Abstract
The abilities of Streptococcus mutans to form biofilms and to survive acidic pH are regarded as two important virulence determinants in the pathogenesis of dental caries. Environmental stimuli are thought to regulate the expression of several genes associated with virulence factors through the activity of two-component signal transduction systems. Yet, little is known of the involvement of these systems in the physiology and pathogenicity of S. mutans. In this study, we describe a two-component regulatory system and its involvement in biofilm formation and acid resistance in S. mutans. By searching the S. mutans genome database with tblastn with the HK03 and RR03 protein sequences from S. pneumoniae as queries, we identified two genes, designated hk11 and rr11, that encode a putative histidine kinase and its cognate response regulator. To gain insight into their function, a PCR-mediated allelic-exchange mutagenesis strategy was used to create the hk11 (Emr) and rr11 (Emr) deletion mutants from S. mutans wild-type NG8 named SMHK11 and SMRR11, respectively. The mutants were examined for their growth rates, genetic competence, ability to form biofilms, and resistance to low-pH challenge. The results showed that deletion of hk11 or rr11 resulted in defects in biofilm formation and resistance to acidic pH. Both mutants formed biofilms with reduced biomass (50 to 70% of the density of the parent strain). Scanning electron microscopy revealed that the biofilms formed by the mutants had sponge-like architecture with what appeared to be large gaps that resembled water channel-like structures. The mutant biofilms were composed of longer chains of cells than those of the parent biofilm. Deletion of hk11 also resulted in greatly diminished resistance to low pH, although we did not observe the same effect when rr11 was deleted. Genetic competence was not affected in either mutant. The results suggested that the gene product of hk11 in S. mutans might act as a pH sensor that could cross talk with one or more response regulators. We conclude that the two-component signal transduction system encoded by hk11 and rr11 represents a new regulatory system involved in biofilm formation and acid resistance in S. mutans.
Two-component signal transduction systems (TCSTSs) function in bacterial adaptation, survival, and virulence by sensing changes in the environment and modulating gene expression in response to a variety of stimuli (12). A typical two-component regulatory system consists of a membrane-associated, histidine kinase sensor protein, which senses a specific environmental condition, and a cytoplasmic response regulator, which enables the cell to respond via regulation of gene expression when this condition varies (29). Upon stimulation by a specific ligand or a signal the histidine kinase sensor protein undergoes autophosphorylation at a conserved histidine residue. The phosphoryl group is then transferred to the cognate response regulator, which can, in turn, activate or repress transcription of target genes. Two-component regulatory systems have been shown elsewhere to regulate diverse metabolic processes, the bacterial cell cycle, cell-cell communication, and virulence factors in a wide range of bacterial species (8). Because of their importance in the regulation of cellular physiology, adaptation to environments, and virulence expression, two-component regulatory systems have been used as targets to develop antimicrobial agents (14, 24).
Streptococcus mutans is a bacterium that has evolved a biofilm lifestyle for survival and persistence in its natural ecosystem, dental plaque (11). Under appropriate environmental conditions, S. mutans can produce sufficient amounts of acid from dietary fermentable carbohydrate and cause an imbalance in the demineralization-remineralization process of tooth enamel, leading to dental caries (23). The ability of S. mutans to initiate dental caries depends on several virulence-associated traits, including (i) initiation of biofilm formation by adherence and accumulation on the tooth surface that is promoted by its synthesis of insoluble, extracellular polysaccharides; (ii) high efficiency in catabolizing carbohydrates and producing acids; and (iii) the ability to grow and continue to metabolize carbohydrates at low pH (25). Environmental factors play important roles in the regulation of these virulence-associated traits in S. mutans (22). Despite many studies demonstrating the importance of environmental stimuli in the regulation of physiology and virulence traits of S. mutans, little is known of the molecular mechanisms by which S. mutans regulates the expression of these virulence traits in response to fluctuations in its environment.
We have recently characterized a quorum sensing signaling system consisting of a two-component regulatory system (ComDE) in S. mutans. This system responds to its native signal peptide pheromone and activates transcription of a number of genes essential for induction of genetic competence, resulting in natural transformation (19). Our previous work has demonstrated that this system appears to play a global regulatory role in genetic competence, biofilm formation, and acid tolerance response (ATR) in S. mutans (18, 20). In this study, we described a novel two-component regulatory system and began to evaluate the role of this system in biofilm formation and acid resistance in S. mutans.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
The strains used in this study and their relevant characteristics are listed in Table 1. S. mutans wild-type (wt) strain NG8 was subcultured routinely on Todd-Hewitt yeast extract (THYE) agar plates (BBL Becton Dickinson, Cockeysville, Md.), whereas the mutants were maintained on THYE agar plus 10 μg of erythromycin/ml. THYE liquid medium was routinely used to grow the strains unless otherwise specified. To grow biofilms, a semidefined minimal (SDM) medium was prepared by a modification of the method described previously (21). The medium contained 58 mM K2HPO4, 15 mM KH2PO4, 10 mM (NH4)2SO4, 35 mM NaCl, and 2 mM MgSO2 · 7H2O and was supplemented with filter-sterilized vitamins (0.04 mM nicotinic acid, 0.1 mM pyridoxine HCl, 0.01 mM pantothenic acid, 1 μM riboflavin, 0.3 μM thiamine HCl, and 0.05 μM d-biotin), amino acids (4 mM l-glutamic acid, 1 mM l-arginine HCl, 1.3 mM l-cysteine HCl, and 0.1 mM l-tryptophan), 0.2% (wt/vol) Casamino Acids, and 20 mM glucose. Biofilms of all strains were developed on polystyrene microtiter plates in SDM medium at 37°C with 5% CO2 for 16 h before quantification and microscopic examination.
TABLE 1.
Bacterial strains, amplicons, and plasmid used in the study
| Strains | Relevant characteristics | Source or reference |
|---|---|---|
| S. mutans strains | ||
| NG8 | wt Ems | A. S. Bleiweis, University of Florida |
| SMHK11 | NG8 Δhk11::PcEm Emr | This study |
| SMRR11 | NG8 Δrr11::PcEm Emr | This study |
| Amplicons | ||
| PcEm | Emr marker with a synthetic promoter amplified from ermAM cassette | 4 |
| aHK11 | HK11-up::PcEm::HK11-dw for allelic replacement of hk11 | This study |
| aHK11 | RR11-up::PcEm::RR11-dw for allelic replacement of rr11 | This study |
| Plasmid | ||
| pDL289 | E. coli-Streptococcus shuttle vector; Kmr | 2 |
Construction of the hk11 and rr11 deletion mutants.
We initiated a search of the S. mutans genome database at the University of Oklahoma OU-ACGT website (http://www.genome.ou.edu/smutans.html) (P. C. Y. Lam and D. G. Cvitkovitch, abstract, J. Dent. Res. 81:2246, 2002) for homologs of the 13 TCSTSs identified in S. pneumoniae (14). A tblastn search using the HK03 and RR03 protein sequences from Streptococcus pneumoniae (14) as queries identified two genes that shared homology with the hk3 and rr3 genes in S. pneumoniae. These two genes, designated hk11 and rr11, respectively encoded a putative histidine kinase and its cognate response regulator in S. mutans. This study focused on the evaluation of the function of this TCSTS, designated HK/RR11, in biofilm formation and acid resistance of S. mutans. We constructed individual deletion mutants of the hk11 and rr11 genes in S. mutans wt strain NG8 by a rapid PCR-based deletion strategy involving restriction-ligation and allelic replacement as described previously (15). The primers used to construct and confirm the gene deletion are listed in Table 2. To construct the hk11 mutant, for example, a 763-bp fragment 5′ from the hk11 start codon (HK11-up) was amplified from S. mutans NG8 genomic DNA by using primers HK11-P1 and HK11-P2 (containing an AscI site at its 5′ end). Another amplicon, designated HK11-dw, was 666 bp 3′ from hk11 and was amplified with HK11-P3 (with an FseI site at the 5′ end) and HK11-P4 primers. An erythromycin resistance marker, PcEm (860 bp), from a synthetic Emr cassette (4) was amplified by using Em cst-P1 and Em cst-P2 primers with AscI and FseI sites engineered into their 5′ ends, respectively. These amplicons were subjected to restriction enzyme digestion and subsequent ligation to produce an HK11-up::PcEm::HK11-dw fragment. The ligated product was directly used for transformation of S. mutans wt strain NG8 with the aid of a synthetic competence-stimulating peptide (CSP) (19). Following double-crossover homologous recombination, the internal region of the hk11 gene was completely replaced by the erythromycin cassette (PcEm). A similar strategy was used to construct the rr11 deletion mutant.
TABLE 2.
Primers used to construct the hk11 and rr11 deletion mutants by PCR restriction-ligation mutagenesis
| Primer | Nucleotide sequence (5′ → 3′)a | Amplicon (bp) |
|---|---|---|
| HK11-P1 | CTGAAGGAAGCCTTATGCG | 763 |
| HK11-P2 | GGCGCGCCCCGGAAATGATGGTAACTGCCG | |
| HK11-P3 | GGCCGGCCAAGTGCTGAAAAGGGGG | 666 |
| HK11-P4 | CATAGACAACGGCTTGGGTC | |
| RR11-P1 | AACGCCACCTCAACCAAA | 887 |
| RR11-P2 | GGCGCGCCGCCTCACCGATAACTTCTACATT | |
| RR11-P3 | GGCCGGCCCCTTCAGCATCATTTAGTGCCAC | 565 |
| RR11-P4 | CACCCACATCCTCAAAGGTC | |
| Em cst-P1 | GGCGCGCCCCGGGCCCAAAATTTGTTTGAT | 860 |
| Em cst-P2 | GCTGGCCGGCCAGTCGGCAGCGACTCATAGAAT |
AscI sites are in boldface; FseI sites are underlined.
The integration sites of the PCR constructs in the mutants were confirmed by PCR. Briefly, genomic DNA was prepared from transformants selected on THYE-erythromycin (10 μg/ml) agar plates by a method described previously (6). The mutant and wt genomic DNAs were then used as templates in PCR with three combinations of primers (P1 and Em cst-P2, P4 and Em cst-P1, and P1 and P4) to verify correct recombination of the construct into the mutant genome based on the predicted size of the products. The wt (NG8) genomic DNA was used as a negative control.
Growth rates.
Strains were grown in both SDM medium and a tryptone-yeast extract (TYE) medium supplemented with 20 mM glucose to assay their growth kinetics with a Bioscreen microbiology reader (Bioscreen C Labsystems, Helsinki, Finland) with multiwell disposable microtiter plates. The Bioscreen was equipped with software that allowed recording and conversion of turbidity readings into growth curves. An aliquot (4 μl) of cell suspension of the same turbidity was inoculated into each well containing 400 μl of fresh medium. Turbidity of the culture was recorded after brief shaking every 15 min for a total of 20 h. Each sample was assayed in triplicate, and three wells without cells were used as blank controls.
Genetic transformation.
To determine if inactivation of hk11 or rr11 had any impact on the development of genetic competence, the mutants were assayed for genetic transformation by using a protocol as described previously (19). Briefly, overnight cultures were diluted with 2 ml of prewarmed, fresh THYE broth supplemented with 5% horse serum to generate 1:20 and 1:40 dilutions. The cultures were incubated at 37°C with 5% CO2 for 2 h to allow turbidities to reach 1.5 to 2.0 units of optical density at 600 nm. Each sample was then divided into two aliquots: one containing 1 μg of transforming plasmid DNA (pDL289, Kmr)/ml (2) and another containing the same concentration of transforming plasmid DNA and freshly made CSP (19) at a final concentration of 500 ng/ml. The cultures were incubated for 2 to 3 h and gently sonicated for 10 s to disperse the streptococcal chains, and an aliquot (100 μl) of cell suspension was spread on THYE plates containing kanamycin (700 ng/ml). An aliquot of the cell suspension, after appropriate dilution, was also spread on THYE plates without antibiotics to determine the total recipient cell number. Transformation of the parent strain NG8 was used as a positive control. Transformation frequency was expressed as the number of transformants divided by the total recipient cells per milliliter of cell suspension.
Biofilm formation and quantification.
All strains were assayed for biofilm formation on a polystyrene surface by the method described previously (20, 21). To facilitate quantification and microscopy, both 96- and 24-well polystyrene microtiter plates were used to develop biofilms. The growth of biofilms was initiated by inoculating 5 μl of suspended cells from an overnight culture into 300 μl of SDM medium in individual wells of a 96-well microtiter plate or 25 μl of cell suspension into 2 ml of SDM medium in 24-well plates. The microtiter plates were then incubated at 37°C with 5% CO2 for 16 h without agitation. After incubation, liquid medium was removed and wells were rinsed once with sterile distilled water. The plates (96 wells) were then air dried and stained with 0.1% (wt/vol) safranin for 10 min. After staining, the plates were rinsed with distilled water to remove excess dye and air dried for 3 h. Biofilms were quantified by measuring the absorbance of stained biofilms at 490 nm with an enzyme-linked immunosorbent assay microplate reader (model 3550; Bio-Rad Laboratories, Richmond, Calif.). Each assay was performed in triplicate, and wells without biofilms were used as blank controls after safranin staining. Biofilms formed in 24-well plates were photographed immediately after removal of planktonic cells before staining.
Adherence assay.
The strains were assayed for their ability to attach to a mucin-coated polystyrene surface to determine the effect of inactivation of individual genes on initial adherence. The surface of the polystyrene microtiter plates was first conditioned with 2 ml of 1% (wt/vol) hog gastric mucin (type III; Sigma) in an adherence buffer (10 mM KPO4, 50 mM KCl, 1 mM CaCl2, 0.1 mM MgCl2, pH 7.0) (17). The plates were incubated at room temperature for 2 h with gentle shaking and air dried after removal of excess mucin solution. Adherence was then initiated by addition of 2 ml of a previously prepared resting cell suspension at a density of 108 cells/ml. The resting cells were prepared by centrifugation of overnight cultures, washed twice, and resuspended in adherence buffer. The plates were incubated at 37°C with gentle shaking for 2 h. After incubation, unattached cells were removed and adherent cells were dissociated into 2 ml of the buffer by gentle sonication. Viable colony counts of both adherent and nonadherent cells were performed to determine percentages of adherent cells.
Acid tolerance assays.
The effect of pH on the growth of the hk11 and rr11 deletion mutants was first evaluated by assessment of growth on THYE agar plates at pH 5.0 and 7.0. Both the mutants and the parent strains were grown in THYE broth (pH 7.0) overnight. One volume of overnight culture was transferred into 9 volumes of fresh medium, and incubation continued for 2 h at 37°C in an atmosphere of 5% CO2. The cultures were gently sonicated for 15 s to disperse the chains of cells prior to serial dilution with 10 mM KPO4 buffer (pH 7.2). An aliquot (20 μl) of cell suspension from each strain was inoculated onto THYE agar plates at both pH 5.0 and pH 7.0. The plates were then incubated at 37°C in an atmosphere of 5% CO2 for 40 h before assessment of acid sensitivity. Sensitivity to low pH was determined by comparison of the growth of parent and mutants on THYE plates at pH 5.0 following a 40-h incubation.
The cultures were also grown in broth to assay the inducible ATR by a method described previously (18). All experiments for ATR were carried out in TYE medium supplemented with 20 mM glucose (TYEG) at pH 7.5, 5.5, and 3.5 prepared with 40 mM phosphate-citrate buffer. Briefly, mid-log-phase cells were prepared by transferring 1 volume of overnight culture into 9 volumes (1:10) of fresh TYEG (pH 7.5) and incubated at 37°C in an atmosphere of 5% CO2 for 2 h. These cells were collected by centrifugation at 10,000 × g for 10 min and resuspended in 2 ml of fresh TYEG (pH 5.5) at a turbidity of 0.6 (A600). The cells were induced for acid adaptation by incubation at 37°C with 5% CO2 for 2 h. The adapted log-phase cells were then exposed to the killing pH of 3.5, which was predetermined by incubating unadapted, mid-log-phase cells in TYEG at pH values from 6.0 to 2.0 for 3 h (18). An aliquot of cell suspension was taken immediately from each sample to determine total viable cell number at zero time, and the cultures were incubated at 37°C with 5% CO2 for 3 h. After incubation an aliquot of the cell suspensions was taken to determine the percentage of survivors by viable cell counts. The ATR was expressed as the percentage of cells to survive the killing pH for 3 h.
14C labeling of cells during acid adaptation.
Changes in protein expression of NG8 and SMHK11 during acid adaptation were assessed by exposing cells to 14C-labeled amino acids followed by protein extraction and separation by two-dimensional (2D) gel electrophoresis. Three independent cultures of NG8 and SMHK11 were grown in a minimal medium comprised of six amino acids (glutamate, serine, cysteine, valine, leucine, and asparagine), 40 mM phosphate-citrate buffer (pH 7.5), and 20 mM glucose (1). Cells were grown to the middle of exponential growth phase (optical density at 600 nm = 0.7), washed twice in glucose- and buffer-free medium, and resuspended to 2 × 108 cells ml−1 in 2.5 ml of fresh minimal medium. The triplicate cultures of each strain were divided into two portions, where one was exposed to pH 5.5 and the other was kept at pH 7.5 in the presence of 150 μCi of a 14C-amino acid mixture. The incubation was carried out for 30 min at 37°C, and protein synthesis was stopped by adding 2 mg of chloramphenicol to each tube. Cells were centrifuged (15,000 × g for 10 min) and washed in 10 mM Tris-HCl, pH 6.8, with 1 mM EDTA and 5 mM MgSO4. The cells were stored at −20°C, and cell protein extracts for 2D gel electrophoresis were prepared by using ultrasonication in the presence of glass beads as previously described (30).
2D gel electrophoresis and image analysis of protein patterns.
2D gel electrophoresis and image analysis of autoradiograms were performed by previously described methods (30). Isoelectric focusing in the first dimension was carried out on linear pH 4 to 7 18-cm immobilized pH gradient gel strips (Pharmacia Biotechnology, Uppsala, Sweden) loaded with 106 cpm, corresponding to 150 μg of cellular protein. The second dimensional separation was performed with 14% polyacrylamide gradient gels (185 by 200 by 1.0 mm), and the dried gels were exposed to X-ray film (Hyperfilm β-max; Amersham, Oakville, Ontario, Canada) for 14 days. Proteins visualized on the autoradiograms were analyzed with the Bio Image software (version 6.1) on a Sun Sparc station. A protein spot was classified as being differently expressed if the relative integrated optical intensity was changed more than twofold in the acid-exposed cells (pH 5.5) compared to the control cells (pH 7.5). Three independent experiments were performed; for each spot, a coefficient of variation was calculated; and those proteins that exhibited a high inherent variation in expression were excluded as being acid stress proteins.
Microscopy.
To examine the spatial distribution and architecture of biofilms by scanning electron microscopy (SEM), biofilms formed on the surface of polystyrene microtiter plates were washed once with 10 mM phosphate-buffered saline, fixed by adding 2 ml of 3.7% formaldehyde in 10 mM phosphate-buffered saline, and incubated at room temperature for 24 h. The samples were then dehydrated through a series of ethanol rinses (30, 50, 70, 95, and 100%) and critical point dried with liquid CO2. The bottom surface of the well was cut off, mounted, and sputter coated with gold. The samples were then examined by SEM (model S-2500; Hitachi Instruments, San Jose, Calif.).
We previously observed that the CSP encoded by the comC gene in S. mutans activated an uncharacterized second pathway that appeared to be related to cell separation or chain formation (20). The second pathway activated by the CSP remains to be identified. To test if the TCSTS encoded by hk/rr11 was potentially the second pathway, we compared the length of chains formed by the hk/rr11 mutants grown in biofilms with or without addition of CSP, by using light microscopy. Briefly, the mutant biofilms were developed in microtiter plates by the same method as described previously (20), with the exception that each well contained a sterilized coverslip as a substrate and cultures were supplemented with 1.0 μg of fresh CSP/ml. Biofilms were grown in the SDM medium for 16 h, and liquid was removed. The biofilms were then stained with 0.1% crystal violet for 1 min before being placed on a microscope slide. Biofilms were then viewed and qualitatively assessed for cell chain length by light microscopy (Olympus CH30RF100; Tokyo, Japan).
RESULTS
Genetic confirmation of the hk11 and rr11 deletion mutants.
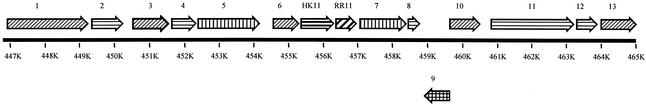
The genetic locus surrounding the hk/rr11 region was annotated by comparing the deduced amino acid sequences of the adjacent open reading frames (ORFs) to the GenBank database by using the blastP algorithm. A map and description of the locus are shown in Fig. 1. The hk11 gene was located at bp 455345 to 456349 in the S. mutans genome database, encoding a hypothetical protein of 334 amino acids with a predicted molecular mass of 38,113 Da. The hk11 ORF shared highest similarity to a putative two-component sensor histidine kinase from Streptococcus pyogenes (accession no. AAK34394) (blast similarity score = 352); a putative histidine kinase, HK03, from S. pneumoniae (CAB54570) (blast score = 244); and a putative histidine kinase, BH1199, from Bacillus halodurans (BAB04918) (blast score = 177). The ORF encoding rr11 was located at bp 456336 to 456983, encoding a hypothetical protein of 215 amino acids having a predicted molecular mass of 24,067 Da. Interestingly the genes overlapped by 12 nucleotides and the response regulator-encoding gene had a promoter-like structure located 5′ from its putative start codon. In this 5′-proximal region the −18 to −10 sequence TACCAACT was very similar to the com-box consensus of S. pneumoniae (16) by a single base pair (TACGAACT). These com-box genes form part of the CSP-mediated regulon. The hk11 gene had putative −10 (−12, TAATGA) and −35 (TGTTATGGA) promoter sequences as well. It did not, however, appear to have a com-box in this vicinity. Similarly to the hk11 gene, rr11 had the highest homology to the respective response regulator gene from S. pyogenes (SPy1621) and the R03 gene from S. pneumoniae but had a higher similarity to hypothetical response regulator gene yvqC from Bacillus subtilis rather than B. halodurans. No putative substrate or signal has been assigned to any of these systems in these organisms.
FIG. 1.
The arrangement of the hk/rr11 genetic locus. Neighboring genes were assigned putative functions based on high blast homology scores with genes for the indicated proteins: 1, primosomal replication factor V (AAK34400); 2, methionyl tRNA transferase Fmt (AAK34399); 3, RNA binding protein SunL (rRNA methyltransferase RsmB) (AAK34398); 4, phosphoprotein Ser/Thr phosphatase PppL (AAK34397); 5, Ser/Thr protein kinase PknB (AAK34396); 6, conserved hypothetical protein (AAK34395); 7, peptidyl-prolyl cis-trans isomerase PpiB (AAK75626); 8, polyribonucleotide nucleotidyltransferase (general stress protein GSP13) (AAK34392); 9, pyruvate formate lyase-activating enzyme PflC (AAK74424); 10, transcriptional regulator RdrA (AAK34719); 11, pyruvate formate lyase 2 PflD (AAK34714); 12, transaldolase-like protein MipB (AAK34713); and 13, glycerol dehydrogenase GldA (AAK74432).
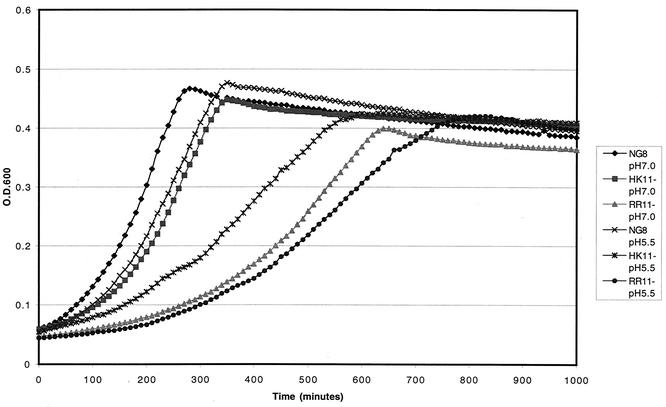
PCR confirmation demonstrated that the target genes of the hk11 and rr11 mutants were correctly replaced by the Erm cassette in their respective mutants (data not shown). The mutants confirmed by PCR were designated SMHK11 and SMRR11, respectively. Growth kinetics showed that both mutants had a decrease in growth rate (increased doubling time [Td]) in SDM medium (Td = 1.47 h for SMHK11 and 1.43 h for SMRR11) compared to the parent strain (Td = 1.27 h). However, the final growth yield of the mutants after 12 h of growth appeared to be the same as that of the parent strain in TYEG (Fig. 2) or SDM medium (data not shown). Both mutants, similar to the parent strain, were able to become genetically competent and were transformed with plasmid DNA with or without addition of CSP, suggesting that the system encoded by hk/rr11 did not affect competence development in this organism.
FIG. 2.
Growth curves of the parent strain S. mutans NG8 and hk/rr11 mutants SMHK11 and SMRR11 grown in TYEG medium at pH 7.0 and 5.5.
Deletion of hk11 or rr11 resulted in defects in biofilm formation.
Deletion of the hk11 or rr11 gene resulted in defects in biofilm formation as illustrated in Fig. 3. Strain SMHK11 had approximately 50% of the biofilm density and strain SMRR11 had about 75% of the density of the parent strain NG8. It is unlikely that the reduction in biofilm density observed with the mutants was caused by their slightly decreased growth rates, especially since their growth yields at neutral pH were nearly the same as that of the parent strain in TYEG (Fig. 2) and SDM medium (data not shown).
FIG. 3.
Biofilm formation and quantification of S. mutans strains. The graph represents the turbidity of the biofilms as reflected by their absorbance after safranin staining. The mean values ± SDs are presented.
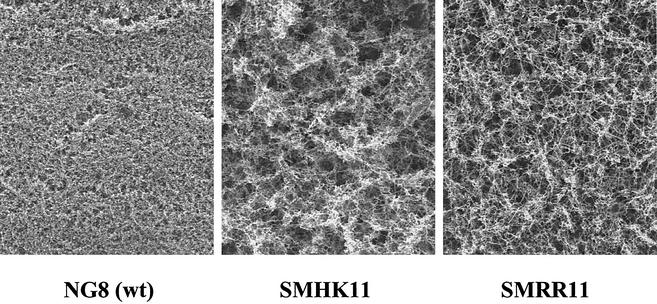
A closer examination of the biofilms by SEM revealed that biofilms formed by the mutants had a very different appearance from the parent biofilm. The mutant biofilms appeared to have sponge-like architecture with what appeared to be large intercellular gaps (Fig. 4). We found that such biofilms formed by both hk11 and rr11 mutants were washed off from the surface more readily than those formed by the parent strain during preparation for the biofilm assay. In addition, the resting cells of the mutants had a reduced ability to attach to the mucin-coated polystyrene surface (percentage of cells attached to the surface ± [standard deviation (SD)]: NG8, 12.08 [2.04]; HK11, 6.77 [1.32]; and RR11, 8.36 [1.53]). Taken together, the apparent defects of the sponge-like architecture and the lower affinity of the cells for adherence to the surface likely contributed to the reduced biomass observed with the mutants. SEM also revealed that both SMHK11 and SMRR11 formed very long chains in comparison to the wt strain when grown as biofilms.
FIG. 4.
Scanning electron micrographs show spatial distribution and architecture of biofilms formed by S. mutans strains.
Since this phenotype was suspected of being linked to the CSP-activated pathway, we examined the impact of CSP on cell chain formation by the hk/rr11 mutants. Addition of CSP to the biofilm cultures did not significantly change the length of chains formed by the SMHK11 and SMRR11 mutants. In an attempt to quantitate the chain lengths from the SEMs, averages were obtained from four independent chains selected randomly. The average numbers of cells per chain (±SD) were as follows: NG8 (wt), 17 (8.04); HK11, 42 (13.8); and RR11, 38 (11.3).
The hk11 mutant is defective in acid tolerance.
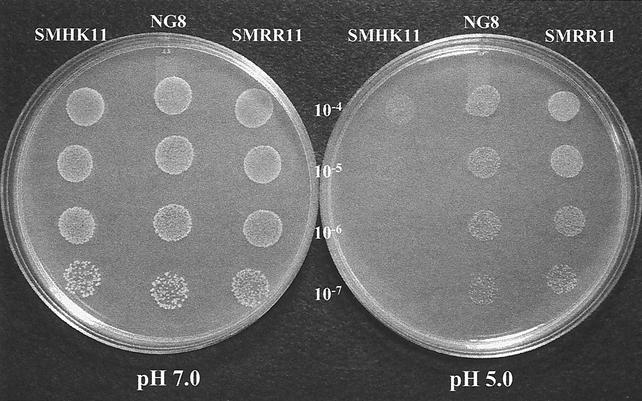
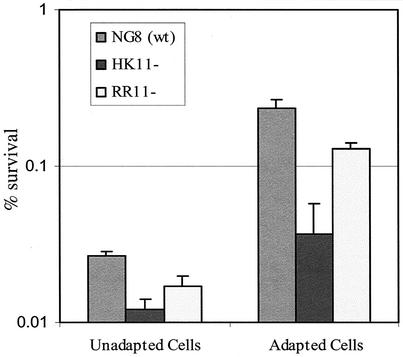
Compared to the parent strain NG8, both SMHK11 and SMRR11 mutants showed decreased growth rates in liquid culture at pH 5.5. NG8 had a Td of 100 ± 1 min. SMHK11 had a Td of 178 ± 3 min, while SMRR11 doubled every 207 ± 9 min (Fig. 2). Mutant SMHK11 also had greatly diminished growth on agar plates at pH 5.0, although it grew as well as the parent strain did on plates at pH 7.0 (Fig. 5) and nearly as well as the parent in broth at pH 7.0 (Fig. 2). Interestingly, we were unable to detect a difference between the growth of the SMRR11 mutant and that of the parent strain NG8 on the pH 5.0 plates. To more closely determine if deletion of the hk11 or rr11 gene affected the inducible ATR, we assayed the log-phase ATR of the mutants grown in liquid cultures by the method described previously (18). The results showed that the SMHK11 mutant had a reduced ATR relative to the parent strain NG8 (Fig. 6). However, the deletion of rr11 resulted in only a slight decrease in inducible ATR as observed with strain SMRR11.
FIG. 5.
Effect of pH on the growth of S. mutans strains. The photographs were taken after 40 h of incubation at 37°C with 5% CO2.
FIG. 6.
Inducible ATR was assayed in log-phase cells of the mutants. The mean percentages of survivors ± SDs from three independent experiments are presented.
The acid stimulon of SMHK11.
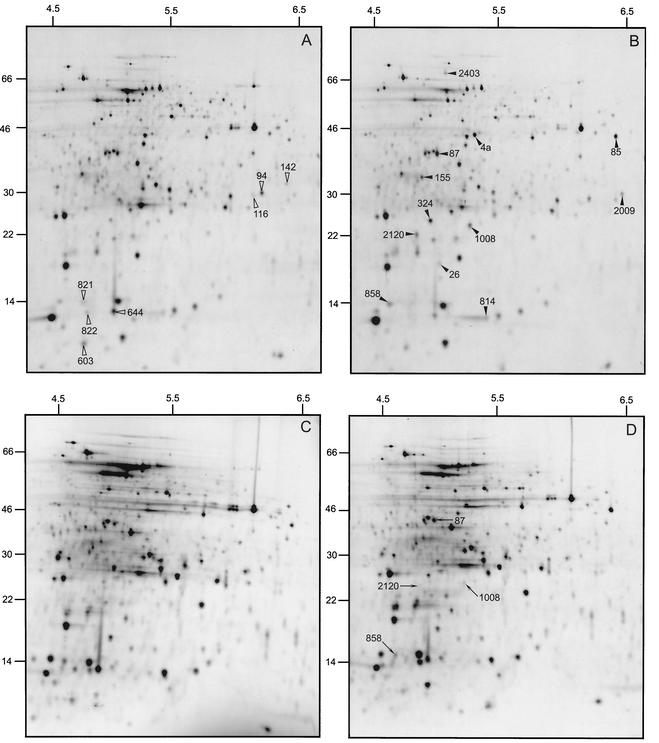
Changes in protein expression underlying the ATR of NG8 and SMHK11 were analyzed by comparative 2D gel electrophoresis of total cellular labeled proteins of cells exposed to pH 7.5 and 5.5 for 30 min. Of 594 proteins monitored in these experiments, all those with differential expression of >2.0 or <0.5 upon acid exposure for 30 min were classified as belonging to the acid stimulon of NG8 and SMHK11, respectively. The acid stimulon of NG8 included 19 proteins, 12 showing increased and 7 showing decreased expression (Fig. 7A and B; Table 3). Interestingly, of the 12 NG8 proteins exhibiting increased expression following the acid shock, four proteins were not induced in the mutant strain (Fig. 7C and D; Table 3). These results confirm that SMHK11 had defects in induction of acid stress proteins relative to the parent strain NG8 and confirm that hk11 plays an important role in the inducible ATR.
FIG. 7.
Autoradiograms of 2D gels obtained with parent strain NG8 (A and B) and mutant HK11 (C and D) of S. mutans exposed to pH 7.5 (A and C) and 5.5 (B and D) for 30 min. Solid and open arrowheads denote proteins that had increased or decreased expression, respectively, in both NG8 and SMHK11 at the acid shock. Note the presence of four protein spots indicated with arrows in panel D that were induced in NG8 but not in SMHK11. Numbers at the top of each panel are pHs; numbers at the left are molecular masses in kilodaltons.
TABLE 3.
Proteins induced or repressed in NG8 and SMHK11
| Protein with >3-fold change, in expression due to acid, stress of NG8 | Common to acid stress of SMHK11 |
|---|---|
| Increaseda | |
| 4a (pyruvate dehydrogenase) | Yes |
| 26 (unknown) | Yes |
| 85 (cysteine synthase) | Yes |
| 87 (exopolyphosphatase) | No |
| 155 (unknown) | Yes |
| 324 (unknown) | Yes |
| 814 (unknown) | Yes |
| 858 (unknown) | No |
| 1008 (histidine kinase candidate) | No |
| 2009 (unknown) | Yes |
| 2120 (unknown) | No |
| 2403 (unknown) | Yes |
| Decreasedb | |
| 94 (unknown) | Yes |
| 116 (unknown) | Yes |
| 142 (unknown) | Yes |
| 603 (unknown) | Yes |
| 644 (unknown) | Yes |
| 821 (unknown) | Yes |
| 822 (unknown) | Yes |
DISCUSSION
Two-component regulatory systems are widespread prokaryotic signal transduction systems that allow regulation of cellular functions in response to changing environments. Although increasing information is available regarding identification and characterization of two-component systems in various species of bacteria, little is known of these systems in S. mutans, a primary etiological agent of dental caries. Genome analyses have revealed many putative TCSTSs in several related gram-positive organisms such as S. pneumoniae (33), S. pyogenes (9), Bacillus spp. (13, 32), and others. Using genome analysis, we have recently described 13 separate TCSTSs in S. mutans and have constructed 25 individual mutants of the 26 genes encoding these systems (Lau and Cvitkovitch, abstract). Since one of our major interests was to identify TCSTSs involved in the expression of virulence factors of S. mutans, we focused our attention on screening the mutants for phenotypes associated with biofilm formation, acid tolerance, and other environmental stresses including ethanol, sodium laurel sulfate (common in dentifrice), and H2O2.
Previous work in our lab has described a quorum sensing-signaling system consisting of a two-component regulatory system (ComDE) that was demonstrated previously to affect genetic competence (19), biofilm formation (20), and ATR (18) in S. mutans. In the present study, we present evidence that a novel two-component regulatory system, HK/RR11, plays an important role as a determinant of biofilm formation and acid resistance phenotypes in S. mutans.
Our results clearly demonstrated that deletion of either hk11 or rr11 resulted in the formation of a biofilm with reduced biomass and a sponge-like architecture (Fig. 3 and 4). One striking feature observed by SEM was that the mutant biofilms appeared to consist of many large intercellular channels relative to the parent biofilm, which had a more confluent appearance. Since water channels in biofilms facilitate exchange of substrates between a biofilm and bulk liquid phase (7), it is possible that the hk/rr11 mutant biofilms were impaired in the transport of a substrate or removal of a metabolic end product.
Another study, by Bhagwhat et al., examined S. mutans mutants defective in the response regulators of six TCSTs (1). One mutant described in this study was analogous to the RR11 mutant (tcek), and its ability to form biofilms was also assayed. Yet, the Bhagwhat group did not describe a defect in biofilm formation by this mutant. Notably, the parent strain and growth and assay conditions were different from ours. Our observation that rr11 deletion did not affect genetic competence was, however, consistent with the observations of Bhagwat et al. These investigators did, however, find that inactivation of tcbR (the comE gene encoding the response regulator of the ComD/ComE TCSTS) resulted in a 10-fold reduction in biofilm formation, which was consistent with our previous findings that comD and comE mutants formed defective biofilms with reduced biomass (20).
Compared to the parent strain, both SMHK11 and SMRR11 biofilms had sponge-like architecture that was composed of cells organized in very long chains, a feature that we previously observed with the biofilm formed by a comC mutant unable to produce the signal peptide pheromone CSP (20). Mutants defective in comD or comE did not, however, have a web- or sponge-like architecture, suggesting that a separate pathway was receptive to CSP. To further support the existence of a second CSP sensor system, we found that exogenous addition of CSP or complementation of the comC mutant with a wt comC gene partially restored the wt phenotype of the comCDE mutant biofilm. Since this mutant was defective in producing the CSP and its cognate receptor (encoded by comD), we hypothesized that there was another receptor(s) that recognized CSP and was involved in cell septation or separation, ultimately affecting biofilm architecture.
Since we suspected that the TCSTS encoded by hk/rr11 might function as the second pathway, we added CSP to the mutant cultures to assess the effect on chain formation by the hk11 and rr11 mutant biofilm cells. The results showed that addition of CSP to the mutant cultures had no observable impact on the length of chains comprising the mutant biofilms (data not shown). This result is consistent with HK11 acting as a CSP receptor but does not provide direct evidence to conclusively assign a role to HK11 as a CSP receptor. A closer examination of the interaction of CSP with the hk/rr11 system is warranted.
Wen and Burne (35) have recently described a gene, designated brpA (biofilm regulatory protein), which encodes a 406-amino-acid protein in S. mutans UA159. Their work also showed that inactivation of brpA resulted in a strain that produced an aberrant biofilm, with the mutant forming longer chains than those of the parent strain. Although the same phenotype was clearly observed here, there are currently no data to link the BrpA-mediated effect to the HK/RR11 system. Future studies will be necessary to examine possible interactions among the HK/RR11 system, brpA, and the comCDE quorum sensing system. Since we suspected that the hk/rr11 genes may encode a peptide sensing system, we searched the region for small ORFs that encoded proteins encompassing potential double GG cleavage sites typical of secreted signal peptides but were unable to identify any candidate genes. The putative function of the surrounding genes does not suggest obvious roles in genetic competence, biofilm formation, or acid tolerance. Although these neighboring genes do not obviously appear to have a role in the phenotypes currently associated with the CSP response, they may aid in optimal existence in a biofilm or a high-cell-density environment. For example, the gene encoding pyruvate formate lyase-activating enzyme, pflC, in S. pneumoniae was recently demonstrated to be activated by the pneumococcal CSP system (28), the gene for which has no apparent relationship to genetic competence. The pflC gene encoding the pyruvate formate lyase-activating enzyme is in close proximity to hk/rr11 genes; it would be interesting to investigate a linkage between these genes, since in S. mutans pyruvate formate lyase is extremely oxygen sensitive and likely functions optimally at high cell density in anaerobic biofilms (31).
Another interesting observation was that the hk11 mutant (SMHK11) was significantly impaired in acid tolerance. SMHK11 was defective both in growth at a low pH and in resistance to acid killing after adaptation to a signal pH (pH 5.5). This suggested that the membrane-associated protein encoded by hk11 might act as a pH sensor involved in activation of one of the many pathways believed to affect the acid-tolerant phenotype of S. mutans. TCSTSs have been shown elsewhere to act as pH sensors: most notably actSR of Rhizobium and lisRK in Listeria monocytogenes (5) are essential for induction of the adaptive ATR (10). Interestingly, only SMHK11 appeared acid sensitive, since we did not observe the same phenotypic effect when the rr11 gene was deleted. Intuitively, one would expect that inactivation or deletion of either of the genes encoding a TCSTS would generate a similar phenotype, since a defect in either the histidine kinase receptor or the cognate response regulator might hinder the input signal from activating genes and pathways controlled by the response regulator (29). In our study, however, the observation that SMHK11 and SMRR11 had different phenotypes suggested that there may have been cross talk between related receptors in which the histidine kinase sensor protein of the hk/rr11 system could pass the pH signal to one or more noncognate response regulators. This phenomenon, called in vivo cross talk, has been described recently by Verhamme et al. (34), who demonstrated interaction among four key two-component systems in Escherichia coli by an in vivo approach. Their results suggested that a functional histidine phosphoryl-transfer (HPt) domain of a sensor kinase appears to be the active participant in physiological cross talk. Further studies will be needed to identify a putative response regulator(s) involved in cross talk occurring via the HK11 sensor protein in SMRR11.
A comparison of the 2D profiles of SMHK11 and NG8 revealed that 14 of the acid-inducible (and -repressible) proteins were conserved between the mutant and parent (Table 3). SMHK11 did, however, have four proteins visible in NG8 that were not detected in the mutants. One of these proteins, 1008, was possibly the histidine kinase HK11 itself that migrated at 30 kDa with a pI of 5.5. The values obtained from the deduced protein sequence of HK11 were as follows: calculated molecular mass of 38,113 Da and an estimated pI of 5.71. Another interesting protein that was not induced in SMHK11 was spot number 87, which represents an exopolyphosphatase. Polyphosphate metabolism has been linked to biofilm formation in many bacteria (3, 26, 27) including S. mutans (30). Polyphosphate likely provides a rapid source of energy needed to cope with environmental fluctuations encountered during biofilm growth.
The identification of promoter-like structures 5′ from rr11 bearing a striking similarity to the com-box of S. pneumoniae is intriguing. Expression of rr11 under CSP-limiting and -inducing conditions could help lead to a deciphering of the S. mutans com-box. We have identified similar structures in proximity to late-competence orthologs found in the S. mutans genome. Deduction of the S. mutans com-box could hasten our unraveling of this regulon, as it would allow us to identify candidate genes by in silico analysis. An understanding of CSP-mediated and other genes involved in expression of the biofilm phenotype will hopefully allow us to discover means to control problematic biofilms.
Acknowledgments
We thank Robert Chernecky for the SEMs.
Our work was supported by PHS grant DE 013230 from the National Institute of Dental and Craniofacial Research and grant MT-15431 from the Canadian Institutes of Health Research and by infrastructure grants from the Canadian Foundation for Innovation and The Ontario Innovation Trust. D.G.C. is supported by a Canada Research Chair.
REFERENCES
- 1.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 2.Buckley, N. D., L. N. Lee, and D. J. LeBlanc. 1995. Use of a novel mobilizable vector to inactivate the scrA gene of Streptococcus sobrinus by allelic replacement. J. Bacteriol. 177:5028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, W., R. J. Palmer, and H. K. Kuramitsu. 2002. Role of polyphosphate kinase in biofilm formation by Porphyromonas gingivalis. Infect. Immun. 70:4708-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 5.Cotter, P. D., N. Emerson, C. G. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cvitkovitch, D. G., D. A. Boyd, T. Thevenot, and I. R. Hamilton. 1995. Glucose transport by a mutant of Streptococcus mutans unable to accumulate sugars via the phosphoenolpyruvate phosphotransferase system. J. Bacteriol. 177:2251-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dziejman, M., and J. J. Mekalanos. 1995. Two-component signal transduction and its role in the expression of bacterial virulence factors, p. 305-317. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 9.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenn, A. R., W. G. Reeve, R. P. Tiwari, and M. J. Dilworth. 1999. Acid tolerance in root nodule bacteria. Novartis Found. Symp. 221:112-126. [PubMed] [Google Scholar]
- 11.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoch, J. A., and T. J. Silhavy (ed.). 1995. Two-component signal transduction. ASM Press, Washington, D.C.
- 13.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 14.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 15.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 16.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, S. F., Y. H. Li, and G. H. Bowden. 1996. Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infect. Immun. 64:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y. H., M. N. Hanna, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y. H., N. Tan, M. B. Aspiras, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh, D. J. 2000. Oral ecology and its impact on oral microbial diversity. Horizon Scientific Press, Wymondham, Norfolk, United Kingdom.
- 23.Marsh, P. D. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8:263-271. [DOI] [PubMed] [Google Scholar]
- 24.Matsushita, M., and K. D. Janda. 2002. Histidine kinases as targets for new antimicrobial agents. Bioorg. Med. Chem. 10:855-867. [DOI] [PubMed] [Google Scholar]
- 25.Quivey, R. G., W. L. Kuhnert, and K. Hahn. 2001. Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:301-314. [DOI] [PubMed] [Google Scholar]
- 26.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 29.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 30.Svensater, G., J. Welin, J. C. Wilkins, D. Beighton, and I. R. Hamilton. 2001. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 205:139-146. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi, N., K. Abbe, S. Takahashi-Abbe, and T. Yamada. 1987. Oxygen sensitivity of sugar metabolism and interconversion of pyruvate formate-lyase in intact cells of Streptococcus mutans and Streptococcus sanguis. Infect. Immun. 55:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 34.Verhamme, D. T., J. C. Arents, P. W. Postma, W. Crielaard, and K. J. Hellingwerf. 2002. Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology 148:69-78. [DOI] [PubMed] [Google Scholar]
- 35.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]