Abstract
The gram-positive, aerobic, moderately halophilic bacterium Halobacillus halophilus is challenged in its environment by frequently changing salt (NaCl) concentrations. Recently, H. halophilus was shown to be the first prokaryote that is dependent on Cl− for growth. In a search for the biological function of Cl− in this prokaryote, we identified different Cl−-dependent processes, which suggests a more general role for Cl− in the metabolism of H. halophilus. To analyze the effect of Cl− in more detail, we concentrated on one model system, the Cl−-dependent production of flagella, and aimed to identify the molecular basis for the Cl− dependence of flagellum production. Here, we report that synthesis of the major subunit of the flagellum, FliC, is dependent on the Cl− concentration of the medium, as determined by Western blot analyses. The gene encoding FliC was cloned and sequenced, and Northern blot as well as reverse transcriptase PCR analyses revealed that expression of fliC is Cl− dependent. FliC is the first protein of known function demonstrated to be synthesized in a Cl−-dependent manner in a prokaryote. Two-dimensional gel electrophoresis of cells grown under different conditions revealed five more Cl−-induced proteins; these were identified by N-terminal sequencing and database searches to be orthologs of proteins involved in stress response in Bacillus subtilis. The data indicate that Cl− is an important environmental signal in this moderate halophile and regulates protein synthesis and gene expression. Furthermore, the data may suggest that Cl− plays a role in the signal transduction involved in salt perception by this bacterium.
Cells are adapted to a certain salinity or salinity range for optimal cellular function. Despite their different salt requirements, living cells have developed only two mechanisms to cope with water stress. To prevent loss of water, some anaerobic bacteria and some archaea (the halobacteria) accumulate salt (KCl) in the cytosol to counterbalance the external salt concentration (11, 22). The majority of prokaryotes accumulate compatible solutes in response to increasing external salt concentrations (14, 28, 31). Compatible solutes are defined as small, neutral, highly soluble organic molecules that do not interfere with cellular metabolism, and they are accumulated by either salt-induced de novo synthesis or salt-induced uptake from the medium (2). The physiological response of living cells to increasing external salt concentrations and the nature of compatible solutes as well as their biosynthesis and uptake mechanisms have been studied in great detail (14, 25, 28, 31).
Of course, the first step in this physiological response is the sensing of osmolality. How do microorganisms sense osmolality, and how is this signal mediated through the cell to gene expression and enzyme activation? The answer to this question might be different for different organisms. There have been model studies with the slightly halotolerant bacteria Escherichia coli and Bacillus subtilis, which can adapt to NaCl concentrations up to 1 M (1, 4, 14). These studies led to the conclusion that the cells sense their own level of turgor. One of the possible mechanisms for this is that membrane-bound sensors record a change in turgor by recognizing alterations in transmembrane helix-helix interactions (12, 41, 42, 45). Recent studies using purified transporters for compatible solutes and a membrane-bound osmosensor involved in potassium uptake in E. coli confirmed this hypothesis but also demonstrated that a certain ionic environment on the “right” side of the membrane is also required (13, 33, 42). Therefore, the question of how salt is sensed by these model organisms is still open, and there might be different strategies in different bacteria.
The moderately halophilic bacteria are a specialized group of organisms which require NaCl for growth and grow over a rather wide range of external salt concentrations (0.5 to 2.0 M), indicating effective salt adaptation mechanisms (43). Recently, we found that the moderately halophilic, aerobic bacterium Halobacillus halophilus requires chloride, one component of common salt, for growth and salt adaptation (30). H. halophilus is the first organism for which a Cl− dependence has been demonstrated. Because H. halophilus accumulates compatible solutes but not KCl to counterbalance the external salt concentration (40), a function of Cl− solely as an intracellular anionic osmolyte is excluded. Furthermore, in addition to growth, endospore germination, activation of transport of the compatible solute glycine betaine, and motility and flagellum production have been identified as Cl−-dependent processes (5, 29, 32). These very different functions of Cl− suggest a more general role for chloride in the physiology of H. halophilus, such as involvement in regulatory processes or interaction with the environment. To test this possibility, we studied on a molecular level the function of chloride in one physiological process, motility, as a paradigm and found evidence that the production of the structural component of the flagellum, flagellin, as well as the expression of the encoding gene, is chloride dependent. In addition, we identified by two-dimensional (2-D) gel electrophoresis five more chloride-induced proteins with very different functions, but most of their orthologs are known to be involved in stress protection in B. subtilis (24, 26). These results point to a possible function of chloride as a signal molecule in salt adaptation in the moderately halophilic bacterium H. halophilus and suggest that this organism senses salinity via the chloride concentration of its environment.
MATERIALS AND METHODS
Organism and cultivation.
Stock cultures of H. halophilus (DSM 2266) were maintained on NB (5 g of peptone/liter, 3 g of beef extract/liter; Difco, Augsburg, Germany) supplemented with 0.05 M MgSO4 and 1 M NaCl. For growth experiments, NaCl and NaNO3 were added as indicated. Motile cells were selected and maintained by several passages over swarm-agar plates (0.3% agar) which contained NB, 0.05 M MgSO4, and 1 M NaCl.
Preparation of flagella.
For preparations of flagella, six cultures, each with 2,000 ml of NB containing 1 M NaCl and 0.05 M MgSO4, were inoculated (1%) from cultures of motile cells maintained on the same medium. Cultures were incubated at 30°C and shaken at 110 rpm. Cells were harvested at the end of the exponential growth phase by centrifugation (6,000 × g, 15 min) and resuspended in 200 ml of 0.05 M Tris (pH 7.5) containing 0.05 M MgSO4 and 1 M NaCl (Tris buffer). To shear off the flagella, this concentrated cell suspension was put into a mixer (Bauknecht, Schorndorf, Germany) and subjected to vigorous mixing for 90 s. Cells were removed by low-speed centrifugation (10,000 × g, 15 min). The supernatant was subsequently subjected to ultracentrifugation (100,000 × g, 1 h), and flagella were sedimented. The resulting pellet was resuspended in 1 ml of Tris buffer, and the flagellum preparation was stored at 4°C until further use.
Electroelution and generation of polyclonal antibodies.
The flagellum preparation was mixed with an appropriate volume of threefold-concentrated denaturing buffer as described previously (38), boiled for 10 min, and subsequently subjected to sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel electrophoresis (PAGE) according to the method previously described (38). The polyacrylamide gel was stained with Coomassie brilliant blue, and the protein was cut out and removed from the gel by electroelution in a Biotrap chamber (Schleicher & Schuell, Dassel, Germany) for 12 h at 100 V. Electroelution was done in a buffer containing 25 mM Tris, 192 mM glycine, and 0.025% SDS. The purified protein was lyophilized, and 300 μg of it was used to raise polyclonal antibodies in rabbits (BioScience, Göttingen, Germany).
SDS-PAGE and immunoblots.
Cells were harvested by centrifugation (10,000 × g, 3 min), resuspended in denaturing buffer as described previously (38), and boiled for 10 min. Proteins were separated by SDS-PAGE on 12.5% gels and transferred to nitrocellulose membranes (Schleicher & Schuell) using a semidry blotting chamber (Bio-Rad, Munich, Germany). Membranes were blocked with 0.5% skim-milk powder in PBST (140 mM NaCl, 10 mM KCl, 6.4 mM Na2HPO4, 2 mM KH2PO4, 0.05% Tween 20) for 1 h, washed three times in PBST for 10 min, and incubated with antisera (4 μg of PBST/ml) for 12 h at room temperature. The membranes were washed again three times with PBST for 30 min and then incubated for 1 h with protein A-horseradish peroxidase conjugate. After three additional washing steps (10 min), luminescence was detected using the chemiluminescence blotting substrate from Roche Molecular Biochemicals (Mannheim, Germany), and signals were detected by autoradiography with Kodak X-OMAT-AR film.
Probe construction and labeling.
Chromosomal DNA of H. halophilus was prepared as described previously (9). A genomic library was constructed by partial digestion of chromosomal DNA with MboI and cloning into the BamHI restriction site of pUC18. fliC and atpD were cloned from this genomic library by colony hybridization (3) using the corresponding genes of B. subtilis and Acetobacterium woodii, respectively, as probes (10, 18). DNA sequencing was performed automatically by using an ABI Prism 310 genetic analyzer (Applied Biosystems, Weiterstadt, Germany) with fluorescence-labeled DNA.
The DNA fragments were radiolabeled with [γ-32P]dATP (Hartmann Analytic GmbH, Braunschweig, Germany) using the Random Primed DNA labeling system (Gibco BRL GmbH, Eggenstein, Germany) (7). Following 32P-labeling, probes were separated from unincorporated nucleotides by using the QIAquick nucleotide removal kit (Qiagen, Hilden, Germany). The specificity of the probes was confirmed by Southern blot analysis as described previously (35).
Preparation of cell extracts and 2-D gel electrophoresis.
Cells were grown to the late exponential growth phase in NB supplemented with 0.05 M MgSO4 and 1 M NaCl or 1 M NaNO3 and harvested by centrifugation (6,000 × g, 15 min). Cells were lysed by resuspension in denaturing buffer (9 M urea, 2% Triton X-100, 130 ml of dithiothreitol, 20 μl of Servalyte 3-10 Iso-Dalt/ml [Serva, Heidelberg, Germany], 8 mM phenylmethylsulfonyl fluoride) and incubation for 2 h at room temperature. Cell debris was removed by centrifugation (10,000 × g, 3 min), and the supernatant was subjected to 2-D gel electrophoresis.
The general method used for 2-D gel electrophoresis was that of O'Farrell (21). One hundred fifty micrograms of protein was subjected to isoelectric focusing by use of 18-cm Immobiline dry strips (Pharmacia, Uppsala, Sweden) with a linear pH gradient from pH 4 to pH 7 at 55 to 60 kV 52 h. In the second dimension, 18- by 18-cm 12.5% polyacrylamide gels were used (38). The fixed gels were Coomassie stained (44) and analyzed using ImageQuant (Molecular Dynamics, Sunnyvale, Calif.) and ImageMaster 2D (Pharmacia) software.
Protein sequencing.
For N-terminal sequencing, the proteins were transferred to polyvinylidene difluoride membranes by using a semidry blotting chamber (Bio-Rad). The protein-containing polyvinylidene difluoride membrane pieces were excised, cut into small pieces (3 by 3 mm), and incubated with 500 μl of 0.2% polyvinylpyrrolidone (PVP 30) in water for 30 min at room temperature. The supernatant was discarded, and the membrane was washed six times with water and incubated with 0.1 M Tris-HCl (pH 8.0), 2 mM CaCl2, 10% acetonitrile, 1% nonylphenoxypolyetoxy ethanol (Tergitol NP-40), and 0.5 μg of endoproteinase LysC (Boehringer, Mannheim, Germany) for 8 h at 37°C.
Northern blots.
Cells were grown to the late exponential growth phase in NB supplemented with 0.05 M MgSO4 and 1 M NaCl or 1 M NaNO3. A 1.5-ml portion of the culture was harvested by centrifugation and resuspended in 100 μl of TE buffer (50 mM Tris, 10 mM EDTA [pH 8.0]). Three hundred micrograms of lysozyme was added, and the cells were lysed by incubation at room temperature for 2 min. Subsequently, total RNA was isolated by using the RNeasy system according to the instructions of the manufacturer (Qiagen). Residual DNA was removed by DNase I treatment (Boehringer). The success of this treatment was checked by PCR. The resulting RNA preparations contained 100 to 200 μg of RNA/ml as determined by spectroscopy. Denaturing agarose gel electrophoresis of RNA in the presence of formaldehyde, transfer to nylon membranes (Amersham Buchler GmbH, Braunschweig, Germany), and Northern blot hybridization were essentially performed as described previously (35). Finally, the blots were visualized using Kodak storage phosphor screens and a Storm 860 laser scanner (both from Molecular Dynamics). Densitometric analyses were performed with ImageQuant software.
RT-PCR.
One microgram of isolated total RNA was used in a reverse transcriptase (RT) reaction using the Omniscript RT kit (Qiagen) and an oligonucleotide as indicated. The RT reaction was performed by following the instructions of the supplier. Subsequently, a PCR with the oligonucleotides indicated was performed using the RT preparations as the template. One microgram of total RNA was subjected to an RT reaction with the oligonucleotide RTFla11 (5′-TCACTAGTTGCTACAGTCCA-3′) as a primer. Subsequently, a PCR was performed with RTFla11 and RTFla21 (5′-TGCAGAAGGAGCGTTGAATG-3′).
Nucleotide sequence accession numbers.
The DNA sequences of fliC and atpD were deposited in GenBank under accession no. AY134860 and AY134861, respectively.
RESULTS
Purification of flagellin of H. halophilus and generation of an antiflagellin antiserum.
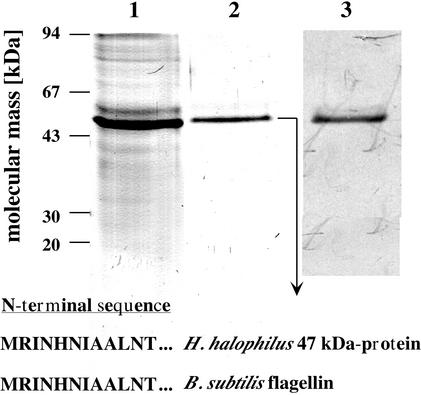
To purify the flagellin of H. halophilus, flagella were sheared off the cell body and prepared by differential centrifugation as described in Materials and Methods. When this sample was subjected to SDS-PAGE, one predominant protein with a molecular mass of 47 kDa was visible (Fig. 1). The 47-kDa protein was eluted out of the gel and subjected to N-terminal sequence analysis. The sequence of the first 12 amino acids determined was 100% identical to that of flagellin of B. subtilis, which is evidence that the 47-kDa protein purified from H. halophilus is indeed flagellin (FliC). An antiserum was generated against FliC of H. halophilus in rabbits; this antiserum was highly specific and reacted with only one protein, which had the expected size of 47 kDa (Fig. 1).
FIG. 1.
Purification of flagellin of H. halophilus and specificity of the antiflagellin antiserum. Flagella were prepared as described in Materials and Methods and subjected to SDS-PAGE (lane 1). The 47-kDa protein was electroeluted (lane 2) and identified as flagellin by its N-terminal amino acid sequence. The specificity of the generated antiserum was determined in a Western blot analysis using crude cell extract of H. halophilus (lane 3).
Chloride-dependent production of flagellin.
To analyze the effect of Cl− on flagellin production, motile cells of H. halophilus were pregrown overnight in NB containing 0.05 M MgSO4 and 1 M NaNO3. It has been shown before that NO3− can be a substitute for Cl− during growth (after some adaptation time) but not in motility (30, 32). This preculture was used to inoculate NB containing 0.05 M MgSO4 and 1 M NaCl or 1 M NaNO3. During growth, samples were taken at the time points indicated in Fig. 2A, motility was monitored by light microscopy, and the cellular level of flagellin was determined with Western blots. Cells grown in NB supplemented with 1 M NaCl were motile during the exponential growth phase (Fig. 2A, time points 1 and 2) and during early stationary phase (Fig. 2A, time point 3), but motility decreased thereafter. This corresponds well to the cellular level of flagellin (Fig. 2A). On the other hand, cells grown in NB supplemented with 1 M NaNO3 had a drastically reduced flagellin content. When analyzed under the same conditions as those for Cl−-grown cells, flagellin was not detectable in a Western blot analysis (Fig. 2B). However, when the amount of protein was increased sixfold, flagellin became detectable. This experiment clearly demonstrated a strong stimulation of flagellin production by Cl−.
FIG. 2.
Synthesis of flagellin of H. halophilus is Cl− dependent. (A) Cells were pregrown in the presence of NO3− and subsequently transferred to medium containing Cl− (▪) or NO3− (○). At the four time points indicated, motility was judged by light microscopy and the cellular flagellin level was determined by Western blots. Twenty-five micrograms of protein was applied to each lane. OD600, optical density at 600 nm. (B) Western blot analysis of different amounts of cell extract hybridized against the antiflagellin antiserum.
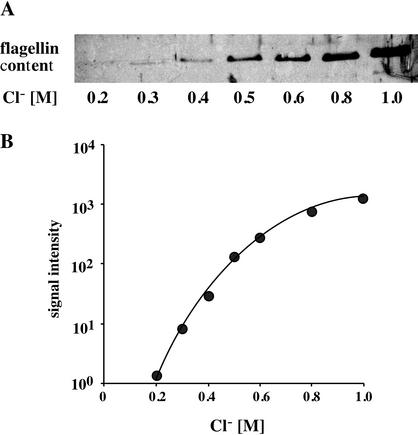
Next, the dependence of the flagellin content of the cells on the Cl− concentration was analyzed quantitatively. Motile cells of H. halophilus were grown in NB with increasing Cl− concentrations ranging from 0.2 to 1 M, while the salt concentration was kept constant at 1 M by appropriate addition of NaNO3. Crude cell extracts were prepared, subjected to SDS-PAGE, and analyzed by immunoblot analysis. Figure 3A shows that there was a negligible flagellin content at a 0.2 M concentration of Cl−, but increasing Cl− concentrations led to increasing amounts of flagellin. A plot of the signal intensities against the Cl− concentrations (Fig. 3B) revealed that the amount of flagellin increased by several orders of magnitude and reached an optimum at external Cl− (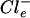 ) concentrations which are also optimal for the growth of H. halophilus (0.8 to 1.0 M) (30).
) concentrations which are also optimal for the growth of H. halophilus (0.8 to 1.0 M) (30).
FIG. 3.
Quantification of the Cl− dependence of the cellular flagellin level of H. halophilus. Cells were grown under isosmotic conditions (the salt concentration of the medium was kept constant at 1 M by the appropriate addition of NaNO3) with the Cl− concentrations indicated. Cells were grown to the late exponential growth phase and harvested, and 25 μg of protein was applied to an SDS-polyacrylamide gel. The flagellin concentration was determined by Western blot analysis (A), and the signal intensity was plotted against the Cl− concentration (B).
Synthesis of flagellin is shut down in the absence of Cl−.
To determine whether the synthesis of flagellin is not only stimulated by the addition of Cl− but also, vice versa, decreased in the absence Cl−, motile cells of H. halophilus grown in the presence of Cl− were harvested in the late exponential growth phase and resuspended in Tris buffer containing 1 M NaCl. This concentrated cell suspension was passaged 20 times through a sterile needle (26 gauge) to shear off the flagella, and cells were then transferred into fresh medium containing 1 M NaNO3 or 1 M NaCl. Cells transferred into Cl−-containing medium produced flagellin shortly (3 h) after the transfer. Similar to what was seen before, cells contained flagellin and were motile over the entire growth curve (Fig. 4). In contrast, flagellum-free cells resuspended in Cl−-free medium could not synthesize flagellin and, consequently, were nonmotile over the entire growth curve. These results demonstrate that in cells initially producing flagellin, the production is stopped as soon as Cl− is replaced by NO3−.
FIG. 4.
Flagellin expression is shut down in the absence of Cl−. Cells were pregrown in the presence of 1 M NaCl, flagella were sheared off, and cells were transferred to medium containing NO3− or Cl−. At four different time points (see the legend to Fig. 2A), samples were taken and analyzed for motility by light microscopy and for flagellin content by Western blot analyses. pc, preculture.
Stability of flagellin.
To prove that the lack of flagellin in cells growing in the absence of Cl− is indeed due to reduced protein synthesis and not due to a lack of stability of the flagella in NO3−-containing medium, motile cells were grown in 1 M NaCl, resuspended in Tris buffer with 1 M NaCl or 1 M NaNO3, and incubated at 30°C and flagella were prepared as described above. After 24 h, these flagellum suspensions were subjected to ultracentrifugation; hence, macromolecular structures were sedimented. The pellets were resuspended in fresh Tris buffer, and aliquots were taken and subjected to SDS-PAGE and immunoblot analysis. The same procedure was repeated after 48 h. The amounts of flagellin sedimented were identical after both 24 and 48 h of incubation (data not shown), indicating that the macromolecular structures of the flagella exhibit the same stability independent of the anion present in the buffer. This was confirmed by electron microscopy, which did not reveal a difference in the lengths of the flagella after incubation for 48 h in Tris buffer containing NO3− or Cl− (data not shown).
Transcription of the gene encoding flagellin, fliC, is chloride dependent.
The results presented so far unequivocally provided evidence that the synthesis of flagellin in H. halophilus is significantly stimulated by chloride. Next, we addressed the question of whether the transcription of the gene encoding flagellin, fliC, is also chloride dependent. For this purpose, part of fliC of H. halophilus was cloned as described in Materials and Methods. This fragment corresponds to bases 190 to 607 of B. subtilis fliC, and the deduced protein fragment is 56% similar to the corresponding fragment of FliC of B. subtilis. This DNA fragment was used as a probe in Northern blots. atpD, encoding the β-subunit of the F1Fo-ATPase of H. halophilus, was cloned as well (see Materials and Methods) and used as a control in Northern blots, since its transcription is expected to be constitutive and unaffected by Cl−.
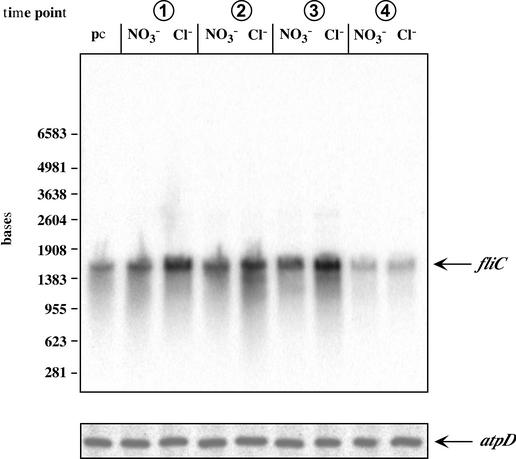
To analyze the transcription of fliC and atpD of H. halophilus, Northern blot analyses were performed. Cells were grown in NB with 1 M NaCl or 1 M NaNO3, and total RNA was isolated at the time points indicated in Fig. 2A and hybridized against fliC or atpD (Fig. 5). The transcription of atpD remained constant during growth, and as expected, there was no difference between cells grown in the presence of Cl− or NO3−. The fliC probe hybridized with one specific RNA fragment of 1,500 bp. This size fits with the molecular mass of 47 kDa for flagellin. It is evident from Fig. 5 that transcription of fliC during the exponential phase and that during the early stationary phase were identical but declined in the late stationary phase. This corresponds nicely to the data obtained by Western blot analyses. Most interestingly, the amount of the fliC transcript was significantly increased when cells were grown in the presence of Cl−. Densitometric analyses revealed a twofold stimulation. To verify the results of the Northern blots, RT-PCR studies were performed as described in Materials and Methods. These analyses confirmed the stimulation of fliC transcription by Cl− (data not shown). Together, these results corroborated the observed Cl−-dependent synthesis of flagellin.
FIG. 5.
Transcription of fliC is Cl− dependent. H. halophilus was pregrown in the presence of NO3− and then transferred to medium containing Cl− or NO3−. Total RNA was isolated at the time points indicated (Fig. 2A) and hybridized against fliC or atpD. pc, preculture.
Identification of additional Cl−-induced proteins in H. halophilus.
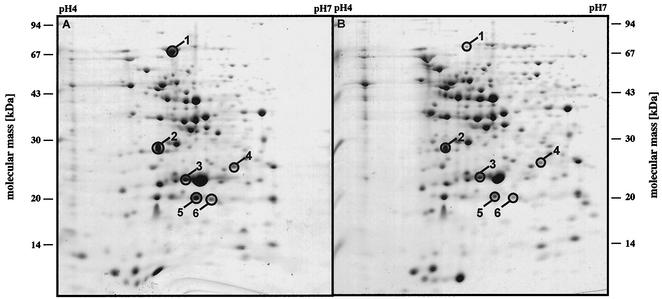
Having determined that the expression of FliC is Cl− dependent, we next addressed the question of whether additional proteins are induced by Cl−. For this purpose, cells were grown in the presence of 1 M Cl− or NO3−, harvested, and resuspended in denaturing buffer, and proteins were separated by 2-D gel electrophoresis. In a total of eight independent experiments, six proteins were reproducibly found in elevated concentrations in cells grown in the presence of Cl− (Fig. 6). Densitometric analyses demonstrated that the level of induction varied from 2.5- to 25-fold at its maximum (Table 1). These proteins were cut out of the gels, and their N-terminal amino acid sequences were determined (Table 1). BLAST searches revealed homologs in other organisms, and the functions of all except one of these homologs are known (Table 1). Protein 1 is similar to an ABC transporter subunit, but the substrate transported cannot be delineated with this approach. Proteins 3, 4, and 5 are all involved in regulatory processes. Protein 5 is similar to LuxS, which is involved in the biosynthesis of AI-2 (39), an autoinducer involved in a quorum-sensing pathway found in gram-negative as well as gram-positive bacteria (15, 34, 39). Remarkably, two of these proteins (SodA and YvyD) belong to the σB regulon of B. subtilis, which forms the core of the general stress response of this organism (24, 26). Additionally, it was shown recently that YvyD modulates the activity of the alternative transcription factor σL (8). Together, these results clearly indicate that Cl− regulates gene expression in H. halophilus.
FIG. 6.
Identification of Cl−-induced proteins by 2-D gel electrophoresis. Cells were grown in the presence of 1 M Cl− (A) or NO3− (B), and crude extracts were subjected to 2-D gel electrophoresis. Indicated proteins were induced at least twofold in eight repetitions of this experiment. The proteins are identified in Table 1.
TABLE 1.
Identification of Cl−-induced proteins of H. halophilus by N-terminal sequence analyses and BLAST searches
| Protein | N-terminal sequence | Induction (fold) | Similaritya | Organism | Function(s) |
|---|---|---|---|---|---|
| 1 | ...ALSDIPDKYASEI... | 25 | Putative protein | Streptomyces coelicolor | ABC transporter, ATP-binding protein |
| 2 | MKVLVVGANGQIGKHLVSTIQESNKLEAKAMI... | 2.5 | YhfK | B. subtilis | Unknown |
| 3 | ...AKFELPELPYAYDALEPTIDKETM... | 10 | SodA | B. subtilis | Superoxide-dismutase |
| 4 | MLQYTIRGENLEVTDSIKDYVEKKVGK... | 6 | YvyD | B. subtilis | σL modulator |
| 5 | MQMNVEVFNLDHTKVKAP... | 3.3 | LuxS | Helicobacter pylori | Autoinducer synthesis |
| 6 | NDb | 5 |
Highest similarity in a BLAST search.
ND, not determined.
DISCUSSION
Organisms like marine (or moderate) halophiles which live in habitats such as seawater, salt marshes, or hypersaline environments are challenged mainly by common salt, NaCl. They respond to changes in external NaCl concentrations, and one can imagine that cells interact with one or both components of common salt, Na+ and/or Cl−. Na+ has been shown before to be used as an environmental signal involved in gene regulation (23, 27). Upon salt stress, Na+ enters the cell and activates expression of genes catalyzing energy-dependent efflux systems of cytotoxic Na+. Intracellular Na+ binds to a transcriptional regulator which then activates expression of Na+/H+ antiporter genes (6). Interestingly, optimal gene expression is achieved at internal Na+ concentrations of ≈20 mM (7, 23), a rather high concentration when compared with that of other ions involved in gene regulation, but this simply reflects the rather high intracellular and extracellular concentrations of Na+ required by microorganisms. The Cl−-dependent expression of genes has been reported previously in a lactic acid bacterium (36, 37), but this organism does not depend on Cl− for growth and the physiological function of the protein induced by Cl− is unknown. The situation is completely different in H. halophilus because this organisms depends on Cl− for growth, indicating an essential function in cellular metabolism. To determine the role of Cl− in the physiology of this moderate halophile, we chose motility as a model system. The data presented here demonstrate that fliC transcription and flagellin production in H. halophilus are very strongly dependent on Cl−. The amount of Cl− required for optimal motility was the same as that required for optimal growth. Since expression of fliC was not completely repressed in the absence of Cl−, additional, Cl−-independent transcriptional activation mechanisms must exist. The regulation of transcription of one gene by various factors is indeed observed very often and reflects the requirement of cross talk between different regulatory networks. In contrast to fliC expression, synthesis of flagellin was strongly stimulated by Cl−, resulting in the absence of flagella in the absence of Cl−. Therefore, different regulatory layers, i.e., transcription, posttranscription, translation, and posttranslation, might be simultaneously and/or differently affected by Cl−. This is also not unusual but has been observed before with other regulatory systems, e.g., synthesis of the E. coli alternative sigma factor σS, whose production is regulated on different levels (16, 17, 19, 20).
Of course, motility itself is not required for salt adaptation per se, but we used it as a model system. The central question is what is the molecular basis for the Cl− dependence of growth? The observation that Cl− is required in H. halophilus for very different functions can result from the fact that Cl− is a key player in regulatory networks. One then has to argue that the additional Cl−-dependent functions in H. halophilus, glycine betaine transport and endospore germination (5, 29), are all part of the same Cl−-dependent regulatory network. This network must also regulate functions essential for growth, most likely including the accumulation of compatible solutes. It is conceivable that cells incubated in the absence of Cl− cannot accumulate compatible solutes due to the lack of the signal molecule. This is corroborated by the fact that not only the growth rates of H. halophilus but also the final cell yields are chloride dependent, indicating that a cellular component is diluted out during growth in the absence of Cl− (30). The presence of a global system responding to Cl− could indeed be demonstrated here by 2-D gel electrophoresis. The proteins induced by Cl− were identified by their N-terminal amino acid sequences. The ABC-type transporter could be involved in the uptake of compatible solutes, but this has to be proven experimentally. Most remarkable are the Cl−-induced superoxide dismutase and YvyD, because both proteins have been identified as components of a stress regulon in a close relative, B. subtilis (24, 26). It is conceivable that a salt stress regulon from a halotolerant was converted to being a regulon conferring salt resistance and, at the same time, a salt requirement for a moderate halophile such as H. halophilus or vice versa. The transformation from salt-stressed halotolerant organism to moderate halophile was accompanied by the change of an inducible stress regulon to a constitutive regulon strictly required for growth; gene expression and protein production can now occur optimally only at “high” salt concentrations (0.8 to 1.0 M in H. halophilus), which results in the strict salt dependence of growth. If the salt concentration falls below a certain threshold, then growth is impaired due to the lack of gene expression. However, the same regulon can be used to adjust the physiological functions of the cell to the wide range of external salt concentrations promoting the growth of H. halophilus.
The Cl−-induced protein, YvyD, of H. halophilus might be a key player in the Cl−-dependent regulatory network. YvyD of B. subtilis has been shown previously to regulate activity of the sigma factor σL (8). Therefore, it might be that YvyD of H. halophilus is involved in Cl−-dependent gene expression either indirectly or directly as a Cl−-activated activator of an alternative sigma factor. A direct interaction of Cl− with YvyD (or any other protein) would, of course, require a certain intracellular Cl− (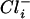 ) concentration in H. halophilus. It has been shown previously that H. halophilus keeps a rather steep Cl− gradient of 10 (
) concentration in H. halophilus. It has been shown previously that H. halophilus keeps a rather steep Cl− gradient of 10 (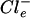 >
> 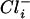 ) across its cytoplasmic membrane at suboptimal external Cl− concentrations. Increasing
) across its cytoplasmic membrane at suboptimal external Cl− concentrations. Increasing 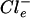 concentrations lead to increased
concentrations lead to increased 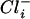 concentrations. At a gradient of 2 and a
concentrations. At a gradient of 2 and a 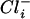 concentration of 400 mM, growth was optimal, and it was speculated that a
concentration of 400 mM, growth was optimal, and it was speculated that a 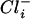 concentration of >400 mM is crucial for optimal growth (30).
concentration of >400 mM is crucial for optimal growth (30).
In summary, the study presented here demonstrates the presence of a Cl−-dependent regulatory network in the moderately halophilic H. halophilus, the first to be identified in a living cell. Cl−-induced proteins were identified, and the expression of a paradigm, fliC, was shown to be stimulated by Cl−. This regulatory network might confer salt dependence (halophily) as well as salt tolerance on this organism. Future studies will be aimed at analyzing this fascinating new network in more detail.
Acknowledgments
We are indebted to J. Kellermann, Max-Planck-Institut für Biochemie, Martinsried, Germany, for N-terminal amino acid sequence analyses and to G. Wanner for performing the electron microscopy.
REFERENCES
- 1.Altendorf, K., M. Gassel, W. Puppe, T. Mollenkamp, A. Zeeck, C. Boddien, K. Fendler, E. Bamberg, and S. Drose. 1998. Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. Suppl. 643:137-146. [PubMed] [Google Scholar]
- 2.Brown, A. D. 1976. Microbial water stress. Bacteriol. Rev. 40:803-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buluwela, L., A. Forster, T. Boehm, and T. H. Rabbitts. 1989. A rapid procedure for colony screening using nylon filters. Nucleic Acids Res. 17:452.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Csonka, L. N., and W. Epstein. 1996. Osmoregulation, p. 1210-1223. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 5.Dohrmann, A. B., and V. Müller. 1999. Chloride dependence of endospore germination in Halobacillus halophilus. Arch. Microbiol. 172:264-267. [DOI] [PubMed] [Google Scholar]
- 6.Dover, N., C. F. Higgins, O. Carmel, A. Rimon, E. Pinner, and E. Padan. 1996. Na+-induced transcription of nhaA, which encodes an Na+/H+ antiporter in Escherichia coli, is positively regulated by nhaR and affected by hns. J. Bacteriol. 178:6508-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dover, N., and E. Padan. 2001. Transcription of nhaA, the main Na+/H+ antiporter of Escherichia coli, is regulated by Na+ and growth phase. J. Bacteriol. 183:644-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drzewiecki, K., C. Eymann, G. Mittenhuber, and M. Hecker. 1998. The yvyD gene of Bacillus subtilis is under dual control of σB and σH. J. Bacteriol. 180:6674-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errington, J. 1984. Efficient Bacillus subtilis cloning system using bacteriophage vector ψ105J9. J. Gen. Microbiol. 130:2615-2628. [DOI] [PubMed] [Google Scholar]
- 10.Forster, A., R. Daniel, and V. Müller. 1995. The Na+-translocating ATPase of Acetobacterium woodii is a F1FO-type enzyme as deduced from the primary structure of its β, γ and ɛ subunits. Biochim. Biophys. Acta 1229:393-397. [DOI] [PubMed] [Google Scholar]
- 11.Galinski, E. A., and H. G. Trüper. 1994. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol. Rev. 15:95-108. [Google Scholar]
- 12.Jung, K., B. Tjaden, and K. Altendorf. 1997. Purification, reconstitution, and characterization of KdpD, the turgor sensor of Escherichia coli. J. Biol. Chem. 272:10847-10852. [DOI] [PubMed] [Google Scholar]
- 13.Jung, K., M. Veen, and K. Altendorf. 2000. K+ and ionic strength directly influence the autophosphorylation activity of the putative turgor sensor KdpD of Escherichia coli. J. Biol. Chem. 275:40142-40147. [DOI] [PubMed] [Google Scholar]
- 14.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 15.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 95:12456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirel, D. B., and M. J. Chamberlin. 1989. The Bacillus subtilis flagellin gene (hag) is transcribed by the σ28 form of RNA polymerase. J. Bacteriol. 171:3095-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the σS subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 20.Muffler, A., D. D. Traulsen, D. Fischer, R. Lange, and R. Hengge-Aronis. 1997. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 179:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 22.Oren, A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padan, E., M. Venturi, Y. Gerchman, and N. Dover. 2001. Na+/H+ antiporters. Biochim. Biophys. Acta 1505:144-157. [DOI] [PubMed] [Google Scholar]
- 24.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poolman, B., and E. Glaasker. 1998. Regulation of compatible solute accumulation in bacteria. Mol. Microbiol. 29:397-407. [DOI] [PubMed] [Google Scholar]
- 26.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 27.Rahav-Manor, O., O. Carmel, R. Karpel, D. Taglicht, G. Glaser, S. Schuldiner, and E. Padan. 1992. NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR), regulates nhaA, the sodium proton antiporter gene in Escherichia coli. J. Biol. Chem. 267:10433-10438. [PubMed] [Google Scholar]
- 28.Roberts, M. F. 2000. Osmoadaptation and osmoregulation in archaea. Front. Biosci. 5:D796-D812. [DOI] [PubMed] [Google Scholar]
- 29.Roeβler, M., and V. Müller. 2001. Chloride dependence of glycine betaine transport in Halobacillus halophilus. FEBS Lett. 489:125-128. [DOI] [PubMed] [Google Scholar]
- 30.Roeβler, M., and V. Müller. 1998. Quantitative and physiological analyses of chloride dependence of growth of Halobacillus halophilus. Appl. Environ. Microbiol. 64:3813-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roeβler, M., and V. Müller. 2001. Osmoadaptation in bacteria and archaea: common principles and differences. Environ. Microbiol. 3:743-754. [DOI] [PubMed] [Google Scholar]
- 32.Roeβler, M., G. Wanner, and V. Müller. 2000. Motility and flagellum synthesis in Halobacillus halophilus are chloride dependent. J. Bacteriol. 182:532-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rübenhagen, R., S. Morbach, and R. Krämer. 2001. The osmoreactive betaine carrier BetP from Corynebacterium glutamicum is a sensor for cytoplasmic K+. EMBO J. 20:5412-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruzheinikov, S. N., S. K. Das, S. E. Sedelnikova, A. Hartley, S. J. Foster, M. J. Horsburgh, A. G. Cox, C. W. McCleod, A. Mekhalfia, G. M. Blackburn, D. W. Rice, and P. J. Baker. 2001. The 1.2 Å structure of a novel quorum-sensing protein, Bacillus subtilis LuxS. J. Mol. Biol. 313:111-122. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 37.Sanders, J. W., G. Venema, and J. Kok. 1997. A chloride-inducible gene expression cassette and its use in induced lysis of Lactococcus lactis. Appl. Environ. Microbiol. 63:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecylsulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:369-379. [DOI] [PubMed] [Google Scholar]
- 39.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 40.Severin, J. 1993. Kompatible Solute und Wachstumskinetik bei halophilen aeroben heterotrophen Eubakterien. Ph.D. thesis. University of Bonn, Bonn, Germany.
- 41.Sugiura, A., K. Hirokawa, K. Nakashima, and T. Mizuno. 1994. Signal-sensing mechanisms of the putative osmosensor KdpD in Escherichia coli. Mol. Microbiol. 14:929-938. [DOI] [PubMed] [Google Scholar]
- 42.van der Heide, T., and B. Poolman. 2000. Osmoregulated ABC-transport system of Lactococcus lactis senses water stress via changes in the physical state of the membrane. Proc. Natl. Acad. Sci. USA 97:7102-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber, K., and M. Osborne. 1969. The reliability of the molecular weight determination by dodecyl sulfate polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 45.Wood, J. M. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63:230-262. [DOI] [PMC free article] [PubMed] [Google Scholar]