5.4 ± 1.5 | 7 ± 1 | |
| MG1363(pED109) | (2.0 ± 0.4) × 10−6 | 4.2 ± 0.9 | 9 ± 2 | |
| u136 | SMQ481(pMIG3) | 1.0 | 100 | 342 ± 28 |
| SMQ481(pED100) | (2.6 ± 1.5) × 10−7 | 4.5 ± 1.2 | 27 ± 6 | |
| SMQ481(pED109) | (2.3 ± 1.9) × 10−7 | 4.3 ± 1.2 | 27 ± 16 |
n ≥ 3.
Effects of AbiT on the phage lytic cycle.
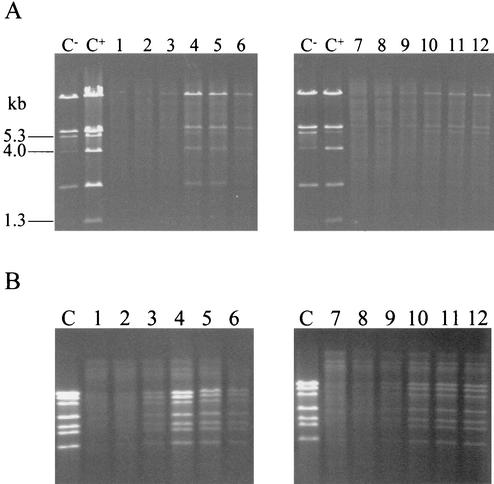
Total DNA was isolated from sensitive and resistant strains at time intervals during infection with phage p2 (936 species) (Fig. 2A) or ul36 (P335 species) (Fig. 2B). For both phages, DNA replication was lowered. In the case of p2, a cos-type phage, DNA was present in its mature form (single unit of the linear phage genome) and immature form (concatemers) in the sensitive strain (Fig. 2A, left). The presence of these two DNA forms was visualized through a 5.3-kb EcoRV fragment, which was the product of the 1.3- and the 4.0-kb EcoRV fragments ligated together through their respective cos termini. The mature form (1.3- and 4.0-kb EcoRV fragments) could not be detected in the AbiT-containing strain (Fig. 2A, right). It appears that AbiT affects DNA replication and also prevents effective encapsidation.
FIG. 2.
DNA replication of phages p2 (A) and ul36 (B) in AbiT− (left) and AbiT+ (right) cells. (A) C−, unheated p2 DNA; C+, p2 DNA heated (10 min at 65°C). Lanes 1 to 6 and 7 to 12, total DNA from infected MG1363(pMIG3) and MG1363(pED109) cells, respectively, at t = 0, 10, 20, 30, 40, and 50 min, heated for 10 min at 65°C. (B) C, ul36 DNA. Lanes 1 to 6 and 7 to 12, total DNA from infected SMQ481(pMIG3) and SMQ481(pED109) cells, respectively, at t = 0, 20, 40, 60, 80, and 100 min. All samples were digested with EcoRV.
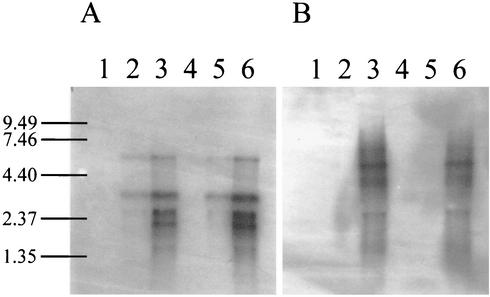
Total RNA was also isolated from sensitive and resistant strains at time intervals during infection with phage p2 (936 species). After gel migration, RNA samples were hybridized with two PCR products. The first probe (Fig. 3A) corresponded to an early gene of phage p2, which is identical to orf35 of phage sk1 (936 species) (10). The second probe (Fig. 3B) matched the late gene that encodes the major capsid protein of phage p2 (34). The early and late transcripts were present in the same amount and at the predicted time points (9) for both strains. This indicates that AbiT does not affect phage gene expression.
FIG. 3.
Hybridization of RNA from p2-infected AbiT− (left) and AbiT+ (right) cells with the orf35 homologue (A) and the mcp gene (B). Lanes 1, 2, and 3, MG1363(pMIG3) at t = 0, 10, and 25 min, respectively; lanes 4, 5, and 6, MG1363(pED109) at t = 0, 10, and 25 min.
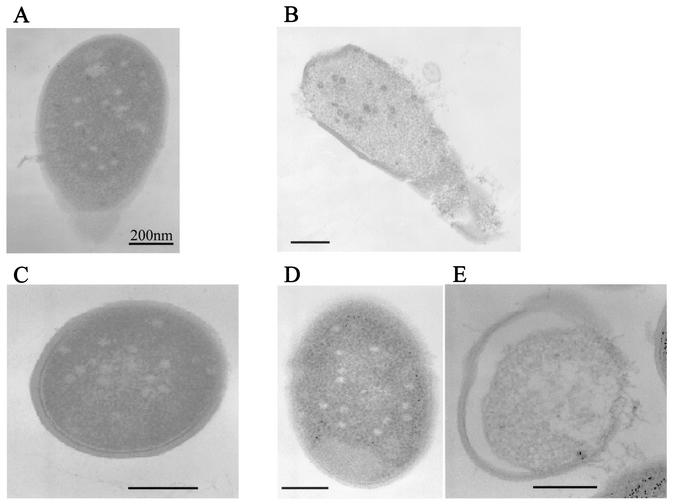
The phage lytic cycle was also monitored at time intervals by electron microscopy in AbiT− and AbiT+ strains. The intracellular course was similar in both infected cells at times 0, 10, and 25 min. At the latter time point, the bacterial chromosome was no longer visible and capsid-like structures were found within the cytoplasm of both strains (Fig. 4A and C). Finally, at later stages the infected AbiT− cells lysed and released progeny phage (Fig. 4B). The fate of the infected AbiT+ cells was less clear, as intact cells with empty capsids (Fig. 4D) and lysed cells could be observed, but the latter did not contain filled phage capsids (Fig. 4E).
FIG. 4.
Monitoring of the lytic cycle by electron microscopy. (A and B) L. lactis AbiT− cells infected with phage sk1 at 25 and 45 min, respectively. (C) L. lactis AbiT+ cells infected with phage sk1 at 25 min. (D and E) L. lactis AbiT+ cells infected at 45 min.
Characteristics of AbiT-insensitive phage mutants.
Four plaques of phage p2 were isolated and amplified on pED1-containing strains. Phage DNA was isolated and digested with at least three restriction enzymes. Restriction profiles of the four p2 mutants insensitive to AbiT were identical to those of the wild-type phage (data not shown). The EOP of the isolated p2 mutants was measured on strains harboring pED109 (low-copy plasmid) and pED209, constructed by cloning the insert of pED109 into the high-copy vector pNZ123. Mutants of p2 were partially insensitive to AbiT, as they had EOP values between 10−5 and 10−6 with pED209 and of 10−1 with pED109.
Similarly, 50 plaques of phage ul36 were also isolated and amplified on pED1-containing strains as well as analyzed by restriction profiles. All these phage isolates were insensitive to AbiT (EOP, 1.0) but presented profiles that significantly differed from those of the wild-type phage (Fig. 5). On the basis of their profiles, DNA-DNA hybridization analyses, PCR, and genome mapping comparisons (data not shown), approximately 20 kb of their genome was nonhomologous to that of ul36 but was related in all mutants. The exchanged region corresponded to the late genes of the phage: the junction points are located between coordinates 13,640 and 15,190 and between 33,051 and 34,755 (35).
FIG. 5.
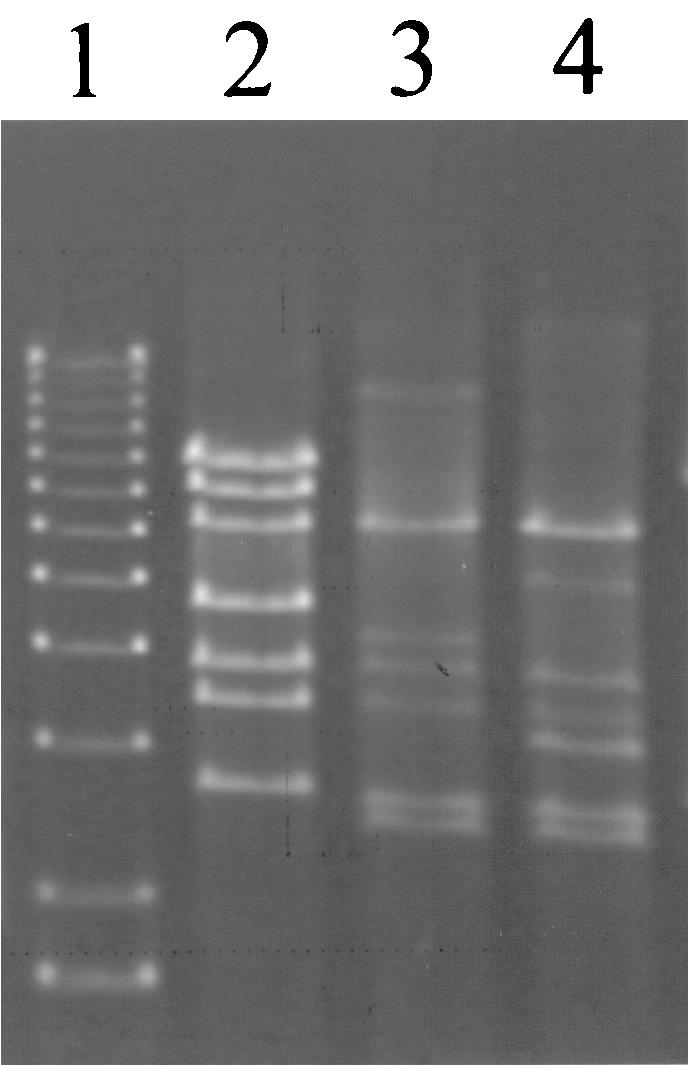
EcoRV restriction profile of AbiT-insensitive mutants of phage ul36. Lane 1, 1-kb DNA ladder (1.6 to 12.2 kb; Invitrogen); lane 2, ul36; lane 3, ul36T1; lane 4, ul36T2 DNA.
DISCUSSION
Since raw milk is a natural environment for L. lactis and its bacteriophages, we hypothesized that the bacterial strains occupying that niche have defense mechanisms against these viruses. We had previously isolated two other plasmids encoding distinct Abi systems from two L. lactis raw milk isolates (23, 24). Here, we isolated another relatively small plasmid with the same approach, pED1, which encodes the AbiT system. Thus, this study confirms that L. lactis strains from raw milk represent a significant source of novel phage resistance mechanisms.
Two genes are responsible for the antiphage activity of AbiT. They are cotranscribed, as observed with other two-component Abi systems (14, 45), and constitutively expressed. All Abi's examined thus far are constitutively transcribed (2, 14, 45). Although the products of abiTi and abiTii do not share similar amino acid sequences with other lactococcal proteins, they are surrounded by regions that have homology, at the DNA level, with plasmids of L. lactis. The homologous 5′ region even includes the promoter from which the abiT genes are expressed. DNA sequence comparisons with lactococcal plasmids indicate that the nonhomologous segment that codes for AbiT (coordinates 961 to 2619) could have been inserted in pED1, since the flanking regions are contiguous in pNZ4000 and pAH82. The AbiT-containing segment has a typical G+C content for L. lactis but comprises, at its 3′ extremity, a region homologous to two transposons of gram-positive bacteria.
AbiT is effective against small isometric-headed 936- and P335-like phages, two of the three most frequently encountered species in the dairy industry. However, a wild-type representative of the P335 species (Q33) and laboratory mutants of ul36 are completely insensitive to AbiT. The restriction profiles of the mutants suggest that homologous recombination events between the phage and its host strain probably occurred as described for mutants that are insensitive to AbiK (7). Our results demonstrate that the gene pool of the P335 quasi-species already includes alleles that confer total insensitivity to this new Abi.
Abi mechanisms include all systems that cause cell death after the penetration of the phage DNA into the host (19). In E. coli, the mode of action of some of these systems has been established. These modes of action share the fact that the presence of a phage element acts as an activator of the Abi and an essential cellular function is blocked before virions are released (44, 51). Little is known about the modes of action of Abi systems in L. lactis, but some can at least be distinguished by their effects on the phage lytic cycle. AbiT allows transcription of early genes and their translation into proteins, since phenomena depending on early protein production are still observed (DNA replication, late transcription). Production of some late proteins also occurs, since procapsid-like structures and activity attributable to lysis proteins are observed in resistant cells. AbiT may block a step of the phage morphogenesis, but it is more likely that one of the late mRNAs or proteins activates the mechanism and rapidly causes premature cell death. This would explain why the normal course of the lytic cycle, including DNA replication, is affected after transcription of late genes (approximately 20 min after the beginning of infection by p2). This hypothesis is also supported by the fact that late genes are heterologous in Q33 and are replaced in insensitive mutants of ul36.
In E. coli, one of the best-characterized two-component Abi mechanisms is the Rex exclusion system from prophage lambda (47, 50, 51). The RexA protein activates the formation of ion channels and the depolarization of the cytoplasmic membrane by RexB, a protein with four transmembrane domains. The putative AbiTi protein has a hydrophobic region, which suggests it could be located in the cell membrane. The introduction of a charged residue that modifies the hydrophobicity profile or a proline that breaks the helix conformation abolishes the Abi phenotype. The efficacy of the Rex system depends on its expression level, but contrary to AbiT, it is also affected by the RexA/RexB ratio. The maintenance transcript contains the rexA and rexB genes, but another difference with AbiT is that rexB is also transcribed independently from the Lit promoter, and the protein ratio would be regulated at this level. Despite a few features common to both antiphage systems, the means by which the AbiT proteins cause Abi still need to be investigated.
Acknowledgments
We are grateful to Diane Montpetit and Hélène Chamberland for the electron micrographs as well as Line Berthiaume for technical assistance.
J.D.B. is a recipient of a graduate scholarship from the Fonds FCAR and the Natural Sciences and Engineering Research Council (NSERC) of Canada. F.B. was supported by an Abitibi-Consolidated scholarship and a Fonds FCAR graduate scholarship. This study was funded by a collaborative research and development grant from NSERC and Agropur.
REFERENCES
- 1.Alatossava, T., and T. R. Klaenhammer. 1991. Molecular characterization of three small isometric-headed bacteriophages which vary in their sensitivity to the lactococcal phage resistance plasmid pTR2030. Appl. Environ. Microbiol. 57:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, G. E., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226. [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST, a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behnke, D., and H. Malke. 1978. Bacteriophage interference in Streptococcus pyogenes. I. Characterization of prophage-host systems interfering with the virulent phage A25. Virology 85:118-128. [DOI] [PubMed] [Google Scholar]
- 5.Bidnenko, E., S. D. Ehrlich, and M. C. Chopin. 1995. Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J. Bacteriol. 177:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 7.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 8.Boucher, I., E. Emond, E. Dion, D. Montpetit, and S. Moineau. 2000. Microbiological and molecular impacts of AbiK on the lytic cycle of Lactococcus lactis phages of the 936 and P335 species. Microbiology 146:445-453. [DOI] [PubMed] [Google Scholar]
- 9.Chandry, P. S., B. E. Davidson, and A. J. Hillier. 1994. Temporal transcription map of the Lactococcus lactis bacteriophage sk1. Microbiology 140:2251-2261. [DOI] [PubMed] [Google Scholar]
- 10.Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 11.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the Dense Alignment Surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 12.Dai, G., P. Su, G. E. Allison, B. L. Geller, P. Zhu, W. S. Kim, and N. W. Dunn. 2001. Molecular characterization of a new abortive infection system (AbiU) from Lactococcus lactis LL51-1. Appl. Environ. Microbiol. 67:5225-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dao, M. L., and J. J. Ferretti. 1985. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl. Environ. Microbiol. 49:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, Y.-M., C.-Q. Liu, and N. W. Dunn. 1999. Genetic organization and functional analysis of a novel phage abortive infection system, AbiL, from Lactococcus lactis. J. Biotechnol. 67:135-149. [DOI] [PubMed] [Google Scholar]
- 15.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vos, W. M. 1987. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Rev. 46:281-295. [Google Scholar]
- 17.Dinsmore, P. K., and T. R. Klaenhammer. 1995. Bacteriophage resistance in Lactococcus. Mol. Biotechnol. 4:297-314. [DOI] [PubMed] [Google Scholar]
- 18.Dinsmore, P. K., and T. R. Klaenhammer. 1997. Molecular characterization of a genomic region in a Lactococcus bacteriophage that is involved in its sensitivity to the phage defense mechanism AbiA. J. Bacteriol. 179:2949-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duckworth, D. H., J. Glenn, and D. J. McCorquale. 1981. Inhibition of bacteriophage replication by extrachromosomal genetic elements. Microbiol. Rev. 45:52-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 21.Durmaz, E., D. L. Higgins, and T. R. Klaenhammer. 1992. Molecular characterization of a second abortive phage resistance gene present in Lactococcus lactis ME2. J. Bacteriol. 174:7463-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durmaz, E., and T. R. Klaenhammer. 2000. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl. Environ. Microbiol. 66:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emond, E., E. Dion, S. A. Walker, E. R. Vedamuthu, J. K. Kondo, and S. Moineau. 1998. AbiQ, an abortive infection mechanism from Lactococcus lactis. Appl. Environ. Microbiol. 64:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emond, E., B. J. Holler, I. Boucher, P. A. Vandenbergh, E. R. Vedamuthu, J. K. Kondo, and S. Moineau. 1997. Phenotypic and genetic characterization of the phage abortive infection mechanism AbiK from Lactococcus lactis. Appl. Environ. Microbiol. 63:1274-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forde, A., and G. F. Fitzgerald. 1999. Bacteriophage defence systems in lactic acid bacteria. Antonie Leeuwenhoek 76:89-113. [PubMed] [Google Scholar]
- 26.Garvey, P., C. Hill, and G. F. Fitzgerald. 1996. The lactococcal plasmid pNP40 encodes a third bacteriophage resistance mechanism, one which affects phage DNA penetration. Appl. Environ. Microbiol. 62:676-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasson, M. J. 1983. Plasmid complements of Streptococcus cremoris NCDO712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill, C., I. J. Massey, and T. R. Klaenhammer. 1991. Rapid method to characterize lactococcal bacteriophage genomes. Appl. Environ. Microbiol. 57:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill, C., L. A. Miller, and T. R. Klaenhammer. 1991. In vivo exchange of a functional domain from a type IIA methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 173:4363-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann, K., and W. Stoffel. 1993. TMBASE—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 31.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. A. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 33.Josephsen, J., and F. K. Vogensen. 1989. Identification of three different plasmid-encoded restriction/modification systems in Streptococcus lactis subsp. cremoris W56. FEMS Microbiol. Lett. 59:161-166. [Google Scholar]
- 34.Labrie, S., and S. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labrie, S., and S. Moineau. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 species of lactococcal phages. Virology 296:308-320. [DOI] [PubMed] [Google Scholar]
- 36.McIntyre, K., H. A. Heap, G. P. Davey, and G. K. Y. Limsowtin. 1991. The distribution of lactococcal bacteriophage in the environment of a cheese manufacturing plant. Int. Dairy J. 1:183-197. [Google Scholar]
- 37.McKay, L. L., K. A. Baldwin, and E. A. Zottola. 1972. Loss of lactose metabolism in lactic streptococci. Appl. Environ. Microbiol. 23:1090-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moineau, S. 1999. Applications of phage resistance in lactic acid bacteria. Antonie Leeuwenhoek 76:377-382. [PubMed] [Google Scholar]
- 39.Moineau, S., M. Borkaev, B. J. Holler, S. A. Walker, J. K. Kondo, E. R. Vedamuthu, and P. A. Vandenbergh. 1996. Isolation and characterization of lactococcal bacteriophages from U.S. buttermilk plants. J. Dairy Sci. 79:2104-2111. [Google Scholar]
- 40.Moineau, S., E. Durmaz, S. Pandian, and T. R. Klaenhammer. 1993. Differentiation of two abortive mechanisms by using monoclonal antibodies directed toward lactococcal bacteriophage capsid proteins. Appl. Environ. Microbiol. 59:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moineau, S., J. Fortier, H.-W. Ackermann, and S. Pandian. 1992. Characterization of lactococcal bacteriophages from Québec cheese plants. Can. J. Microbiol. 38:875-882. [Google Scholar]
- 42.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moineau, S., S. A. Walker, E. R. Vedamuthu, and P. A. Vandenbergh. 1995. Cloning and sequencing of LlaDCHI restriction and modification genes from Lactococcus lactis and relatedness of this system to the Streptococcus pneumoniae DpnII system. Appl. Environ. Microbiol. 61:2193-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molineux, I. J. 1991. Host-parasite interactions: recent developments in the genetics of abortive phage infection. New Biol. 3:230-236. [PubMed] [Google Scholar]
- 45.O'Connor, L., M. Tangney, and G. F. Fitzgerald. 1999. Expression, regulation, and mode of action of the AbiG abortive infection system of Lactococcus lactis subsp. cremoris UC653. Appl. Environ. Microbiol. 65:330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. Rapid mini-prep isolation of high quality plasmid DNA from Lactococcus lactis and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parma, D. H., M. Snyder, S. Sobolevski, M. Nawroz, E. Brody, and L. Gold. 1992. The Rex system of bacteriophage λ: tolerance and altruistic cell death. Genes Dev. 6:497-510. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sanders, M. E., and T. R. Klaenhammer. 1980. Restriction/modification in group N streptococci: effect of heat on development of modified lytic bacteriophage. Appl. Environ. Microbiol. 40:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schinedling, S., D. Parma, and L. Gold. 1987. Wild-type bacteriophage T4 is restricted by the lambda rex genes. J. Virol. 61:3790-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snyder, L. 1995. Phage exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol. Microbiol. 15:415-420. [DOI] [PubMed] [Google Scholar]
- 52.Twomey, D. P., P. J. De Urraza, L. L. McKay, and D. J. O'Sullivan. 2000. Characterization of AbiR, a novel multicomponent abortive infection mechanism encoded by plasmid pKR223 of Lactococcus lactis subsp. lactis KR2. Appl. Environ. Microbiol. 66:2647-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van de Guchte, M., J. M. B. M. van der Vossen, J. Kok, and G. Venema. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Heijne, G. 1992. Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 55.Wells, J. M., P. W. Wilson, and R. W. F. LePage. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]