Abstract
Genomic rearrangements (duplications and inversions) in enteric bacteria such as Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12 are frequent (10−3 to 10−5) in culture, but in wild-type strains these genomic rearrangements seldom survive. However, inversions commonly survive in the terminus of replication (TER) region, where bidirectional DNA replication terminates; nucleotide sequences from S. enterica serovar Typhimurium LT2, S. enterica serovar Typhi CT18, E. coli K12, and E. coli O157:H7 revealed genomic inversions spanning the TER region. Assuming that S. enterica serovar Typhimurium LT2 represents the ancestral genome structure, we found an inversion of 556 kb in serovar Typhi CT18 between two of the 25 IS200 elements and an inversion of about 700 kb in E. coli K12 and E. coli O157:H7. In addition, there is another inversion of 500 kb in E. coli O157:H7 compared with E. coli K12. PCR analysis confirmed that all S. enterica serovar Typhi strains tested, but not strains of other Salmonella serovars, have an inversion at the exact site of the IS200 insertions. We conclude that inversions of the TER region survive because they do not significantly change replication balance or because they are part of the compensating mechanisms to regain chromosome balance after it is disrupted by insertions, deletions, or other inversions.
The order of orthologous genes on the chromosomes of enteric bacteria such as Escherichia coli K12 and Salmonella enterica serovar Typhimurium LT2 was shown by classical genetic exchange methods to be strongly conserved (24, 37), and this has now been confirmed by nucleotide sequence data (6, 30). This conservation was retained even after the genera diverged over 100 million years ago (32) and in spite of base pair divergence of orthologues, which averages about 15%. In addition, lateral genetic transfer has inserted nonorthologous genes that comprise about 30% of the total genetic structure, resulting in mosaic chromosomes. This conservation is surprising, because during growth in culture, chromosome rearrangements such as duplications occur at high frequencies (10−3 to 10−5) (3, 19) and some inversions and translocations, especially those with end points in the rrn operons, are common (36). These rearrangement types must have been selected against in evolution, because they are rarely detected in wild-type strains from nature. In contrast to this conservation within the enteric bacteria, gene order in most bacteria is not conserved during evolution (10, 21, 31), though species of Chlamydia also show striking conservation (35).
However, even in the enteric bacteria, two types of genomic rearrangements are often observed. Firstly, inversions and translocations due to homologous recombination between the seven rrn operons are common in species such as Salmonella enterica serovar Typhi; among 127 wild-type strains there were 21 different genome types, based on differences in order of the fragments between the rrn operons (28). Rearrangements were also found in some other serovars, such as S. enterica serovar Pullorum and S. enterica serovar Gallinarum, which are “host specialized,” but most species, especially those that are “host generalists,” do not show these types of rearrangements (29). Secondly, some wild-type strains show inversions which cover the terminus of replication (TER) region; this was first detected by genetic analysis as an inversion between E. coli K12 and S. enterica serovar Typhimurium LT2 (11, 38). This inversion was confirmed using pulsed-field gel electrophoresis (PFGE); the same technique detected inversions in the TER region involving S. enterica serovar Typhimurium, S. enterica serovar Enteritidis, and S. enterica serovar Typhi (27).
In the past few years, the complete nucleotide sequences have been published for E. coli K12 (6), E. coli O157:H7 (E. coli O157:H7 strain EDL933 and E. coli O157:H7 strain Sakai-VT2) (17, 34), S. enterica serovar Typhimurium LT2 (30), and S. enterica serovar Typhi CT18 (33). Pulsed-field gel electrophoresis showed that serovar Typhi Ty2 differs from S. enterica serovar Typhimurium LT2 by an inversion over the TER region (27). We have made sequence comparisons which show that the same inversion in serovar Typhi strain CT18 is due to homologous recombination between two of the 25 IS200 elements in serovar Typhi CT18. PCR analysis of representative strains revealed that all S. enterica serovar Typhi strains tested, but no other Salmonella strains, have an inversion at that point.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
The strains, which are maintained in 15% glycerol at −70°C in the collection of the Salmonella Genetic Stock Center (www.ucalgary.ca/∼kesander), are shown together with their sources in Table 1. Single colony isolates were used in all experiments. All strains were grown at 37°C in Luria-Bertani medium; 1.5% agar was added for solid media.
TABLE 1.
Production of PCR products, using DNA of strains of Salmonella as template, with different combinations of primers
| SGSC no.c | Strain | Serovar or speciesa | Sub-species | Source or reference | Size of PCR products (bp) with primer combination:
|
|||
|---|---|---|---|---|---|---|---|---|
| 1F + 1R | 2F + 2R | 1F + 2F | 1R + 2R | |||||
| 1412 | LT2 | S. enterica serovar Typhimurium | I | 540 | 520 | Nb | Nb | |
| 2181 | SARA1 | S. enterica serovar Typhimurium | I | 4 | 1,500 | 520 | ||
| 2182 | SARA2 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2183 | SARA3 | S. enterica serovar Typhimurium | I | 4 | 1,500 | 520 | ||
| 2184 | SARA4 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2185 | SARA5 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2186 | SARA6 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2187 | SARA7 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2188 | SARA8 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2189 | SARA9 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2190 | SARA10 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2191 | SARA11 | S. enterica serovar Typhimurium | I | 4 | 540 | 1,800 | ||
| 2192 | SARA12 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2193 | SARA13 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2194 | SARA14 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2195 | SARA15 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2196 | SARA16 | S. enterica serovar Typhimurium | I | 4 | 1,500 | 520 | ||
| 2197 | SARA17 | S. enterica serovar Typhimurium | I | 4 | 1,500 | 520 | ||
| 2198 | SARA18 | S. enterica serovar Typhimurium | I | 4 | 1,500 | 520 | ||
| 2199 | SARA19 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2200 | SARA20 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| 2201 | SARA21 | S. enterica serovar Typhimurium | I | 4 | 540 | 520 | ||
| CT18 | S. enterica serovar Typhimurium | I | 4 | N | N | 1,100 | 1,200 | |
| 2408 | TY2-b | S. enterica serovar Typhi | I | N | N | 1,100 | 1,200 | |
| 3124 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3126 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3127 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3128 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3129 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3130 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3131 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3132 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3133 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 3134 | S. enterica serovar Typhi | I | LCDC | 1,100 | 1,200 | |||
| 2770 | Malaysia | S. enterica serovar Typhi | I | T. Pangd | 1,100 | 1,200 | ||
| 2771 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2772 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2773 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2774 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2775 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2776 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2777 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2778 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2779 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2780 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2781 | Malaysia | S. enterica serovar Typhi | I | T. Pang | 1,100 | 1,200 | ||
| 2473 | SARB16 | S. enterica serovar Enteritidis | I | 7 | 540 | 520 | ||
| 2474 | SARB17 | S. enterica serovar Enteritidis | I | 7 | 1,500 | 520 | ||
| 2475 | SARB18 | S. enterica serovar Enteritidis | I | 7 | 540 | 520 | ||
| 2476 | SARB19 | S. enterica serovar Enteritidis | I | 7 | 540 | 1,800 | ||
| 2508 | SARB51 | S. enterica serovar Pullorum | I | 7 | 540 | 520 | ||
| 2509 | SARB52 | S. enterica serovar Pullorum | I | 7 | 540 | 520 | ||
| 3029 | SARC1 | S. enterica serovar Typhimurium | I | 8 | 540 | 520 | ||
| 3036 | SARC2 | S. enterica serovar Typhi | I | 8 | 1,100 | 1,200 | ||
| 3039 | SARC3 | S. enterica serovar Typhi | II | 8 | N | 520 | ||
| 3047 | SARC4 | S. enterica serovar Typhi | II | 8 | N | 520 | ||
| 3061 | SARC5 | S. enterica serovar Arizonae | IIIa | 8 | N | 520 | ||
| 3063 | SARC6 | S. enterica serovar Arizonae | IIIa | 8 | N | 520 | ||
| 3068 | SARC7 | S. enterica serovar Arizonae | IIIb | 8 | N | 520 | ||
| 3069 | SARC8 | S. enterica serovar Arizonae | IIIb | 8 | N | 520 | ||
| 3074 | SARC9 | S. enterica serovar Arizonae | IV | 8 | N | 520 | ||
| 3086 | SARC10 | S. enterica serovar Arizonae | IV | 8 | N | 520 | ||
| 3100 | SARC11 | S. bongori | V | 8 | N | 520 | ||
| 3103 | SARC12 | S. bongori | V | 8 | N | 520 | ||
| 3116 | SARC13 | S. bongori | VI | 8 | N | 520 | ||
| 3118 | SARC14 | S. bongori | VI | 8 | N | 520 | ||
| 3120 | SARC15 | S. bongori | VII | 8 | N | 520 | ||
| 3121 | SARC16 | S. bongori | VII | 8 | N | 520 | ||
Most strains are the serovars of S. enterica. S. bongori is a separate species.
N indicates that no PCR product was produced.
SGSC, Salmonella Genetic Stock Center.
Kuala Lumpur, Malaysia.
Enzymes and chemicals.
Taq polymerase and deoxynucleoside triphosphates were obtained from Pharmacia Biotech. All other chemicals, including LB media and agarose, were obtained from Gibco BRL.
Primers.
The following primers were designed from S. enterica serovar Typhimurium LT2 sequences and synthesized by the University Core DNA Services (Health Science Centre, University of Calgary): F1 (5′ TTCTGTCTGCGGAGATGATG 3′), R1 (5′ GCC TTGTAGAAG AGCAAATAAAGC 3′), F2 (5′ CGGGCAATGAATCTGTTCTT 3′), and R2 (5′ GGTCAGGTGACCGAGCTG 3′) (see Fig. 2D). Their locations on the genome are illustrated below (see Fig. 2D).
FIG.2.
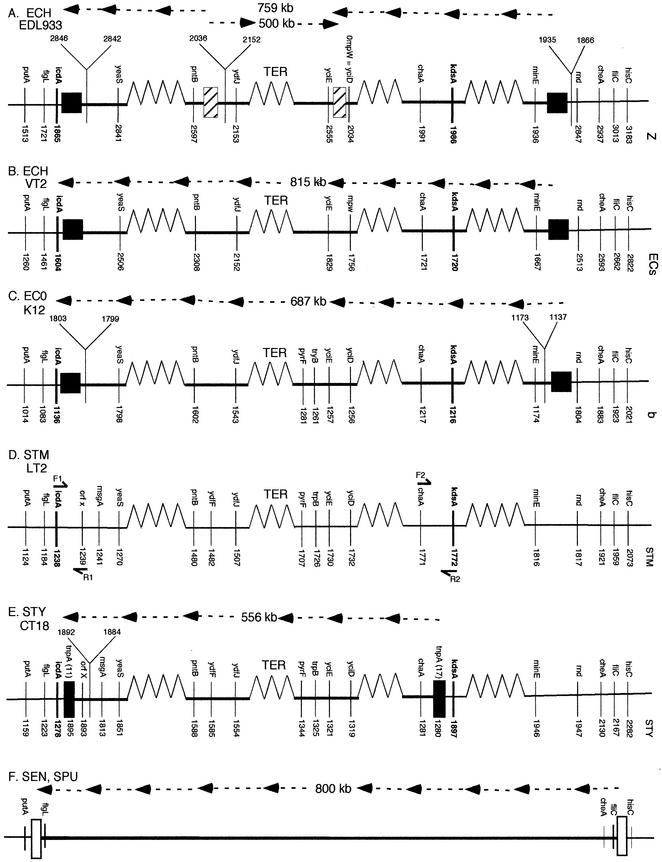
Segments of the chromosome covering the TER region in strains of E coli and Salmonella, showing inverted regions (not drawn to scale). The named genes are indicated above the lines, and the number for each gene (not the kilobase number but the serial number of the gene, based on sequence numbering from thrA) is indicated below the line. Numbered genes above the line on a “Y” formation indicate an island present in one strain but not in others. The order of genes from left to right in S. enterica serovar Typhimurium is considered to be the ancestral order (see text). Orthologous genes in other species are shown, for convenience, in the same order as in serovar Typhimurium, but the actual order on their chromosomes is revealed by the order of their gene numbers. Inverted segments are shown as dotted lines above the maps with arrows from right to left, with the size of the inversion indicated in kilobases. Panels A to E are based on nucleotide sequence data, and panel F is based on PFGE data. (A) E. coli O157:H7 EDL933 (ECH EDL933). The filled squares indicate the ends of the large inverted segment; the squares containing diagonal lines indicate the ends of the inverted segment within it. (B) E. coli O157:H7 Sakai-VT2 (ECH VT2). The filled squares indicate the ends of the large inverted segment. (C) E. coli K12 (ECO K12). The filled squares indicate the ends of the large inverted segment. (D) S. enterica serovar Typhimurium LT2 (STM LT2). The short arrows above and below the map show PCR primers F1, R1, F2, and R2. (E) S. enterica serovar Typhi CT18 (STY CT18). The two filled rectangles indicate the positions of IS200 -11 and -17 elements (containing the tnpA genes); the 556-kb segment between them is inverted in serovar Typhi CT18 compared with serovar Typhimurium LT2. (F) Salmonella enterica serovar Enteritidis (SEN) and Salmonella enterica serovar Pullorum (SPU). The open rectangles indicate sites of inversion, as determined from PFGE data.
PCR amplification and agarose gel electrophoresis.
DNA templates were prepared by boiling a small amount of cells (obtained on the tip of a toothpick from a single colony) in 250 μl of double-distilled H2O for 5 min and then rapidly cooling on ice. A total of 2 μl of the template was used for each reaction. PCR was carried out with a Techne Gene E thermal cycler according to the instructions accompanying the Taq polymerase, with 30 cycles of 1 min of denaturation (94°C), 1 min of annealing (57°C), and 1 min of extension (72°C), followed by a final extension at 72°C for 10 min.
The PCR products were electrophoresed on a 1% agarose gel in 0.5× TBE buffer (1× TBE buffer contains 90 mM Tris, 90 mM boric acid, and 2 mM EDTA [pH 8.0]) with 0.5 μg of ethidium bromide per ml.
Computer methods.
The alignments were tested using the Gap program from GCG, ClustalX, BLAST programs (1), and the MUMmer program from TIGR (www.tigr.org/).
RESULTS
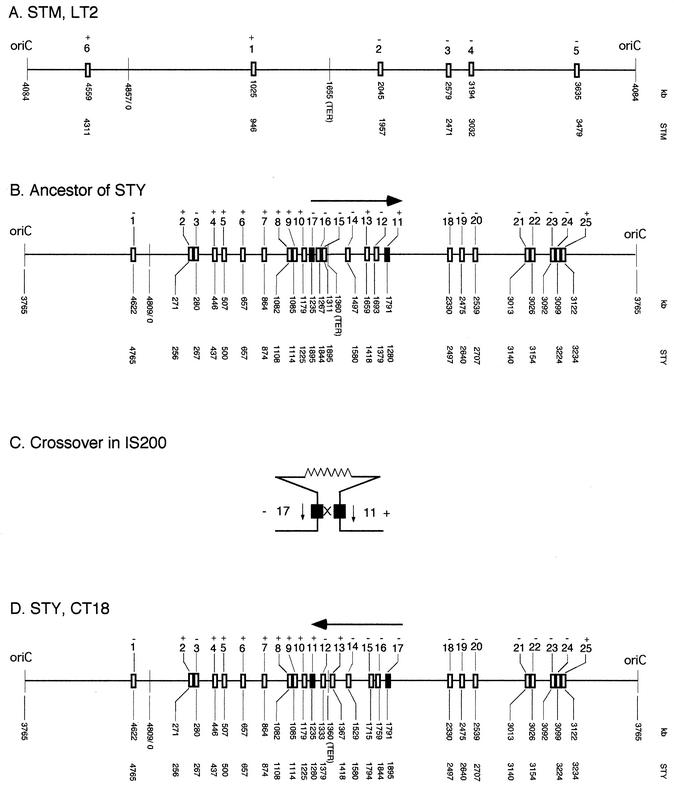
Examination of the genes and proteins of S. enterica serovar Typhimurium LT2 and of S. enterica serovar Typhi CT18 revealed that a segment which includes all the genes between STY1280 tnpA (at kb 1235) and STY1895 tnpA (at kb 1791) (a 556-kb segment) (Fig. 1D; shown in detail in Fig. 2E) is inverted in serovar Typhi CT18. These two tnpA genes for IS200 transposase are oriented in opposite directions. We postulate that an inversion occurred as follows. Since serovar Typhimurium strains are able to grow on a variety of animal hosts and are heterogeneous in many genetic traits while serovar Typhi strains are limited to humans as their only natural host and are very homogeneous in most genetic traits, we assume that serovar Typhi is of recent origin. Sequence data from housekeeping genes also show homogeneity of serovar Typhi and indicate that it originated only about 50,000 years ago (Mark Achtman, personal communication). Serovar Typhimurium LT2 has only six IS200 elements (Fig. 1A) (25, 39). We therefore propose that there evolved from some strain of Salmonella a hypothetical ancestor of serovar Typhi CT18 in which IS200s increased in number and transposed to new positions (Fig. 1B); this was followed by a reciprocal crossover between IS200-11 and -17 (Fig. 1C), which resulted in inversion of the 556-kb segment (Fig. 1D), forming the strain serovar Typhi CT18.
FIG. 1.
Part of the chromosome of S. enterica serovar Typhimurium LT2 is inverted in Salmonella enterica serovar Typhi CT18 between IS200-11 and -17. The horizontal lines indicate the chromosome of each strain, as determined from their nucleotide sequences, from oriC at the left proceeding clockwise around the chromosome through the TER region and ending with the oriC on the right. The rectangles indicate the tnpA gene within each IS200 element. The horizontal arrows indicate the inverted segments. The numbers immediately below the line indicate the positions in kilobases, based on published sequences (30, 33). The numbers below position numbers indicate the coding sequences for either serovar Typhimurium or serovar Typhi CT18, which are numbered from the gene thrA. The numbers above the boxes have been assigned in order to each IS200 element. + and − indicate the orientation of the tnpA genes and thus of the IS200. (A) Salmonella serovar Typhimurium strain LT2 (STM), showing six IS200 elements. (B) Derivative of serovar Typhimurium, postulated to become the ancestor of Salmonella serovar Typhi (STY). The order of orthologous genes on the chromosome is the same as that in serovar Typhimurium LT2. The number of IS200 elements has increased to 25, presumably due to transposition. IS200 -11 and -17, shown as filled rectangles, are postulated to recombine (see below). (C) Postulated crossover between IS200 -11 and -17. (D) Salmonella serovar Typhi strain CT18 (STY, CT18). The segment between IS200 -11 and -17 is inverted.
The chromosome segments from putA to hisC, which constitute about 20% of the chromosome and include the TER region, are shown in Fig. 2; selected orthologous genes of different strains are shown in the same order, but the actual order of each gene on the chromosome is revealed by its gene number (e.g., STM LT2 number or STY number) and by boxes which indicate inversions. For example, in Fig. 2D and E, the icdA gene of serovar Typhimurium (STM1238) and its orthologue in S. enterica serovar Typhi CT18 (STY1278) are to the left of the inverted region. In serovar Typhimurium, icdA is adjacent to STM1239 (an unnamed putative cytoplasmic protein designated orfX), while orfX in serovar Typhi CT18 is not adjacent to icdA but is displaced to STY1893; thus, this gene is in the inverted segment in serovar Typhi CT18. Similarly, chaA (STM1771), as well as its orthologue STY1281, is within the inverted region in serovar Typhi CT18. chaA is adjacent to kdsA in serovar Typhimurium (STM1772), and its orthologue is STY1897; this is at the right end of this region and is not inverted in serovar Typhi CT18. Thus, the IS200 elements, with their tnpA genes, transposed between icdA and orfX (IS200-11) and between kdsA and chaA (IS200-17) during the evolution of the ancestor of serovar Typhi, followed by reciprocal homologous recombination between the two IS200 elements, inverting the orfX-to-chaA segment, to yield serovar Typhi CT18 (Fig. 1C and D). Nucleotide sequence comparisons of serovar Typhimurium and serovar Typhi CT18, using the programs BLASTN, GCG Gap, and ClustalX, confirmed the inversion and revealed that the inverted segment ends exactly at the IS200 elements.
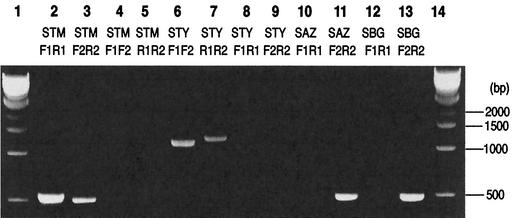
We used PCR to determine whether this inversion is present in other Salmonella strains. Primer pair F1 and R1 and primer pair F2 and R2, designed from the S. enterica serovar Typhimurium LT2 sequence (shown by short arrows in Fig. 2D), yielded the expected products of about 500 bp when DNA from serovar Typhimurium LT2 (SGSC1412, a strain of LT2) was used as template (Fig. 3, lanes 2 and 3; Table 1), but no product was produced when S. enterica serovar Typhi CT18 was used as template (Fig. 3, lanes 8 and 9). When primer pair F1 and R1 and primer pair F2 and R2 were used with serovar Typhimurium DNA as template, products were neither expected nor observed (Fig. 3, lanes 4 and 5), but when serovar Typhi CT18 DNA was used as template, PCR products of about 1,200 bp were produced (Fig. 3, lanes 6 and 7); the increased size of the product is due to the ca. 700-bp size of the IS200 element (5) which is present between these primer sites in serovar Typhi CT18. We used PCR analysis to determine if the inversion is present only in Typhi CT18, in all Typhi strains, or in many other Salmonella strains (Table 1). Twenty-one wild-type strains of serovar Typhimurium (SARA1 to -21 from the Salmonella reference A set) (4) and four strains of Salmonella enterica serovar Enteritidis as well as two of Salmonella enterica serovar Pullorum from the Salmonella reference B set (7) all yielded PCR products with primer pair F1 and R1 and primer pair F2 and R2, indicating that inversion at this specific site had not occurred in any of these strains. In most cases the size of the PCR product is indistinguishable from that detected when LT2 DNA was used as template, but in a few cases the PCR product increased in size from about 500 bp to over 1 kb, suggesting an unknown insertion in specific strains. When primer pair F1 and F2 or primer pair R1 and R2 was used for testing, DNAs from 22 wild-type strains of serovar Typhi all gave PCR products of the same size as that of the DNA of serovar Typhi CT18, but when F1 and R1 or F2 and R2 were used for testing, none of the tested strains gave PCR products. These data show that all tested strains of serovar Typhi have an inversion at the same site as CT18. Further, they indicate that all strains have IS200 at the inversion sites, since the size of the product was indistinguishable from that of the product obtained when the template was CT18.
FIG. 3.
PCR analysis of inversions in Salmonella. PCR was carried out using template DNA from different strains of Salmonella and using the primer pairs indicated (primers F1, F2, R1, and R2; see Fig. 2C for locations); the PCR products were electrophoresed in 1% agarose. Lanes 1 and 14, size markers; lanes 2 to 5, Salmonella enterica serovar Typhimurium LT2 template DNA (STM); lanes 6 to 9, S. enterica serovar Typhi CT18 template DNA (STY); lanes 10 and 11, S. enterica serovar Arizonae strain SARC5 (SAZ); lanes 12 and 13, S. bongori strain SARC11 (SBG).
We tested the 16 strains of Salmonella reference set C (SARC) which represent the seven subspecies of S. enterica (subsp. I, II, IIIa, IIIb, IV, VI, and VII) and S. bongori (previously subsp. V). All strains yielded PCR products of about 500 bp with primers F2 and R2, indicating that they are not inverted in the chaA-kdsA region. SARC1 (S. enterica serovar Typhimurium) and SARC2 (S. enterica serovar Typhi), both members of subsp. I, gave PCR products with primers F1 and R1 and with primers F1 and F2, respectively, as expected, but all the other strains in the SARC set failed to give amplification with these primers. This suggests that there may be an insertion of DNA between these primer sites or deletion of one of the primer sites; an inversion at this site is also possible but not proven.
The nucleotide sequence of E. coli K12 has an inversion relative to S. enterica serovar Typhimurium which begins to the right of icdA (b1136) (Fig. 2). Genes b1137 to b1173 represent an “island” which is present in E. coli K12 and missing from serovar Typhimurium, part of which is the e14 defective prophage, but minE (b1174) has an orthologue in serovar Typhimurium at STM1816. The island containing genes b1799 to b1803 is missing from serovar Typhimurium, but the orthologue of yeaS (b1798) in E. coli K12 is STM1270. These data indicate that serovar Typhimurium and E. coli K12 differ by an inversion which includes the segment from b1137 to b1803, which is 687 kb of E. coli K12 DNA. We have not detected any repeated sequences which might serve as a target for homologous recombination in the way IS200 serves to produce inversion in serovar Typhi CT18, but prophages are commonly found at the points of inversion (b1137 to b1159, e14 prophage; z1866 to z1934, 933X prophage; z2036 to z2151, 933O prophage); integrases or other phage sequences might have a role in these inversions. The left end of the inversion site resembles that of serovar Typhi CT18, for it is close to icdA, although it is not identical to that in serovar Typhi CT18, since it does not involve an IS200; however, the inversion site is clearly further to the right than in serovar Typhi CT18. The nucleotide sequence of E. coli O157:H7 (E. coli O157:H7 Sakai-VT2) shows the same order of orthologues as in E. coli K12; therefore, like E. coli K12, it differs from serovar Typhimurium by an inversion of 870 kb (including the genes ECs1667 to ECs2506) (Fig. 2B). The order of most genes of EDL933, an independent wild-type strain of E. coli O157:H7, is the same as that in E. coli O157:H7 Sakai-VT2, with an inversion with respect to serovar Typhimurium (759 kb, including the genes z1866 to z2846), but there is an additional inversion of 500 kb (z2036 to z2555) within this inversion (Fig. 2A) (34).
We feel that our decision to treat S. enterica serovar Typhimurium as the uninverted state and serovar Typhi CT18 as the inverted state is justified because S. enterica serovar Typhi CT18 evolved more recently and because the inverted segment in serovar Typhi CT18 has repeated IS200s at its junctions. However, treating serovar Typhimurium LT2 as the uninverted state and the strains of E. coli as the inverted state is more arbitrary; we justify it by the fact that with this assumption, all the strains analyzed in Fig. 2 can be derived from the others by a series of single-inversion steps.
DISCUSSION
Some types of inversions, such as the site-specific mechanisms for flagellar phase variation in Salmonella (hin), are common in bacteria, but they invert small segments of DNA for on-off switches (23). Inversions and genome downsizing in Mycobacterium leprae result from recombination between dispersed units of repetitive elements which resemble insertion sequences (12). However, the most common large inversions in bacteria are quasisymmetrical over oriC or TER, like the inversions in Fig. 2. For example, the inversion in Lactococcus lactis, resulting from homologous recombination between a pair of IS905 elements, was nearly symmetrical over the oriC-TER regions (13). In addition, the tufA and tufB genes of Salmonella enterica serovar Typhimurium, which appear on opposite sides of oriC, have coevolved by gene conversions and chromosomal inversions (20). Inversions are more common in some eukaryotes, such as Saccharomyces cerevisiae and Candida albicans, than in bacteria; these inversions are often small, inverting only a single gene (22).
Extensive analysis in E. coli K12, selecting for inversions between homologous regions (such as Tn10s) inserted into the chromosome, has shown that many inversions in regions flanking TER are deleterious or not detected; these are called nondivisible zones (15). The region close to TER, including pyrC (b1062) to pyrF (b1281) (see Fig. 2C), was shown to be a patchwork of inversion-refractory and inversion-tolerant segments in close proximity (16). The inversion sites we have detected by sequence analysis (Fig. 2) must fall into the inversion-tolerant segments.
However, at most sites the frequency of inversions in independent wild-type strains of Salmonella and E. coli is very low. Three reasons for failure to detect inversions are chromosome balance, gene dosage, and transcription orientation.
(i) Chromosome balance.
Replication of the chromosome begins at oriC and proceeds bidirectionally to TER, on the opposite side of the circular chromosome; thus, there are two replichores, replicating CW or CCW. Theoretically, oriC and TER should be exactly opposite, which leads to chromosome balance with replication in both directions reaching TER at about the same time; inversions could disrupt this balance (2, 18, 19).
(ii) Gene dosage.
Inversions with both endpoints within a single replichore do not alter chromosome balance but change gene dosage, because genes which are closer to oriC are present in more cellular copies during replication than those further away and hence have increased expression. Gene expression level may be adaptive, and changes may be selected against (28, 40).
(iii) Transcription orientation.
Genes such as those for rRNA are normally transcribed at a high level in the same direction as that in which they are replicated, which reduces collision of RNA polymerase and DNA polymerase (9). Inversions within the same replichore would be selected against in evolution, because the inversion would switch the direction of transcription with respect to replication.
These models may explain the failure to detect many classes of inversions in wild-type strains, just as many classes of inversions are forbidden in experimental selections from strain LT2 (41). All the inversions described in this study (Fig. 2) would not be selected against by any of the above mechanisms, because they are approximately symmetrical over the TER region. It is possible, however, that none of the inversions over oriC or TER so far detected in bacteria is exactly symmetrical. Inversions may serve as compensating mechanisms to regain the balance between oriC and TER after the balance is disrupted by insertions or deletions (26).
A study of the relative positions of unique orthologous pairs of genes, in comparisons of pairs of closely related bacterial genomes from the genus Chlamydia as well as those from Mycobacterium or from Helicobacter, reveals that although most pairs of orthologous genes plotted starting from oriC are on a diagonal line with a slope of about 1 (indicating conservation of gene location), a segment comprising a surprising number of pairs of orthologous genes is on a perpendicular diagonal line with a slope of −1 that intersects the first line near the site of TER (42), indicating an inversion over TER. A similar observation was made by Eisen et al. (14) and can be tested with the MUMmer program (http://www.tigr.org/tigr-scripts/CMR2/webmum/mumplot). The interpretation of Tillier and Collins (42) is that in these genera, rearrangements in gene order occur due to recombination between sites determined by the positions of replication forks, which results in translocation of blocks of genes across the replication axis. However, our observations for Salmonella and E. coli do not reveal the same features: we saw only single reciprocal exchanges between replichores which result in inversions, whereas they observed double exchanges which translocate blocks of genes between replichores; all the exchanges we saw were close to TER, whereas they saw translocated genes over the whole replichore; and in our study of serovar Typhi CT18, the inversion was via homologous recombination between IS200 elements, while in their studies, homologous recombination had no role.
The chromosome of S. enterica serovar Typhimurium LT2 has only 6 IS200 elements (25, 30, 39), but S. enterica serovar Typhi CT18 has 25 (33); only 1 of these is in the same site in both serovar Typhimurium LT2 and serovar Typhi CT18, indicating that serovar Typhi CT18 diverged from a different source than strain LT2. The transposase of IS200, tnpA, is transcribed at extremely low rates, and in addition, a hairpin which could act as a transcription terminator, located near the left-hand end of the element, might prevent transcription from external promoters (5). However, it is surprising that in these tnpA genes with low transcription rate, there is a strong tendency for transcription to be oriented in the same direction as replication in both serovar Typhimurium LT2 and serovar Typhi CT18 (Fig. 1). Such biased orientation is common in genes with a high transcription rate but is seldom seen in genes with a low transcription rate (9).
Acknowledgments
This work was supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a grant from the National Institute of Allergy and Infectious Diseases of the NIH (AI34829) to K.E.S. and by an operating grant from NSERC to S.-L.L.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, P., and J. Roth. 1981. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc. Natl. Acad. Sci. USA 78:3113-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. P., and J. R. Roth. 1978. Tandem chromosomal duplications in Salmonella typhimurium: fusion of histidine genes to novel promoters. J. Mol. Biol. 119:147-166. [DOI] [PubMed] [Google Scholar]
- 4.Beltran, P., S. A. Plock, N. H. Smith, T. S. Whittam, D. C. Old, and R. K. Selander. 1991. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137:601-606. [DOI] [PubMed] [Google Scholar]
- 5.Beuzon, C. R., and J. Casadesus. 1997. Conserved structure of IS200 elements in Salmonella. Nucleic Acids Res. 25:1355-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, E. F., F.-S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, E. F., F. S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewer, B. J. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53:679-686. [DOI] [PubMed] [Google Scholar]
- 10.Casjens, S. 1998. The diverse and dynamic structure of bacterial genomes. Annu. Rev. Genet. 32:339-377. [DOI] [PubMed] [Google Scholar]
- 11.Casse, F., M. C. Pascal, and M. Chippaux. 1973. Comparison between the chromosomal maps of Escherichia coli and Salmonella typhimurium. Length of inverted segment in the trp region. Mol. Gen. Genet. 124:253-257. [DOI] [PubMed] [Google Scholar]
- 12.Cole, S. T., P. Supply, and N. Honore. 2001. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr. Rev. 72:449-461. [PubMed] [Google Scholar]
- 13.Daveran-Mingot, M. L., N. Campo, P. Ritzenthaler, and P. Le Bourgeois. 1998. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J. Bacteriol. 180:4834-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, J. A., J. F. Heidelberg, O. White, and S. L. Salzberg. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1:RESEARCH0011.1-RESEARCH0011.9. [Online.] http://www.genomebiology.com/2000/1/6/research/0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francois, V., J. Louarn, J. Patte, J. E. Rebollo, and J. M. Louarn. 1990. Constraints in chromosomal inversions in Escherichia coli are not explained by replication pausing at inverted terminator-like sequences. Mol. Microbiol. 4:537-542. [DOI] [PubMed] [Google Scholar]
- 16.Guijo, M. I., J. Patte, M. del Mar Campos, J. M. Louarn, and J. E. Rebollo. 2001. Localized remodeling of the Escherichia coli chromosome: the patchwork of segments refractory and tolerant to inversion near the replication terminus. Genetics 157:1413-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 18.Hill, C. W., and J. A. Gray. 1988. Effects of chromosomal inversion on cell fitness in Escherichia coli K-12. Genetics 119:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes, D. 2000. Co-evolution of the tuf genes links gene conversion with the generation of chromosomal inversions. J. Mol. Biol. 297:355-364. [DOI] [PubMed] [Google Scholar]
- 21.Huynen, M. A., and P. Bork. 1998. Measuring genome evolution. Proc. Natl. Acad. Sci. USA 95:5849-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynen, M. A., B. Snel, and P. Bork. 2001. Inversions and the dynamics of eukaryotic gene order. Trends Genet. 17:304-306. [DOI] [PubMed] [Google Scholar]
- 23.Kolsto, A. B. 1997. Dynamic bacterial genome organization. Mol. Microbiol. 24:241-248. [DOI] [PubMed] [Google Scholar]
- 24.Krawiec, S., and M. Riley. 1990. Organization of the bacterial chromosome. Microbiol. Rev. 54:502-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam, S., and J. R. Roth. 1986. Structural and functional studies of insertion element IS200. J. Mol. Biol. 187:157-167. [DOI] [PubMed] [Google Scholar]
- 26.Liu, G.-R., A. Rahn, W.-Q. Liu, K. E. Sanderson, R. N. Johnston, and S.-L. Liu 2002. The evolving genome of Salmonella enterica serovar Pullorum. J. Bacteriol. 184:2626-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S. L., and K. E. Sanderson. 1995. Genomic cleavage map of Salmonella typhi Ty2. J. Bacteriol. 177:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S. L., and K. E. Sanderson. 1996. Highly plastic chromosomal organization in Salmonella typhi. Proc. Natl. Acad. Sci. USA 93:10303-10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, S. L., and K. E. Sanderson. 1998. Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella. FEMS Microbiol. Lett. 164:275-281. [DOI] [PubMed] [Google Scholar]
- 30.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 31.Mushegian, A. R., and E. V. Koonin. 1996. Gene order is not conserved in bacterial evolution. Trends Genet. 12:289-290. [DOI] [PubMed] [Google Scholar]
- 32.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26:74-86. [DOI] [PubMed] [Google Scholar]
- 33.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 34.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 35.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth, J. R., N. Benson, T. Galitski, K. Haack, J. G. Lawrence, and L. Miesel. 1996. Rearrangements of the bacterial chromosome: formation and applications, p. 2256-2276. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 37.Sanderson, K. E. 1976. Genetic relatedness in the family Enterobacteriaceae. Annu. Rev. Microbiol. 30:327-349. [DOI] [PubMed] [Google Scholar]
- 38.Sanderson, K. E., and C. A. Hall. 1970. F-prime factors of Salmonella typhimurium and an inversion between S. typhimurium and Escherichia coli. Genetics 64:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanderson, K. E., P. Sciore, S. L. Liu, and A. Hessel. 1993. Location of IS200 on the genomic cleavage map of Salmonella typhimurium LT2. J. Bacteriol. 175:7624-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid, M. B., and J. R. Roth. 1987. Gene location affects expression level in Salmonella typhimurium. J. Bacteriol. 169:2872-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segall, A., M. J. Mahan, and J. R. Roth. 1988. Rearrangement of the bacterial chromosome: forbidden inversions. Science 241:1314-1318. [DOI] [PubMed] [Google Scholar]
- 42.Tillier, E. R., and R. A. Collins. 2000. Genome rearrangement by replication-directed translocation. Nat. Genet. 26:195-197. [DOI] [PubMed] [Google Scholar]