Abstract
Although Enteroccus faecalis is the paradigm for biochemical studies on the arginine deiminase (ADI) pathway of fermentative arginine catabolism, little genetic information exists on this pathway in this organism. We fill this important gap by characterizing, in an 8,228-bp region cloned from a λgt11 genomic library of E. faecalis, a five-gene cluster forming a transcriptional unit (revealed by Northern blots and primer extension in E. faecalis) that corresponds to the ADI operon. Four additional genes in the opposite DNA strand and one in the same DNA strand are also identified. Studies on the protein products, including heterologous expression and/or sequence comparisons, allow us to ascertain or propose functions for all but 1 of the 10 genes. The ADI operon genes, arcABCRD, encode, respectively, ADI, ornithine transcarbamylase, carbamate kinase, a putative Crp/Fnr-type regulator (ArcR), and a putative ornithine-arginine antiporter (ArcD). Arginine induces the expression of arcABCRD, most likely by means of two homologous ArgR/AhrC-type regulators encoded by two genes, argR1 and argR2, that precede arcABCRD in each DNA strand and that are transcribed monocistronically, their transcription being influenced differentially by glucose and arginine. Potential ArgR1/ArgR2 (double and single) binding sequences are found in the promoter regions of arcA and of argR1/argR2 themselves. In addition, putative binding sequences for ArcR and for CcpA are found, respectively, in the argR1/argR2 and arcA promoter regions. Of the three other genes identified, two form a transcriptional unit and encode a putative metal-sensitive transcriptional regulator (ArsR) and a cysteine protease.
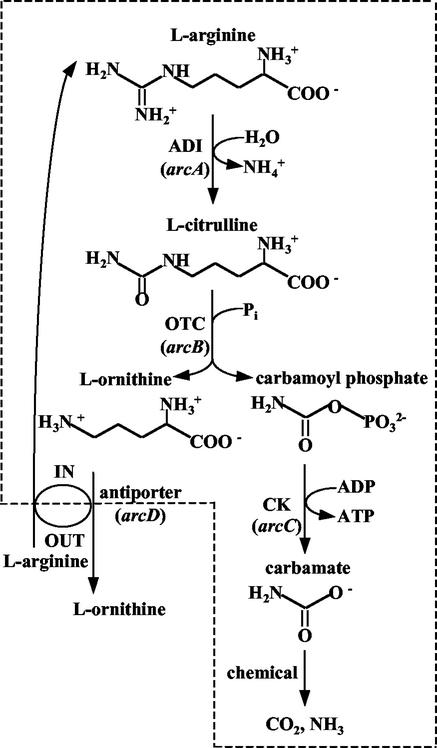
A number of microorganisms generate ATP fermentatively from arginine by a route (1) (Fig. 1) in which arginine is deiminated by arginine deiminase (ADI; EC 3.5.3.6), the resulting citrulline is phosphorolyzed by ornithine transcarbamylase (OTC; EC 2.1.3.3), giving ornithine and carbamoyl phosphate, and finally the carbamoyl phosphate is used to phosphorylate ADP in a reaction catalyzed by carbamate kinase (CK; EC 2.7.2.2), yielding ATP. This route, called the ADI pathway, operates in many eubacteria (1, 10, 12, 64) and in some archaea (53) and unicellular eukaryotes (59, 71), but it has not been found in multicellular organisms.
FIG. 1.
Arginine catabolism in E. faecalis. The broken line denotes the cell boundary, and IN and OUT denote intra- and extracellular medium. The relevant genes are given in italics.
Possibly no microbial species has been more important for the biochemical characterization of the ADI pathway than Enterococcus faecalis (formerly Streptococcus faecalis). The ADI pathway was originally demonstrated and characterized in this microorganism (22, 25, 62), and its enzymes were purified (40, 41, 50) and shown to be coordinately induced by arginine (61). Recently the first three-dimensional structure of a CK was determined (38) on the enzyme isolated from what was believed to be Enterococcus faecium (6) but that we presently show to be E. faecalis (called here E. faecalis SD10).
Despite the abundant information derived from E. faecalis on the biochemistry of the ADI pathway, there was no other genetic information about the corresponding genes of this organism than the observation that mutants that cannot use arginine as an energy source are of four types (61). Mutant types II and III lack ADI or OTC activity, respectively, and may carry mutations in the genes for these enzymes. Type I mutants show low levels of activity for the three ADI pathway enzymes and were postulated (61) to be deficient in arginine transport. Indeed, E. faecalis has an arginine-ornithine antiporter that is induced by arginine and that is linked to the operation of the ADI pathway (13, 51). Type IV mutants show little, if any, activity of the three ADI pathway enzymes and were hypothesized (61) to carry either a deletion of the genes for the three ADI pathway enzymes or an inactivating mutation in a controlling gene.
The organization of the genes encoding the ADI pathway was characterized first in Pseudomonas aeruginosa (4, 5, 33, 34), in which the genes for the three pathway enzymes (arcA, arcB, and arcC, encoding ADI, OTC, and CK, respectively) were found to be clustered and ordered as the enzymatic steps in the pathway (Fig. 1), forming an operon that also includes a fourth gene, arcD, that precedes arcA and that encodes an arginine-ornithine antiporter (69). Somewhat different gene cluster organizations, in some cases not including an antiporter gene or including other genes, have been reported more recently for the ADI pathway of Halobacterium salinarium (53), Clostridium perfringens (48), Rhizobium etli (12), Lactobacillus sake (72), Bacillus licheniformis (36, 37), and Oenococcus oeni (67). Except for that of P. aeruginosa, none of the ADI pathways of these microorganisms have been characterized biochemically and functionally so thoroughly as E. faecalis. In P. aeruginosa, an organism with four routes of arginine catabolism, the ADI pathway is induced under anaerobic conditions, with an auxiliary induction effect by exogenous arginine (16, 18, 32, 49). In contrast, in E. faecalis, which is only known to degrade arginine by the ADI pathway, arginine is a strong inducer irrespective of the state of aeration, and aeration is a poor repressor of ADI pathway expression unless glucose is added (61).
To provide further insights about the ADI pathway of E. faecalis, we have cloned and sequenced an 8,228-bp genomic region of this microorganism based on the previously characterized arcC gene encoding CK (39). The ADI operon was found to be composed of five genes whose expression in the presence of arginine seems to be mediated by the homologous products of two divergent genes, each one localized in a different DNA strand, preceding the ADI operon and encoding putative arginine regulatory proteins resembling those found in enteric bacteria or Bacillus subtilis (11, 35).
MATERIALS AND METHODS
Primers.
The following primers (primer names are followed by the starting and ending nucleotide numbers) correspond to the deposited genomic sequence (see below): R2, 154 to 178; P3, 565 to 592; A1, 1666 to 1695 (nucleotide 1675 replaced by T); A, 1900 to 1922; B1, 2932 to 2951, preceded at the 5′ end by the sequence CTCACTGCAT; CK5, 4070 to 4087; CK7, 4669 to 4691; D, 6625 to 6647; I, 7311 to 7333. The following primers correspond to the sequence complementary to that deposited (nucleotide numbering is for the deposited sequence): P2, 1009 to 985; R1R, 1376 to 1351; P1, 1761 to 1737; AR, 2607 to 2583; A1R, 2951 to 2920; BR, 3956 to 3941, preceded at the 5′ end by the sequence CTTGAGTGCTCA; P4, 4159 to 4131; CK6R, 4691 to 4675; C1R, 4997 to 4973, preceded at the 5′ end by the sequence TCTGCTCAGC; P5, 5090 to 5066; E1R, 5596 to 5572; P6, 5854 to 5829; DR, 7207 to 7184.
Cloning strategy and DNA sequencing.
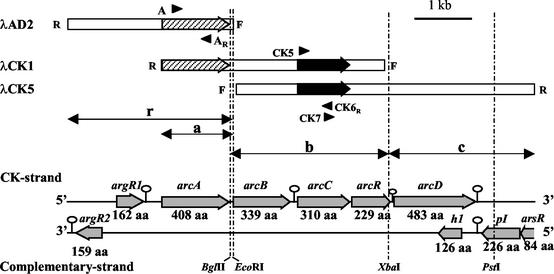
Standard protocols were used for transformation of Escherichia coli and for isolation, digestion, and ligation of DNA (3, 56). An E. faecalis ATCC 29212 chromosomal DNA phage λgt11 library (2) was used for the isolation of two clones (λCK1 and λCK5) (Fig. 2) testing positive in the screening with a monoclonal antibody (MAbCK3) against CK (43) and sharing an insert region of 2,773 bp (Fig. 2, fragment b), as previously reported (39). The fragments a (EcoRI-EcoRI ends; 1,269 bp), b (BglII-XbaI ends; 2,773 bp; overlapping 40 bp with fragment a), and c (XbaI-EcoRI ends; 2,556 bp; consecutive to fragment b) were isolated from their inserts and subcloned in pGEM-3Z using T4 DNA ligase and E. coli DH5α cells, yielding the plasmids pEF-A1, pEF-B1 (previously called p2ABX2 [38, 39]), and pEF-C1, respectively. A second round of library screening (3) using 27,000 plaques and a radioactive probe consisting of fragment a labeled with 32P (see below) identified nine positive phagic clones. Their inserts were characterized by PCR and restriction analysis, and one of these clones (λAD2) was used to isolate a 2,939-bp insert (Fig. 2, fragment r) encompassing all of fragment a preceded by 1,636 extra base pairs and followed by a tail of 40 bp overlapping with fragment b. This insert was subcloned in pGEM-3Z, yielding plasmid pEF-R1. Fragments a, c, and r were also subcloned in pBluescript S/K+ to provide appropriate flanking restriction sites for sequencing of both DNA strands with the directional digestion strategy used (3) by use of the double-strand nested deletion kit of Amersham Pharmacia. The complete sequence of the cloned DNA fragments was determined for both strands (57).
FIG. 2.
Cloning of arcC and the surrounding genes in E. faecalis and gene organization of the sequenced region. The bars at the top illustrate to scale the inserts in isolated λgt11 clones, indicating the positions of arcA (striped arrow) and arcC (solid arrow) and the orientation of the inserts with respect to the flanking forward (F) and reverse (R) sequences of the cloning site in λgt11. Insert-specific primers used to characterize the constructions are shown as solid arrowheads and are named as described in Materials and Methods. The broken vertical lines give relevant restriction sites, some of which were used for subcloning of fragments r, a, b, and c (double-headed arrows). The bottom arrows schematize the gene organization of the two strands of the sequenced region, with indications of the number of amino acid (aa) residues expected for each gene product. The positions of the six predicted stem-loops (see Materials and Methods) are indicated by the open circles.
Probe labeling.
Double-stranded probes labeled with [α-32P]dATP (3,000 Ci/mmol) were prepared (approximately 108 cpm/μg; Cerenkov radiation) by random priming (3), using either fragment a or the electrophoretically purified PCR products generated with template genomic DNA from E. faecalis SD10 and the primer pairs A and AR (706-bp probe for arcA) (Fig. 2) and B1 and BR (1,024-bp probe for arcB). Single-stranded probes (106 to 107 cpm/μg; 396 to 938 bases) were prepared (54) by asymmetric PCR (35 cycles) using a single primer complementary to the sense strand of the target gene (R2 for argR2; R1R for argR1; AR for arcA; BR for arcB; C1R for arcC; E1R for arcR; DR for arcD; I for pI; D for h1), either [α-32P]dATP or [α-32P]dCTP (800 Ci/mmol), and, as templates, appropriate restriction fragments of plasmid inserts purified from agarose gels.
For primer extension assays, primers P1 to P6 were 5′ labeled by use of T4 polynucleotide kinase (3) with [γ-33P]ATP (3,000 Ci/mmol) and were freed from unbound radioactivity by centrifugal gel filtration (3).
Cloning and expression in E. coli of arcA and arcB.
arcA and arcB were PCR amplified from templates pEF-R1 and genomic DNA from E. faecalis SD10, respectively, by utilizing a high-fidelity thermostable DNA polymerase (Deep Vent; New England Biolabs) and the primer pairs A1 and A1R (for arcA) and B1 and BR (for arcB). These primers were designed to introduce a BspHI or NdeI site at the initiator ATG codon and a BlpI site at the end of the amplified region, 34 (arcA) or 5 (arcB) nucleotides downstream of the stop codon. The PCR products, digested with BspHI-BlpI or NdeI-BlpI, were inserted directionally in the NcoI-BlpI or NdeI-BlpI sites of plasmid pET 15b (arcA) or pET 22b (arcB) behind the promoter recognized by T7 DNA polymerase. The resulting plasmids, pADI and pOTC, isolated from transformed E. coli DH5α cells grown in Luria-Bertani (LB) medium containing 70 μg of ampicillin per ml were used to transform E. coli BL21(DE3) cells. After growth of the cells at 37°C in liquid LB medium supplemented with 70 μg of ampicillin per ml until a turbidity at 600 nm of 0.6 to 0.7 was attained, 1 mM isopropyl β-d-thiogalactopyranoside was added and the culture was continued for three to four additional hours. Subsequent steps were carried out at 0 to 4°C. The cells were harvested by centrifugation, suspended in 1/5 of the original culture volume of 10 mM Tris-HCl (pH 8), and sonicated (four pulses of 30 s each; MSE Soniprep 150 at 12-μm amplitude of the exponential probe), and the sonicate was centrifuged at 15,800 × g for 10 min. The supernatant was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and for assays of ADI (50), agmatine deiminase (60), OTC (41), and putrescine transcarbamylase (60) activities.
E. faecalis SD10 characterization and use.
Slade's strain (62) of what was previously believed to be E. faecium (6) (provided by S. Grisolia, Instituto de Investigaciones Citológicas, Valencia, Spain) was shown to actually be E. faecalis by using the diagnostic system Viteck GPI (Biomerieux, Hazelwood, Mo.), and it will be called E. faecalis SD10 in this paper. The arrangement and sizes of the genes and the intergenic distances in the sequenced region are the same in E. faecalis SD10 and in E. faecalis ATCC 29212, as shown by the identical sizes (judged by agarose gel electrophoresis; results not illustrated) of the overlapping PCR products covering most of the sequenced region (nucleotides 154 to 7207) that were generated in parallel using as templates the genomic DNA from E. faecalis SD10 and the cloned fragments of the genomic library of E. faecalis ATCC 29212.
E. faecalis SD10 was grown at 37°C without shaking in medium A (61) supplemented with either 50 mM arginine or 25 mM glucose. Oxidative metabolism was induced by aeration (200 strokes per min in an orbital incubator) of cultures in medium A supplemented with 25 mM glucose and 20 mg of hematin per liter (61). Cell extracts were prepared as for E. coli (except for the use of 14 pulses of 30 s each in the sonication step).
Northern blots and primer extension experiments.
Total RNA (56) obtained from E. faecalis SD10 grown under different conditions (specified with individual experiments) was loaded (10 to 30 μg per lane, as indicated) in agarose-formaldehyde denaturing gels (9 by 11 cm) and, after electrophoresis, was blotted onto Hybond-N nylon membranes (3, 56). The blot quality and RNA integrity (judged from the rRNA pattern) and the position of RNA markers (0.24- to 9.5-kb RNA ladder) were determined by methylene blue staining. Hybridization with radioactive probes (106 cpm/ml) was carried out at 65°C, using Denhardt's solution (54) for the washings.
For primer extension assays, 5′-33P-labeled primers (0.6 × 106 cpm per assay) P1 to P6, 11 to 16 μg of total E. faecalis SD10 RNA, and the retrotranscriptase Thermoscript RT-PCR were used according to a standard protocol (56). After extension for 1 h at 47°C the products were separated in a polyacrylamide sequencing gel, together with the products of sequencing reactions carried out with the same labeled primers (56). The radioactivity in the gels was determined autoradiographically using Hyperfilm MP.
Bioinformatics tools.
Unless indicated, sequences are identified by their accession numbers in the Swissprot-TrEMBL database of Expasy (http://us.expasy.org). The program packages PC/GENE (Intelligenetics), Gene Runner 3.05 (Hastings Software Inc.), and Proteomics tools from Expasy were used for nucleic acid and protein sequence analysis. Stem-loops were localized with the program REPEATS (PC/GENE package) and their ΔG values were calculated with the program MFOLD (RNA mfold, version 2.3, server [http://bioinfo.math.rpi.edu]). Transmembrane helices were predicted with the program TMpred accessed through the Proteomics tools of Expasy. Database searches were performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) by using the BLAST network service. Multiple sequence alignments were carried out with ClustalW, version 1.7 (66), using defect values. Alignment editing and highlighting were performed with GeneDoc 2.3000 (47).
Nucleotide sequence accession number.
The annotated genomic sequence has been deposited to the EMBL nucleotide sequence database (http://www.ebi.ac.uk) with the accession number AJ312276. Nucleotide numbering used throughout this work refers to that of the deposited sequence.
RESULTS
Overall analysis of the genomic region sequenced.
Screening of a genomic library of E. faecalis ATCC 29212 by use of an anti-CK monoclonal antibody (39) as an initial probe allowed the cloning and sequencing of 8,228 bp of enterococcal genome centered in the CK gene (arcC) (Fig. 2; EMBL-EBI accession number AJ312276). Six complete nonoverlapping open reading frames (ORFs) are found in the CK-encoding strand (Fig. 2), leaving ORF-free regions of 890 and 1,023 bp at the 5′ and 3′ ends. The five more distal ORFs form a cluster in which intergenic distances are short (27 to 105 bp). This cluster, in which arcC is the third gene, is preceded and followed by highly stable potential hairpins (nucleotides 1379 to 1424 and 7255 to 7300; estimated ΔG values for RNA, −170 and −146 kJ/mol, respectively) with the characteristics of protein-independent transcription terminators (15), suggesting that this five-gene cluster is an independent transcriptional unit. Weaker stem-loops (nucleotides 3976 to 4030 and 5722 to 5745; ΔG = −84 and −53 kJ/mol, respectively) are predicted after the second and fourth genes of the cluster, although only the strongest one complies with the characteristics of transcription terminators (15) and appears to act as a partial terminator (see below).
In the complementary strand, one incomplete and three complete ORFs are found, three of them mapping at the 5′ end and the other mapping towards the other end, leaving an ORF-free central region of 5,863 bp that corresponds to most of the coding region of the CK strand (Fig. 2). The incomplete ORF, at the strand 5′ end, overlaps by 4 nucleotides the next ORF. The latter ends with a highly stable (ΔG = −134 kJ/mol) potential transcription termination hairpin (nucleotides 7298 to 7241). Another such hairpin (nucleotides 104 to 71) of lower predicted stability (ΔG= −78 kJ/mol) (Fig. 2) follows the fourth ORF of this strand.
Overall, the base composition (37.5% G+C) is characteristic for enterococci (46, 58), with a strong bias towards the use of codons terminating in A and T (46). Only 1,004 (12.2%) of the sequenced 8,228 bp are noncoding. Shine-Dalgarno ribosome binding sequences (19, 21) precede all ORFs (sequence no. AJ312276 [http://www.ebi.ac.uk]), and only one ORF (the first in the CK strand) begins with GTG instead of ATG.
The first three ORFs of the five-gene cluster encode ADI, OTC, and CK.
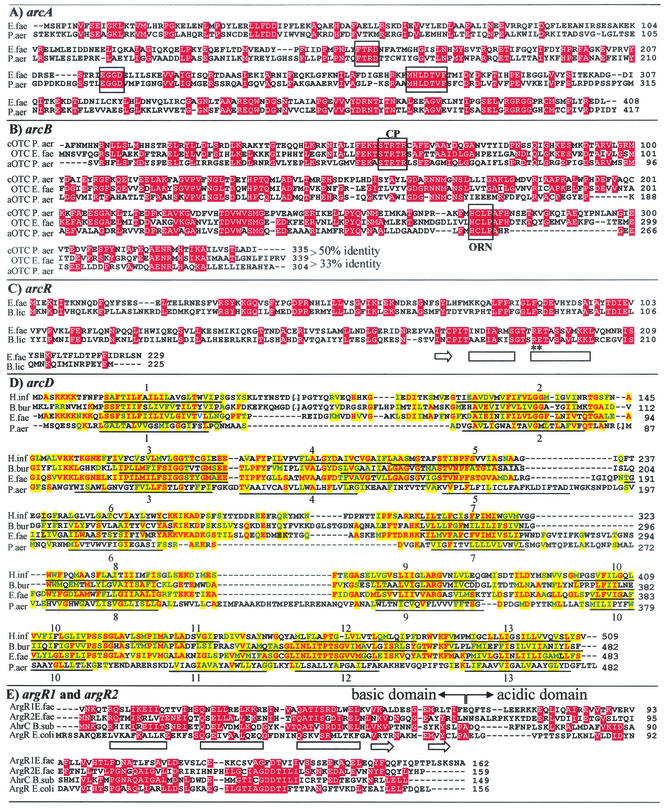
The enzyme CK is encoded by the third gene (ORF spanning nucleotides 4050 to 4989) of the five-gene cluster of the CK strand, as shown previously by cloning and sequencing of this gene and by its expression in E. coli (39). The first and second genes (ORFs spanning nucleotides 1677 to 2903 and 2932 to 3951, respectively) of this five-gene cluster encode polypeptides of 408 and 339 residues, respectively, resembling in length and sequence the P. aeruginosa ADI pathway enzymes ADI (4) (32% sequence identity) (Fig. 3A; characteristic ADI motifs [27] are boxed) and catabolic OTC (5) (50% sequence identity) (Fig. 3B; the conserved carbamoyl phosphate and ornithine binding motifs [29] are boxed). Thus, these three ORFs would appear to correspond to the arcABC genes that in P. aeruginosa encode the three enzymatic steps of the ADI pathway (4, 5, 34). However, alternatively, since E. faecalis also catabolizes agmatine by a highly similar route involving three analogous enzymes, agmatine deiminase, putrescine transcarbamylase, and CK (60), these three ORFs might encode the enzymes of the agmatine-catabolizing pathway. This possibility is rendered untenable by the evidences given in the next paragraph.
FIG. 3.
Alignment of homologous genuine or putative proteins with the protein products of E. faecalis arcA, arcB, arcR, arcD, argR1, and argR2. Red shading, identity; yellow shading, either identity (letters in red) or conservative replacement (green letters; I≈V≈L≈M≈F, F≈W≈Y, A≈G≈S, S≈T, D≈E, D≈N, E≈Q, N≈Q, K≈R) with respect to the enterococcal sequence. Horizontal arrows and bars under the sequences represent β strands and α helices, respectively. (A) E. faecalis arcA gene product aligned with ADI from P. aeruginosa (P13981). The boxes highlight signature ADI sequences (27). (B) E. faecalis arcB product aligned with catabolic (P08308) and anabolic (P11724) OTCs from P. aeruginosa. The percent identity is indicated after the sequences. CP and ORN denote signature carbamoyl phosphate and ornithine bindingsequences (29). (C) E. faecalis arcR product, aligned with ArcR from B. licheniformis (Y17554). The two helices of the DNA-binding motif and the preceding β strand in the homologous protein Crp are indicated. Two conserved residues that are involved in DNA contacts are indicated with asterisks (28). (D) Alignment of the putative E. faecalis arcD product with ArcD of P. aeruginosa (P18275) and with homologous putative proteins of H. influenzae (P44023) and B. burgdorferi (O51783). Underlining indicates transmembrane helices as annotated in Swissprot for P44023 and P118275, numbered according to these annotations, or, for the other two proteins in the alignment, as predicted by TMpred (see Materials and Methods). (E) Alignment of the putative E. faecalis argR1 and argR2 products with AhrC from B. subtilis (P17893) and ArgR from E. coli (P15282). The basic and acidic domains are indicated, and the secondary structure elements of the winged helix-turn-helix DNA binding domain are shown under the sequence of the E. coli protein (65).
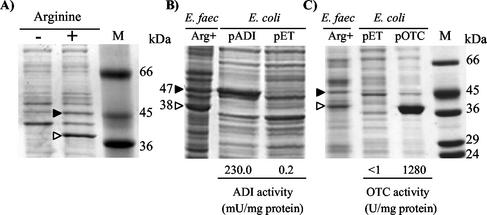
Arginine induces in E. faecalis the ADI pathway enzymes but not the agmatine deiminase pathway enzymes (60, 61). Correspondingly, SDS-PAGE of E. faecalis grown in the presence of arginine, but not in its absence, reveals (Fig. 4A) two prominent bands, of 47 and 38 kDa, approximating the expected masses for the putative ADI and OTC products (46,735 and 38,043 Da, respectively) of the first and second ORFs and coinciding, in the case of the 38-kDa band, with prior mass estimates (42) for the genuine E. faecalis OTC polypeptide. The cloning of the first ORF of the cluster, and its overexpression in E. coli, led to the appearance (Fig. 4B) of the 47-kDa band and of ADI activity (>100-fold the activity in E. coli transformed with the parental pET vector), without any agmatine deiminase activity. Similarly, the cloning and overexpression in E. coli of the second ORF led to the appearance (Fig. 4C) of the 38-kDa band and a high level of OTC activity, without any putrescine transcarbamylase activity. Thus, the first and second ORFs truly encode ADI and OTC. In accordance with the arginine catabolic role of the ADI pathway (1, 10, 64), the E. faecalis OTC sequence resembles more the sequence of the catabolic OTC (5) (50% identity) than that of the anabolic OTC (23) (35.5% identity) of P. aeruginosa (Fig. 3B). Regarding the product of the cloned third ORF of the five-gene cluster, it was shown (38, 39) to be indistinguishable enzymologically, immunologically, chromatographically, and in terms of mass (determined by mass spectrometry), amino acid sequence, and three-dimensional structure from the CK purified from E. faecalis SD10 grown on arginine. Therefore, these evidences firmly establish that these three ORFs encode the three enzymatic steps of the ADI pathway and thus that they correspond to the genes arcA, arcB, and arcC.
FIG. 4.
Arginine induction of the expression of arcA and arcB in E. faecalis (A) and expression in E. coli of plasmid-cloned arcA (B) and arcB (C), monitored by SDS-PAGE of cell extracts. M, protein markers. (A) E. faecalis SD10 was grown to an optical density at 600 nm of 1.4 either in the presence of 50 mM arginine (+) or with arginine replaced by 25 mM glucose (−). The induced bands of 47 and 38 kDa are indicated with a filled and an open arrowhead, respectively. In panels B and C, the left lanes show E. faecalis grown in arginine-containing medium, with indication of the 47- and 38-kDa bands corresponding to the putative arcA and arcB products. Shown in the right two lanes are IPTG-induced (1 mM, 3 h) E. coli BL21 cells transformed as indicated with either the parental pET plasmid having no insert or the pADI (B) or pOTC (C) plasmids carrying the E. faecalis arcA or arcB genes, respectively. The level of ADI or OTC activity of the sonicated E. coli extracts is given below the E. coli lanes.
In contrast with the observation of prominent ADI and OTC bands, no prominent CK band (33 kDa) was detected in Coomassie-stained SDS gels of arginine-grown E. faecalis SD10 (Fig. 4A), although the induction of CK was detected by the more sensitive immunoblotting technique and by enzyme activity assays (5 U/mg of protein). Thus, the amount of CK protein that accumulates in the cells is considerably lower than the amounts of the other two ADI pathway enzymes. In agreement with this conclusion, a molar OTC/CK ratio of approximately 8 can be estimated under conditions of arginine induction from the enzyme activity data (61), using the specific activities of pure E. faecalis OTC (70) and CK (40). The low amount of CK protein suggests more efficient transcription of arcA and arcB than of arcC, in agreement with the prediction of a stem-loop after arcB that may act as a partial terminator (see above), even if arcA, -B, and -C belong to the same operon. Alternatively, CK might be less stable than ADI or OTC. However, this possibility is not favored by the parallel increase in the specific activity of the three ADI pathway enzymes in the presence of arginine (61). An important difference in stability between the different enzymes should result in a different time course for the accumulation of each individual enzyme, in contrast with the observations reported (61).
The fourth ORF of the five-gene cluster encodes ArcR, a putative transcriptional regulator of the Crp/Fnr family.
In B. licheniformis, the gene that follows arcC, arcR, encodes a transcriptional regulator of the Crp/Fnr family (20, 28) which is essential for anaerobic expression of the ADI pathway (36). In E. faecalis, arcR also follows arcC, since the ORF (nucleotides 5017 to 5706) found 27 bp downstream of arcC encodes a putative protein of 229 amino acids that is only four residues longer than ArcR of B. licheniformis (36) and exhibits 33% sequence identity with it (Fig. 3C). The identities include Arg192 and Glu193 (see asterisks in Fig. 3C), two residues that are also invariant in E. coli Crp (Arg180 and Glu181; data not shown), where they were shown to make specific bonds with DNA (28), and 16 additional residues among the 28 residues of the predicted helix-turn-helix DNA binding motif and the preceding β strand (36) (indicated in Fig. 3C; prediction based on sequence alignment with Crp [28]), strongly suggesting that the DNA sequences recognized by the B. licheniformis and E. faecalis forms of ArcR are similar.
In B. licheniformis a nearly perfectly palindromic ArcR binding site, 5′-TGTGAAATATATCACG-3′, is found upstream of arcA (36), resembling the E. coli Crp binding site [5′TGTGA(N6)TCACA3′] (30). No putative ArcR binding sequence is found in E. faecalis upstream of arcA, but such a putative site, 5′-731TGTACAACGTTTTACT746-3′ (underlined bases are palindromic), is found near the transcription initiation point of argR2 (Fig. 5J ), the gene (see below) for a potential mediator of arginine induction of the ADI pathway in E. faecalis, rendering feasible ADI pathway regulation by ArcR through changes in the expression of argR2.
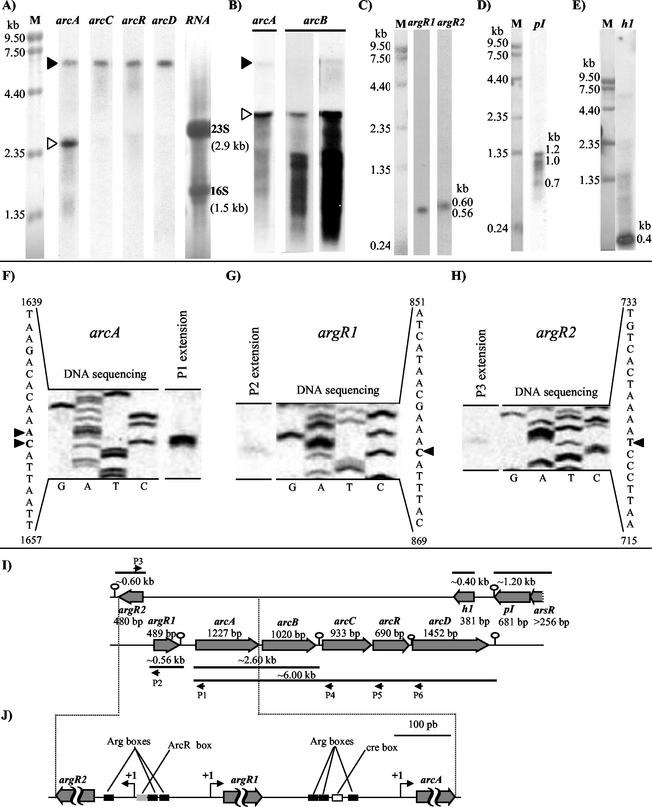
FIG. 5.
Expression at the RNA level of the sequenced genes (A to E), transcription initiation points (F to H), and schematic interpretation of the results (I and J). (A to E) Northern blots of total RNA (10 μg per lane, except in panels B and E, where it was 30 μg) isolated from E. faecalis grown to an optical density at 600 nm of 1.4 in the presence of either 50 mM arginine (A, B, C, and E) or 25 mM glucose and 20 mg of hematin per liter, with aeration (D). Lanes labeled with gene names are autoradiograms hybridized with 32P-labeled probes for the indicated genes, and lanes labeled M or RNA denote methylene blue staining of RNA size markers (sizes indicated on the side) or of total RNA (the 23S and 16SrRNAs are indicated on the side), respectively. The arcB lanes of panel B illustrate the autoradiographic results after short and long film exposition times. (F to H) Extension of 33P-labeled primers complementary to the mRNA for arcA (primer P1), argR1 (primer P2), and argR2 (primer P3). The positions of these primers, relative to the corresponding genes, and of primers P4 to P6, which yielded no discrete extension products, are shown in panel I. Total RNA (11 μg) extracted from arginine-grown, late-exponential-phase cultures was used in the extension reactions. Sequence reactions with the same labeled primers using plasmid pEF-R1 (which carries, as an insert, fragment r; see Fig. 2) as template were performed and subjected to electrophoresis in a sequencing gel next to the corresponding primer extension products. (I) Scheme of the genomic region sequenced with the genes identified, illustrating the correspondence with the observed mRNA products. (J) The region encompassing argR1, argR2, and arcA is expanded to indicate the transcription initiation points and potential control elements.
The fifth ORF of the cluster encodes the putative arginine-ornithine antiporter ArcD.
The last ORF of the five-gene cluster (nucleotides 5754 to 7205) encodes a putative protein sharing the following with the ornithine-arginine antiporter ArcD of P. aeruginosa, which in this organism is the product of the first gene of the ADI operon (33): a nearly identical sequence length (483 versus 482 residues) and the prediction of a large number of transmembrane helices (11 in E. faecalis; 13 annotated in Swissprot for ArcD of P. aeruginosa [P18275]). Although the sequence similarity of the two proteins is modest (11% identity and 31% similarity; Fig. 3D), there is solid functional evidence for the existence in E. faecalis (13, 51), as in P. aeruginosa (7, 69), of an ornithine-arginine antiporter linked to the operation of the ADI pathway. Higher identity exists with hypothetical transmembrane proteins O51783 of Borrelia burgdorferi (30% identity) and P44023 of Haemophilus influenzae (28% identity) (Fig. 3D), which are encoded by genes that map next to putative arcB and arcC genes, respectively (TIGR database [http://www.tigr.org]), and for which 11 and 13 transmembrane helices are predicted at approximately the same sites as in E. faecalis or even in P. aeruginosa ArcD (Fig. 3D).
The five-gene cluster is transcribed in the presence of arginine as a polycistronic mRNA.
Northern blots showed that, in the presence of arginine, an mRNA of 6 kb (Fig. 5A and B, filled arrowheads; Fig. 6, band of 6 kb) is produced that is revealed with probes for each of the five genes of the five-gene cluster, arcABCRD, but not with the probe for the other gene found in the same strand (see below), argR1 (Fig. 5C), and that must correspond to the transcript of the entire arcABCRD cluster (transcript schematized as the thick line at the bottom of Fig. 5I). Thus, this cluster constitutes an operon. However, the probes for arcA and arcB, but not those for the other genes, also constantly detected a very abundant mRNA of 2.6 kb (indicated with open arrowheads in Fig. 5A and B and by giving the size in kilobases in Fig. 6) that we interpret (see scheme of Fig. 5I) as the bicistronic arcAB mRNA that would be produced if transcription of the arcABCRD operon stopped frequently at the stem-loop that follows arcB. Indeed, the abundance of this transcript agrees with the higher production of the ADI and OTC proteins than of CK.
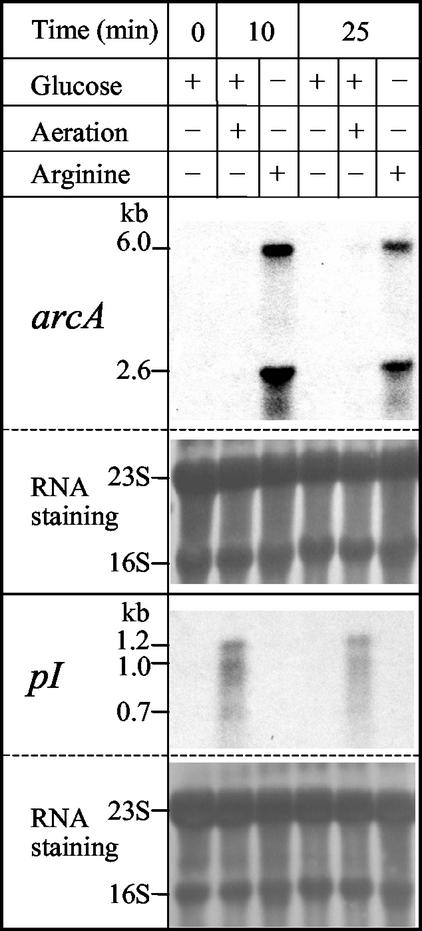
FIG. 6.
Arginine induction of the ADI operon and expression of the pI gene under glucose oxidation conditions. Northern blots with 10 μg of total RNA, either stained with methylene blue (in the panels designated “RNA staining” [the 23S and 16S rRNAs are indicated]) or hybridized with 32P-labeled probes for arcA and pI as indicated, are shown. Cells were grown in 50 ml of medium A supplemented with 25 mM glucose in 50-ml stoppered tubes without an air chamber (no aeration) to an optical density at 600 nm of 0.6 (measured after thorough mixing) and then were centrifuged and resuspended in 50 ml of fresh medium A containing the indicated additions. Except when aeration is indicated, the incubation was continued at 37°C in the same stoppered tubes without an air chamber for the indicated times before cells were centrifuged again and RNA was extracted. When aeration is indicated, the 50-ml culture also contained 0.02 mg of hematin per ml and was placed in a 250-ml Erlenmeyer flask and shaken at 200 rpm.
Extension of primer P1, which is complementary to arcA, yielded (Fig. 5F) two discrete products differing in size by a single base and corresponding to two contiguous points of initiation, nucleotides 1649 and 1650, 27 to 28 bases upstream of the arcA translation initiation codon and 11 to 12 residues downstream of the first nucleotide of the potential −10 promoter sequence, 1638TATTCT. No discrete extension products were observed with primers P4, P5, and P6 (Fig. 5I), which are complementary to arcC, arcR, and arcD, respectively. These results are expected if arcABCRD constitutes an operon in which transcription begins only upstream of arcA.
When E. faecalis SD10 was grown in the presence of glucose and the absence of arginine there was no detectable transcription of the arcABCRD operon, monitored in Northern blots using the arcA probe (Fig. 6, arcA panel, 0 min), as expected from the low levels of activity reported for the enzymes of the ADI pathway under these conditions (61). Transfer of the cells to fresh medium lacking glucose and containing 50 mM arginine resulted in the rapid appearance of the 6- and 2.6-kb RNAs corresponding to the arcABCRD and arcAB transcripts (observed already after 10 min; Fig. 6, arcA panel). These mRNA species were not observed if the fresh medium contained glucose instead of arginine, irrespective of whether fermentative or oxidative metabolism (8) (hematin present with aeration) prevailed (Fig. 6, arcA). A control was provided by the gene pI (a complementary strand gene; see below), because it was transcribed only under conditions of glucose oxidation, not during fermentation of either glucose or arginine (Fig. 6, bottom panel). Thus, as expected from the induction by arginine of the ADI pathway enzymes (61), the transcription of the arcABCRD operon is strongly induced by arginine.
The sequenced region includes two genes for putative arginine regulators of the ArgR/AhrC family.
Given the induction by arginine of the arcABCRD operon, a transcriptional regulator sensitive to arginine would be expected to control the expression of this operon in E. faecalis by binding to its promoter region. The first gene of the CK strand (ORF spanning nucleotides 891 to 1379), which precedes arcA (Fig. 2), encodes a putative protein called here ArgR1 that resembles the arginine regulatory proteins characterized from enteric bacteria and B. subtilis, ArgR/AhrC (11, 35, 44). Surprisingly, a second gene encoding another nonidentical copy of the ArgR regulator, called here ArgR2, is found near the 3′ end of the complementary DNA strand (nucleotides 648 to 169) (Fig. 2). The divergent argR1 and argR2 genes leave an intervening ORF-free region of 241 bp where their promoters and controlling elements must be found (Fig. 2). ArgR1 and ArgR2 exhibit 30.4% amino acid sequence identity between themselves and 26 to 36% identity with AhrC of B. subtilis (Fig. 3E) and present the characteristic traits of the members of the ArgR/AhrC family, including the polypeptide length (approximately 160 residues), a number of invariant residues and highly conserved residue blocks (Fig. 3E), a basic DNA binding domain (65) (estimated pI for ArgR1, 9.69, and for ArgR2, 9.34), and an acidic C domain (68) (estimated pI for ArgR1, 4.71, and for ArgR2, 4.96). In E. coli ArgR, Ser47 and Arg48 are involved in recognition of the target DNA (35). They are conserved (Fig. 3E) in ArgR1, but in ArgR2 the arginine is replaced by glycine, perhaps resulting in altered DNA recognition. Two residues, Asp128 and Asp129, that in E. coli ArgR interact with the effector arginine are conserved in ArgR2, but in ArgR1 Asp128 is replaced by a phenylalanine, raising concerns about the capacity of ArgR1 to bind arginine.
argR1 and argR2 are transcribed monocistronically, and glucose and arginine influence argR1 and argR2 transcription.
Northern blots of RNA extracted from E. faecalis SD10 grown in the presence of arginine and using probes for argR1 and argR2 revealed bands of 0.56 and 0.60 kb (Fig. 5C), respectively, indicating monocistronic expression of these two genes (schematized in Fig. 5I), in agreement with the prediction of terminator hairpins after argR1 and argR2 (see above). Primer extension experiments (Fig. 5G and H) with 33P-labeled probe P2, for argR1, and P3, for argR2 (probe positions are schematized in Fig. 5I), yielded discrete labeled products for the two genes corresponding to transcription initiation for argR1 at G863, 28 bp upstream of the argR1 translation start codon, and to transcription initiation for argR2 in the nucleotide corresponding to the complementary strand to base 722 of the CK strand, 75 bp upstream of the argR2 translation start codon. In the region between argR1 and argR2, putative −10 and −35 promoter sequences are found at nucleotides 851TAGTAT856 and 825TTCTAT830 for argR1 and at nucleotides 734TACAGT729 and 757TTGCAC752 for argR2 (the sequences of the argR2 coding strand but CK strand numbering are shown).
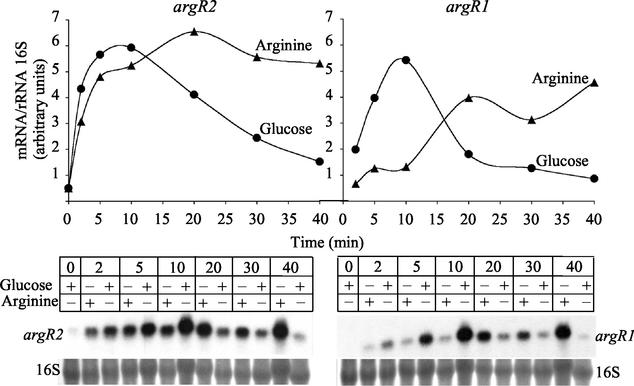
Cells grown to early exponential phase in the presence of glucose, in the absence of added arginine, and without aeration exhibited little mRNA for either argR1 or argR2, but the two genes were highly expressed transiently (with a maximum at approximately 10 min) when the cells were placed in fresh medium containing glucose (Fig. 7). When arginine replaced glucose, argR2 was expressed similarly but its expression was not transient, reaching a plateau, whereas the mRNA for argR1 increased slowly, reaching high levels after longer times (Fig. 7). Thus, arginine appears to influence the expression of these two genes.
FIG. 7.
Changes in the mRNA levels for the argR2 (left) and argR1 (right) genes with time of incubation in the presence of 25 mM glucose or 50 mM arginine. The graphs depict estimates of the mRNA contents (in arbitrary units) determined by densitometry from Northern blots of 10 μg of total E. faecalis RNA hybridized with the gene-specific 32P-labeled probes (bottom) and normalized versus the amounts of 16 S rRNA in the same lane, also determined by densitometry after methylene blue staining (also shown at the bottom of the figure). Conditions were as described for Fig. 6, without aeration. The time given is that after resuspension of the cells in fresh medium.
Additional ORFs found in the complementary strand.
The incomplete ORF (256 bp) at the 5′ end of the complementary strand (arsR; Fig. 2) encodes an 84-residue sequence resembling that of the metallothionein repressor SmtB (9), a metal-sensitive 122-residue transcription factor of the ArsR family (Pfam database no. PF01022). The similarity includes the DNA recognition helix and proposed metal binding residues of SmtB, suggesting a similar function of E. faecalis ArsR. The next ORF, pI (nucleotides 7976 to 7296; Fig. 2), overlaps arsR by 4 nucleotides. These two genes may form a bicistronic transcriptional unit ending in the terminator stem loop at the end of pI (see above), since a 1.2-kb band (in addition to 1- and 0.7-kb bands) revealed in Northern blots by use of a pI probe (Fig. 5D and 6, pI panel; interpretation in Fig. 5I) can accommodate the transcripts of the two genes, if ArsR has a similar size to SmtB. Regulation of pI expression differs diametrically from that of arcABCRD expression, since pI is expressed under oxidative growth in glucose, but not in arginine or when using glucose under oxygen restriction. pI encodes a putative 226-residue acidic (pI = 5.45) protein that may have a function of a cysteine protease, since its sequence resembles that of cysteine proteinase I of Pyrococcus horikoshii (14), with conservation of the Cys-His of the catalytic triad (as part of the invariant motif I/VCHG) and with conservative replacement by Asp108 of the glutamate of the triad. The next gene of this strand, h1 (nucleotides 6892 to 6512; Fig. 2), encodes a basic (pI = 10.3), lysine-rich (15% of the residues) putative protein that exhibits 20% identity with histone H1 of Tetrahymena thermophila (52) and which appears to be transcribed monocistronically (mRNA size, 0.4 kb, revealed with a single-stranded probe; Fig. 5E) in the presence of arginine.
DISCUSSION
In the present work we have identified for E. faecalis the genes for the three enzymes of the ADI pathway of arginine catabolism (61) and we have excluded the possibility that these genes might encode the enzymes of the homologous pathway of agmatine catabolism (60). In agreement with the observation for E. faecalis of coordinate changes in the levels of the three arginine catabolic enzymes (61) and of functional linkage between the operation of the ADI pathway and ornithine-arginine antiporter activity (13), the genes for the three enzymes and for a putative membrane protein that may encode the ornithine-arginine antiporter have been found clustered together. The cluster also includes a gene resembling arcR of B. licheniformis (36), which in the latter organism also belongs to the ADI gene cluster and encodes a transcriptional regulator of the Crp/Fnr family. The arcABCRD cluster of E. faecalis is expressed as a polycistronic mRNA from a single transcription initiation point upstream of arcA, thus constituting an operon. Nevertheless, the activity levels reached by the three ADI pathway enzymes and the accumulation of the corresponding polypeptides in extracts of E. faecalis grown in the presence of arginine indicate that ADI and OTC are more abundant than CK, in agreement with our finding of a long and a short mRNA encoding ADI and OTC, of which only the long one also encodes CK, ArcR, and ArcD. The production of the shorter, more abundant, mRNA could be due to the existence of a weak internal terminator hairpin predicted after arcB (see above) that may reduce the efficiency of transcription downstream of arcB, although we cannot exclude completely the possibility that the three enzymes have different stabilities in vivo. A very stable hairpin after arcD appears to mark the end of the operon.
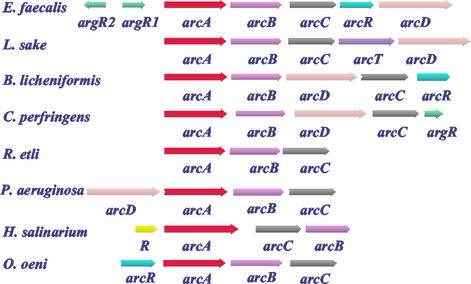
Among the studied ADI pathway gene clusters, those of B. licheniformis (36, 37) and E. faecalis resemble each other the most, since they appear to be constituted by the same genes, although the gene order is not identical, being in E. faecalis arcABCRD and in B. licheniformis arcABDCR. Figure 8 schematizes the ADI pathway gene clusters of the organisms for which it has been well studied. In all cases except H. salinarium (53), arcAB forms a fixed nucleus that is followed by arcC, either alone or together with arcD and with an extra gene that in B. licheniformis and E. faecalis is arcR, in L. sake (72) encodes a product resembling a transaminase in sequence, and in C. perfringens (48) resembles argR. arcD is missing from three of the clusters shown in Fig. 8, and in the gene clusters in which it is present it has a variable position, either preceding or following the arcABC nucleus or being interposed between arcB and arcC. There is evidence that, as is the case with E. faecalis, the gene cluster or at least part of it constitutes an operon in P. aeruginosa (17), R. etli (12), L. sake (72), and B. licheniformis (36), whereas in H. salinarium (53) each gene appears to have an independent promoter.
FIG. 8.
Comparison of the gene composition and organization of the ADI operon in E. faecalis with those of other ADI gene clusters reported for the following microorganisms: L. sake (72), B. licheniformis (36), C. perfringens (48), R. etli (12), P. aeruginosa (33), H. salinarium (53), and O. oeni (67).
A peculiarity of E. faecalis is the presence of two nonidentical putative genes for the arginine regulator, ArgR, preceding the ADI operon, each in a DNA strand and thus in divergent directions. These genes constitute independent transcriptional units that are expressed as monocistronic mRNAs. The E. faecalis ADI operon is highly induced in the presence of arginine, and tandemly arranged and solitary putative ArgR binding sequences, or Arg boxes [E. coli consensus, (t/a)NTGAAT(t/a)(t/a)(a/t)(a/t)ATTCAN(a/t), where lowercase letters indicate incomplete conservation (10, 35)], are found in E. faecalis upstream of the transcription initiation point of the arcABCRD operon (Fig. 5J). Two of them (5′-1485AATGCATTTTTTTTATTT1502-3′ and 5′-1506TATGCATTATTTTTTTAG1523-3′; bold letters denote matches to the consensus) are separated by three intervening residues, as is characteristic for repressor Arg boxes (10), and are located 164 to 147 and 143 to 126 residues, respectively, upstream of the transcriptional start of arcABCRD. The third Arg box (5′-1558TATGCATAAGAAAGAATA1575-3′) is alone, as is characteristic for activator boxes for AhrC of B. subtilis (44), and is found 90 to 73 nucleotides upstream of the arcA transcription start point, similarly to the lone Arg box found in B. licheniformis 116 to 99 residues upstream of the arcA transcription initiation point (37). These boxes might act as binding sites for ArgR1, ArgR2, or both. The presence of multiple Arg boxes in a promoter region has already been reported recently (26) for the E. coli arginine catabolic operon ast (encoding another pathway of arginine catabolism). Suggestive evidence has been provided (26) for the simultaneous binding of two ArgR hexamers to four of these Arg boxes. E. faecalis ArgR1 might not be the arginine transducer if it cannot bind arginine, as is likely because of the replacement in ArgR1 of the Asp128 residue (E. coli ArgR numbering) that is essential for arginine binding (68) by phenylalanine. ArgR2 appears to be a good candidate for mediating the induction by arginine, since its sequence evidences no amino acid change that might interfere with the binding of arginine or with the formation of hexamers that is characteristic of ArgR/AhrC regulators (35, 44). If ArgR2 were the arginine signal transducer, the regulated ADI operon and the regulating argR2 gene would be close and divergent, as was also found to be the case (45) for the P. aeruginosa gene involved in arginine catabolism, gbuA (encoding guanidinobutyrase), and its regulator gene gbuR, although these genes are considerably closer than argR2 and arcA, sharing an intergenic promoter region of only 206 bp.
Also potentially important for regulation is the presence among the genes of the ADI operon of a homolog of arcR of B. licheniformis. This organism has oxidative and fermentative arginine catabolic routes (37), and ArcR, by binding near the promoter of the ADI operon, is essential for expression of the fermentative route under anerobiosis (36). For E. faecalis the ADI pathway is the only known route of arginine catabolism and, as already indicated, aeration has little effect on the expression of the ADI pathway enzymes unless glucose is present (61). No putative Crp/Fnr-binding sequence is found upstream of arcA in E. faecalis, but a characteristic Crp/Fnr box (28, 63) overlaps with the putative −10 component of the argR2 promoter, suggesting that ArcR may influence argR2 expression and, indirectly, the expression of the ADI operon.
In agreement with arginine also having an effect on the expression of argR1 and argR2, double and lone putative Arg boxes are identified in the intervening region between these two genes (Fig. 5J). The double Arg box (5′-751TGTGCAAGTCTCACTTTT768-3′ and 5′-772AATGCGAAATATCCGACA789-3′; bold letters denote matches with the consensus [10, 35]) is only 5 nucleotides from the Crp/Fnr box, at −112 to −95 and −91 to −74 bp with respect to the transcription initiation point of argR1 and at −67 to −50 and −46 to −29 bp with respect to the corresponding point of argR2, overlapping fully with the putative −35 element of the argR2 promoter and resembling the promoter-operator topology reported for the arginine-inducible gdhB gene of P. aeruginosa, in which the operator for ArgR (in P. aeruginosa ArgR is an AraC/XylS type regulator) consists of two repeats, with the proximal half site overlapping the −35 region of the promoter (31). The lone third putative Arg box (5′-672CATGCAAAAAAACATCATT690-3′; bold letters, sequence matches; an extra base with respect to the consensus is underlined) is at −191 to −174 bp from the transcription start point of argR1 and at −24 to −42 bp from the argR2 start codon, being encompassed within the untranslated leader region of the mRNA for argR2. In addition, a putative cre box (55, 72), 5′-1532TGAAAGCGCATTCT1545-3′ (underlined nucleotides correspond to the cre consensus sequence), is found between the double and single Arg boxes preceding arcA (Fig. 5J), opening the possibility of the regulation of the ADI operon by CcpA (30), the protein that mediates catabolite repression in gram-positive organisms. For L. sake the expression of the ADI pathway genes was shown to be subjected to catabolite repression, and a putative cre box was also identified in the promoter region for the ADI operon (72). The modest negative effect of glucose on the arginine induction of the ADI pathway in E. faecalis (61) might result from binding of CcpA to the putative cre sequence found upstream of arcA. The potentiation by aeration of this negative effect of glucose (61) suggests cumulative repression by CcpA and by ArcR if the latter is sensitive to the oxidative state, as is characteristic for other Fnr family members (20, 24). In any case, the presence in E. faecalis of three different putative transcriptional regulators (ArgR1, ArgR2, and ArcR), of putative double and solitary Arg boxes and of a putative ArcR box in the region preceding argR1 and argR2, and of putative double and solitary Arg boxes and a putative cre box in the region preceding the ADI operon suggest a complex regulation of the ADI operon that warrants further investigation.
Acknowledgments
This work was supported by the Spanish Ministerio de Educación, Ciencia y Cultura (grant PM97-0134-C02-01 and fellowship to B.B.A.). A.M. was a fellow of the Generalitat Valenciana.
We thank R. N. Perham (University of Cambridge) for providing the library of genomic DNA from E. faecalis; J. Cervera and S. Grisolía (Instituto de Investigaciones Citológicas, Valencia, Spain) for supplying MAbCK3 and E. faecalis SD10, respectively; R. Borrás (Valencia University) for typing E. faecalis SD10; D. Barettino (IBV-CSIC) for advice, and A. Chawla, E. Pertegaz, and E. Grau for help with sequencing. We warmly acknowledge the technical help of Filo Bote and Inma Micó.
REFERENCES
- 1.Abdelal, A. T. 1979. Arginine catabolism by microorganisms. Annu. Rev. Microbiol. 33:139-168. [DOI] [PubMed] [Google Scholar]
- 2.Allen, A. G., and R. N. Perham. 1991. Two lipoyl domains in the dihydrolipoamide acetyltransferase chain of the pyruvate dehydrogenase multienzyme complex of Streptococcus faecalis. FEBS Lett. 287:206-210. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 4.Baur, H., E. Luethi, V. Stalon, A. Mercenier, and D. Haas. 1989. Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of Pseudomonas aeruginosa. Eur. J. Biochem. 179:53-60. [DOI] [PubMed] [Google Scholar]
- 5.Baur, H., V. Stalon, P. Falmagne, E. Luethi, and D. Haas. 1987. Primary and quaternary structure of the catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa. Eur. J. Biochem. 166:111-117. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, S. H., and S. Grisolía. 1966. Crystalline carbamate kinase. Biochim. Biophys. Acta 118:211-218. [DOI] [PubMed] [Google Scholar]
- 7.Bourdineaud, J. P., D. Heierli, M. Gamper, H. J. Verhoogt, A. J. Driessen, W. N. Konings, C. Lazdunski, and D. Haas. 1993. Characterization of the arcD arginine:ornithine exchanger of Pseudomonas aeruginosa. Localization in the cytoplasmic membrane and a topological model. J. Biol. Chem. 268:5417-5424. [PubMed] [Google Scholar]
- 8.Bryan-Jones, D. G., and R. Whittenbury. 1969. Haematin-dependent oxidative phosphorylation in Streptococcus faecalis. J. Gen. Microbiol. 58:247-260. [DOI] [PubMed] [Google Scholar]
- 9.Cook, W. J., S. R. Kar, K. B. Taylor, and L. M. Hall. 1998. Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins. J. Mol. Biol. 275:337-346. [DOI] [PubMed] [Google Scholar]
- 10.Cunin, R., N. Glansdorff, A. Piérard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czaplewski, L. G., A. K. North, M. C. M. Smith, S. Baumberg, and P. G. Stockley. 1992. Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol. Microbiol. 6:267-275. [DOI] [PubMed] [Google Scholar]
- 12.D'Hooghe, I., C. Vander Wauven, J. Michiels, C. Tricot, P. de Wilde, J. Vanderleyden, and V. Stalon. 1997. The arginine deiminase pathway in Rhizobium etli: DNA sequence analysis and functional study of the arcABC genes. J. Bacteriol. 179:7403-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driessen, A. J. M., E. J. Smid, and W. N. Konings. 1988. Transport of diamines by Enterococcus faecalis is mediated by an agmatine-putrescine antiporter. J. Bacteriol. 170:4522-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du, X., I. G. Choi, R. Kim, W. Wang, J. Jancarik, H. Yokota, and S. H. Kim. 2000. Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-Å resolution. Proc. Natl. Acad. Sci. USA 97:14079-14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 16.Galimand, M., M. Gamper, A. Zimmermann, and D. Haas. 1991. Positive fnr-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 173:1598-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamper, M., B. Ganter, M. R. Polito, and D. Haas. 1992. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J. Mol. Biol. 226:943-957. [DOI] [PubMed] [Google Scholar]
- 18.Gamper, M., A. Zimmermann, and D. Haas. 1991. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 173:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold, L., D. Pribnow, T. Schneider, S. Shinedling, B. S. Singer, and G. Stormo. 1981. Translational initiation in prokaryotes. Annu. Rev. Microbiol. 35:365-403. [DOI] [PubMed] [Google Scholar]
- 20.Green, J., B. Bennett, P. Jordan, E. T. Ralph, A. J. Thomson, and J. R. Guest. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartz, D., D. S. McPheeters, and L. Gold. 1991. Influence of mRNA determinants on translation initiation in Escherichia coli. J. Mol. Biol. 218:83-97. [DOI] [PubMed] [Google Scholar]
- 22.Hills, G. M. 1940. Ammonia production in pathogenic bacteria. Biochem. J. 34:1057-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh, Y., L. Soldati, V. Stalon, P. Falmagne, Y. Terawaki, T. Leisinger, and D. Haas. 1988. Anabolic ornithine carbamoyltransferase of Pseudomonas aeruginosa: nucleotide sequence and transcriptional control of the argF structural gene. J. Bacteriol. 170:2725-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, F., B. Mannervik, and B. Bergman. 1997. Evidence for redox regulation of the transcription factor NtcA, acting both as an activator and a repressor, in the cyanobacterium Anabaena PCC 7120. Biochem. J. 327:513-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, M. E., and F. Lipmann. 1960. Chemical and enzymatic synthesis of carbamyl phosphate. Proc. Natl. Acad. Sci. USA 46:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiupakis, A. K., and L. Reitzer. 2002. ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J. Bacteriol. 184:2940-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knodler, L. A., E. O. Sekyere, T. S. Stewart, P. J. Schofield, and M. R. Edwards. 1998. Cloning and expression of a prokaryotic enzyme, arginine deiminase, from a primitive eukaryote, Giardia intestinalis. J. Biol. Chem. 273:4470-4477. [DOI] [PubMed] [Google Scholar]
- 28.Kolb, A., S. Busby, H. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749-795. [DOI] [PubMed] [Google Scholar]
- 29.Kraus, J. P., P. E. Hodges, C. L. Williamson, A. L. Horwich, F. Kalousek, K. R. Williams, and L. E. Rosenberg. 1985. A cDNA clone for the precursor rat mitochondrial ornithine transcarbamylase: comparison of rat and human leader sequences and conservation of catalytic sites. Nucleic Acids Res. 13:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leboeuf, C., L. Leblanc, Y. Auffray, and A. Hartke. 2000. Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by CcpA. J. Bacteriol. 182:5799-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, C. D., and A. T. Abdelal. 2001. The gdhB gene of Pseudomonas aeruginosa encodes an arginine-inducible NAD(+)-dependent glutamate dehydrogenase which is subject to allosteric regulation. J. Bacteriol. 183:490-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, C. D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lüthi, E., H. Baur, M. Gamper, F. Brunner, D. Villeval, A. Mercenier, and D. Haas. 1990. The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene 87:37-43. [DOI] [PubMed] [Google Scholar]
- 34.Lüthi, E., A. Mercenier, and D. Haas. 1986. The arcABC operon required for fermentative growth of Pseudomonas aeruginosa on arginine: Tn5-751-assisted cloning and localization of structural genes. J. Gen. Microbiol. 132:2667-2675. [DOI] [PubMed] [Google Scholar]
- 35.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maghnouj, A., A. A. Abu-Bakr, S. Baumberg, V. Stalon, and C. Vander Wauven. 2000. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 191:227-234. [DOI] [PubMed] [Google Scholar]
- 37.Maghnouj, A., T. F. de Sousa Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster encoding the arginine deiminase pathway of Bacillus licheniformis and its activation by the arginine repressor ArgR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marina, A., P. M. Alzari, J. Bravo, M. Uriarte, B. Barcelona, I. Fita, and V. Rubio. 1999. Carbamate kinase: new structural machinery for making carbamoyl phosphate, the common precursor of pyrimidines and arginine. Protein Sci. 8:934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marina, A., M. Uriarte, B. Barcelona, V. Fresquet, J. Cervera, and V. Rubio. 1998. Carbamate kinase from Enterococcus faecalis and Enterococcus faecium. Cloning of the genes, studies on the enzyme expressed in Escherichia coli, and sequence similarity with N-acetyl-l-glutamate kinase. Eur. J. Biochem. 253:280-291. [DOI] [PubMed] [Google Scholar]
- 40.Marshall, M., and P. P. Cohen. 1966. Kinetic study of the mechanism of crystalline carbamate kinase. J. Biol. Chem. 841:4197-4208. [PubMed] [Google Scholar]
- 41.Marshall, M., and P. P. Cohen. 1972. Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. I. Isolation and subunit structure. J. Biol. Chem. 247:1641-1653. [PubMed] [Google Scholar]
- 42.Marshall, M., and P. P. Cohen. 1980. Ornithine transcarbamylases. Ordering of S-cyano peptides and location of characteristically reactive cysteinyl residues within the sequence. J. Biol. Chem. 255:7287-7290. [PubMed] [Google Scholar]
- 43.Mierendorf, R. C., C. Percy, and R. A. Young. 1987. Gene isolation by screening λgt11 libraries with antibodies. Methods Enzymol. 152:458-469. [DOI] [PubMed] [Google Scholar]
- 44.Miller, C. M., S. Baumberg, and P. G. Stockley. 1997. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol. Microbiol. 29:37-48. [DOI] [PubMed] [Google Scholar]
- 45.Nakada, Y., and Y. Itoh. 2002. Characterization and regulation of the gbuA gene, encoding guanidinobutyrase in the arginine dehydrogenase pathway of Pseudomonas aeruginosa PAO1. J. Bacteriol. 184:3377-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBO News 4:14. [Google Scholar]
- 48.Ohtani, K., M. Bando, T. Swe, S. Banu, M. Oe, H. Hayashi, and T. Shimizu. 1997. Collagenase gene colA is located in the 3′-flanking region of the perfringolysin O pfo A locus in Clostridium perfringens. FEMS Microbiol. Lett. 146:155-159. [DOI] [PubMed] [Google Scholar]
- 49.Park, S. M., C. D. Lu, and A. T. Abdelal. 1997. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 179:5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrack, B., L. Sullivan, and S. Ratner. 1957. Behavior of purified arginine deiminase from S. faecalis. Arch. Biochem. Biophys. 69:186-197. [DOI] [PubMed] [Google Scholar]
- 51.Poolman, B., A. J. Driesen, and W. N. Konings. 1987. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J. Bacteriol. 169:5597-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth, S. Y., I. G. Schulman, R. Richman, R. G. Cook, and C. D. Allis. 1988. Characterization of phosphorylation sites in histone H1 in the amitotic macronucleus of Tetrahymena during different physiological states. J. Cell Biol. 107:2473-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruepp, A., and J. Soppa. 1996. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J. Bacteriol. 178:4942-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sagerströn, C. G., and H. L. Sive. 1996. RNA blot analysis, p. 83-103. In P. A. Krieg (ed.), A laboratory guide to RNA: isolation, analysis and synthesis. Wiley-Liss, Inc., New York, N.Y.
- 55.Saier, M. H., Jr., S. Chauvaux, J. Deutscher, J. Reizer, and J. J. Ye. 1995. Protein phosphorylation and regulation of carbon metabolism in gram-negative versus gram-positive bacteria. Trends Biochem. Sci. 20:267-271. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 57.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schleifer, K. H., and R. Kilpper-Bälz. 1987. Molecular and chemotaxonomic approaches to the classification of streptococci, enterococci and lactococci: a review. Syst. Appl. Microbiol. 10:1-19. [Google Scholar]
- 59.Schofield, P. J., M. R. Edwards, J. Matthews, and J. R. Wilson. 1992. The pathway of arginine catabolism in Giardia intestinalis. Mol. Biochem. Parasitol. 51:29-36. [DOI] [PubMed] [Google Scholar]
- 60.Simon, J. P., and V. Stalon. 1982. Enzymes of agmatine degradation and the control of their synthesis in Streptococcus faecalis. J. Bacteriol. 152:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon, J. P., B. Wargnies, and V. Stalon. 1982. Control of enzyme synthesis in the arginine deiminase pathway of Streptococcus faecalis. J. Bacteriol. 150:1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slade, H. D., and W. C. Slamp. 1952. The formation of the arginine dihydrolase by Streptococci and some properties of the enzyme system. J. Bacteriol. 64:455-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spiro, S. 1994. The FNR family of transcriptional regulators. Antonie Leeuwenhoek 66:23-36. [DOI] [PubMed] [Google Scholar]
- 64.Stalon, V. 1985. Evolution of arginine metabolism, p. 277-308. In H. Schelider and H. Stackebrandt (ed.), Evolution of prokaryotes. Academic Press, Inc., London, United Kingdom.
- 65.Sunnerhagen, M., M. Nilges, G. Otting, and J. Carey. 1997. Solution structure of the DNA-binding domain and model for the complex of the multifunctional hexameric arginine repressor with DNA. Nat. Struct. Biol. 4:819-826. [DOI] [PubMed] [Google Scholar]
- 66.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tonon, T., J. P. Bourdineaud, and A. Lonvaud-Funel. 2001. The arcABC gene cluster encoding the arginine deiminase pathway of Oenococcus oeni, and arginine induction of a CRP-like gene. Res. Microbiol. 152:653-661. [DOI] [PubMed] [Google Scholar]
- 68.Van Duyne, G., G. Ghosh, W. K. Maas, and P. B. Sigler. 1996. Structure of oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol. 256:377-391. [DOI] [PubMed] [Google Scholar]
- 69.Verhoogt, H. J., H. Smit, T. Abee, M. Gamper, A. J. Driessen, D. Haas, and W. N. Konings. 1992. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine-ornithine exchanger. J. Bacteriol. 174:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wargnies, B., N. Lauwers, and V. Stalon. 1979. Structure and properties of the putrescine carbamoyltransferase of Streptococcus faecalis. Eur. J. Biochem. 101:143-152. [DOI] [PubMed] [Google Scholar]
- 71.Yarlett, N., M. P. Martinez, M. A. Moharrami, and J. Tachezy. 1996. The contribution of the arginine dihydrolase pathway to energy metabolism by Trichomonas vaginalis. J. Mol. Biochem. Parasitol. 78:117-125. [DOI] [PubMed] [Google Scholar]
- 72.Zúñiga, M., M. Champomier-Verge, M. Zagorec, and G. Pérez-Martínez. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]