Abstract
Leptin controls feeding behavior and insulin secretion from pancreatic β-cells. Insulin stimulates the production of leptin, thereby establishing an adipoinsular axis. Earlier we identified leptin receptors on pancreatic β-cells and showed leptin-mediated inhibition of insulin secretion by activation of ATP-sensitive potassium channels. Here we examine transcriptional effects of leptin on the promoter of the rat insulin I gene in rodent β-cells. A fall in levels of preproinsulin mRNA is detected in vivo in islets of ob/ob mice 24 h after a single injection of leptin, in isolated ob/ob islets treated with leptin in vitro and in the β-cell line INS-1 on leptin exposure when preproinsulin mRNA expression is stimulated by 25 mM glucose or 10 nM glucagon-like peptide 1. Under these conditions, transcriptional activity of −410 bp of the rat insulin I promoter is inhibited by leptin, whereas transactivation of a 5′-deleted promoter (−307 bp) is not. The −307 sequence contains the known glucose-responsive control elements (E2:A3/4). Constitutive activation of ATP-sensitive potassium channels by diazoxide does not alter leptin inhibition of preproinsulin mRNA levels. Distinct protein–DNA complexes appear on the rat insulin I promoter sequences located between −307 and −410 with nuclear extracts from ob/ob islets in response to leptin, including a signal transducer and activator of transcription (STAT)5b binding site. These results indicate that leptin inhibits transcription of the preproinsulin gene by altering transcription factor binding to sequences upstream from the elements (307 bp) that confer glucose responsivity to the rat insulin I gene promoter. Thus leptin exerts inhibitory effects on both insulin secretion and insulin gene expression in pancreatic β-cells, but by different cellular mechanisms.
Uncertainty exists regarding the nature of the earliest biochemical changes that culminate in the development of obesity-associated diabetes. The diabetes is preceded by an extended period of hyperinsulinemia believed to occur in compensation for insulin resistance that develops in response to obesity (1). In human obesity, however, insulin resistance is less frequent than insulin hypersecretion, leading to the hypothesis that hyperinsulinemia of obesity may be the result of both compensatory (to insulin resistance) and primary hypersecretion of insulin (2). This notion is supported by studies demonstrating that hyperinsulinemia may even precede alterations in insulin sensitivity in humans (3). The metabolic and/or hormonal signals that contribute to the development of obesity-associated hyperinsulinemia are unknown. It is feasible, however, that molecular mechanisms that directly alter the function of the pancreatic β-cells may lead to the induction of insulin hypersecretion.
A primary candidate for linking the status of body fat to the function of the pancreatic β-cell is the obesity-associated hormone leptin (4). Leptin is produced and secreted predominantly by white adipose tissue, and serum levels of leptin correlate directly with body-fat mass (5). Leptin exerts its effects on appetite and thermogenesis centrally on centers located in the hypothalamus (6). In obese individuals, however, leptin fails to exert its physiological role in the reduction of appetite and induction of energy expenditure despite the presence of markedly elevated serum levels. Thus defective leptin reception by hypothalamic centers seems to be operative in the development of obesity (6). At the cellular level, leptin acts by means of specific leptin receptors (OB-R) structurally related to the family of cytokine receptors (7). OB-R activates intracellular signaling cascades including the Janus kinase-signal transducer and activator of transcription (STAT) pathway of signal transduction (8).
The ob/ob mouse bears a mutation in the gene encoding leptin and provides a model of adipogenic type 2 diabetes mellitus with a phenotype of antecedent hyperinsulinemia and insulin resistance followed by β-cell failure (9). We and others have demonstrated earlier the presence of leptin receptors on pancreatic β-cells (10, 11) and have shown that leptin inhibits insulin secretion by acting on ATP-sensitive potassium channels (KATP) (12, 13), a mechanism later reported to mediate leptin action in the hypothalamus (14). Inhibition of insulin secretion and preproinsulin mRNA expression by leptin was found by several other laboratories (11, 15–26).
In this study, we examine regulatory mechanisms of leptin on the expression of the preproinsulin gene in the pancreatic β-cell line INS-1 and ob/ob islets. We find that leptin reduces the transcriptional activity of the rat insulin I gene promoter and alters binding of distinct proteins including STAT5b complexes to upstream sequences within the 5′-promoter region of the rat insulin I gene. These results establish a molecular mechanism by which leptin affects insulin biosynthesis in pancreatic β-cells at the level of preproinsulin gene transcription.
MATERIALS AND METHODS
Leptin Inhibition of Insulin Secretion in ob/ob Mice.
Ten week-old ob/ob mice were injected intraperitoneally with recombinant murine leptin (PeproTech, Rocky Hill, NJ) at 1 μg/g body weight or vehicle (n = 6 per group) and serum values for glucose and insulin determined after a 24-h fasting period.
Detection of Preproinsulin mRNA Levels in ob/ob Islets by Semiquantitative Reverse Transcription–PCR (RT-PCR).
Islets from leptin-injected ob/ob mice were isolated and semiquantitative RT-PCR for preproinsulin mRNA and β-actin was performed (27). PCR was performed for 18 cycles to cover the linear range of the amplification kinetics, and linearity was shown for a relative cDNA input across one order of magnitude (Fig. 1B Bottom graphs). For the in vitro study, islets from noninjected ob/ob mice were isolated and cultured for 24 h in RPMI medium 1640 supplemented with 0.1% BSA and 6.25 nM (100 ng/ml) murine leptin or vehicle at 13.2 mM glucose for 24 h before RT-PCR analysis. The results are derived from two independent experiments, each performed in triplicate.
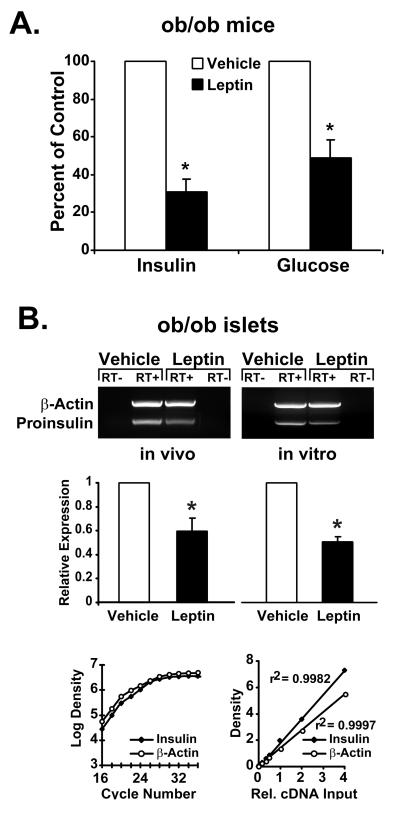
Figure 1.
Leptin reduces serum insulin levels and preproinsulin mRNA expression in pancreatic islets of ob/ob mice. (A) Effects of a single intraperitoneal injection with leptin (1 μg/g body weight) or vehicle (n = 6) on plasma insulin and serum glucose concentrations in ob/ob mice. ∗, P < 0.05. (B) Semiquantitative RT-PCR for preproinsulin expression in pancreatic islets from leptin- or vehicle-injected ob/ob mice (Left, in vivo) and islets from noninjected ob/ob mice incubated in culture with leptin (6.25 nM) for 24 h (Right, in vitro). Densitometric values are derived from two independent experiments, each performed in triplicate. ∗, P < 0.05. PCR amplification was performed for 18 cycles, and linearity of the assay with respect to amplification kinetics and relative cDNA input is demonstrated (bottom graphs). Values are means ± SD.
Detection of OB-R mRNA in INS-1 Cells.
RT-PCR was performed by using primer oligonucleotides detecting specifically the long form of the leptin receptor B isoform (OB-RB) (sense primer: 5′-ATGAAGTGGCTTAGAATCCCTTCG-3′; antisense primer: 5′-ATATCACTGATTCTGCATGCT-3′). RNA extracted from rat hypothalamus served as positive control. PCR products were hybridized to an end-labeled oligonucleotide internal to the target sequence (5′-ACACTGTTAATTTCACACCAGAG-3′).
Regulation of Preproinsulin mRNA Levels by Leptin in INS-1 cells.
INS-1 cells were incubated at 25 mM or 11.1 mM glucose for 6, 16, and 24 h with murine leptin at concentrations of 6.25 nM (100 ng/ml) and/or 10−8 M glucagon-like peptide (GLP)-1 (7–36) amide (Peninsula Laboratories). Diazoxide was used at a concentration of 100 μM. Northern RNA analysis was performed by using a double-stranded labeled DNA probe encoding 400 bp of the rat insulin I cDNA (28). Figures show representative blots of at least two independently performed experiments each.
Regulation of the Transcriptional Activity of the Rat Insulin I Gene Promoter by Leptin in INS-1 Cells.
The plasmid −410rINS1 and −307rINS1 contain 410 bp or 307 bp of the 5′ untranscribed (promoter) region of the rat insulin I gene cloned into the luciferase reporter vector pGL3 (Promega). INS-1 cells were preincubated at 11.1 mM glucose for 24 h with 6.25 nM (100 ng/ml) leptin or vehicle followed by transfection of the reporter vector by liposomal DNA transfer (Lipofectamine, Life Technologies, Gaithersburg, MD) and medium change to 5.6 mM or 25 mM glucose, supplemented with leptin and/or GLP-1 or diazoxide. Luciferase expression was determined (29) 48 h after transfection. Results represent the average of five to six independent transfection experiments ± SEM.
DNA-Binding Activity in Nuclear Extracts of Leptin-Treated Pancreatic Islets of ob/ob Mice.
Pancreatic islets from ob/ob mice were incubated for 24 h with 6.25 nM (100 ng/ml) murine leptin. Electrophoretic mobility shift assay was performed (28) by using the following oligonucleotides: INSTAT, 5′-GATCCAACTGCAACTTTCTGGGAAATGAGGTGGAA-3′; INS1A, 5′-GATCTGAGCTAAGAATCCA GCTATCAATAGAAACTATGAAACAG-3′; INS1B, 5′-GATCGAAACAGTTCCAGGGACAAAGATACCAGGTCCCCAACAAC-3′. For competition experiments, an oligonucleotide was used in which the STAT-binding site had been mutated, mutINSTAT (competitor with mutated STAT binding site) 5′-GATCCAACTGCAACTTCAGTTTAAATGAGGTGGAA-3′.
Statistics.
All values are expressed as means ± SD. Statistical analysis was performed by means of the Student’s t test for paired and unpaired values.
RESULTS AND DISCUSSION
When 10-week old ob/ob mice (n = 6 per group) were injected intraperitoneally with recombinant murine leptin at a dose of 1 μg/g body weight, a substantial lowering of elevated plasma insulin levels (75%) and serum glucose (55%) occurred 24 h after the injection (Fig. 1A). These findings indicate a substantial action of leptin on glucose homeostasis in this diabetic animal model in vivo and support our previous findings of inhibitory actions of leptin on insulin secretion in ob/ob islets analyzed in vitro (13).
In pancreatic islets of leptin-treated ob/ob mice, steady-state levels of preproinsulin mRNA were reduced by 40% as compared with vehicle-treated animals (Fig. 1B Left, in vivo). To test whether this effect of leptin is mediated by means of indirect mechanisms such as signaling from the CNS or by means of direct actions on pancreatic β-cells, ob/ob islets from noninjected animals were exposed to leptin in vitro. Similar to the results obtained from the in vivo experiment, preproinsulin mRNA levels are reduced by 50% in ob/ob islets incubated with leptin for 24 h (Fig. 1B Right, in vitro). The reductions in preproinsulin mRNA levels observed in both the in vivo and the in vitro experiments suggest that leptin exerts direct inhibitory effects on the expression of the preproinsulin gene in pancreatic islets.
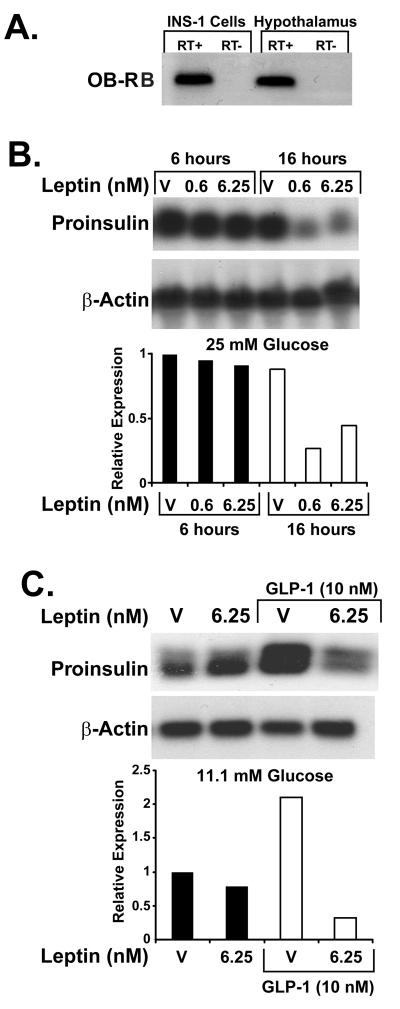
To examine further the effects of leptin on preproinsulin gene regulation in pancreatic β-cells, we used a β-cell culture model (INS-1 cells) that is glucose-responsive and responds to leptin as do pancreatic islets (30). Expression of the mRNA for the active signaling isoform of the leptin receptor (OB-RB) is detected in INS-1 cells by RT-PCR and subsequent Southern hybridization of a 350-bp product comigrating with a RT-PCR fragment obtained from rat hypothalamus as a positive control (Fig. 2A).
Figure 2.
Regulation of preproinsulin mRNA expression by leptin in INS-1 β-cells. (A) Southern detection of OB-RB RT-PCR products in INS-1 cells and rat hypothalamus (positive control). (B) Representative Northern blot for rat preproinsulin mRNA in INS-1 cells treated for 6 h and 16 h with leptin (0.6 or 6.25 nM) or vehicle at 25 mM glucose. (C) Representative Northern blot for rat preproinsulin mRNA in INS-1 cells treated for 16 h with 6.25 nM leptin or vehicle at 11.1 mM glucose in the absence or presence of 10 nM GLP-1. For each study condition, the experiments have been performed at least twice.
For examination of leptin effects on steady-state levels of preproinsulin mRNA INS-1 cells were exposed to 0.6 nM and 6.25 nM recombinant leptin for 6 and 16 h. At an elevated concentration of glucose (25 mM) in the incubation medium, an average reduction of preproinsulin mRNA expression to 60% was observed in the leptin-treated cells after 16 h of incubation (Fig. 2B). In contrast, no suppression of preproinsulin mRNA was seen at time points before 16 h (Fig. 2B Left, and data not shown). The reduction at 16 h was observed independently from the leptin concentrations of 0.6 nM (10 ng/ml) or 6.25 nM (100 ng/ml). Of note, leptin did not reduce preproinsulin mRNA in INS-1 cells at glucose concentrations of 5.6 mM (data not shown) or 11.1 mM (Fig. 2C), indicating a dependence of leptin-mediated expression of preproinsulin mRNA on glucose augmentation.
The insulinotropic hormone GLP-1 appears to override the inhibitory effect of leptin on insulin secretion (13), and GLP-1 has been reported to both augment glucose-dependent insulin secretion and stimulate the expression of preproinsulin mRNA in pancreatic β-cells (31). Thus we tested the effect of leptin on GLP-1-stimulated expression of preproinsulin mRNA. At glucose concentrations of 5.6 mM (not shown) and 11.1 mM (Fig. 2C), which did not support leptin-mediated reduction of insulin mRNA levels in the absence of GLP-1, the presence of 10 nM GLP-1 stimulated the expression of preproinsulin mRNA 2-fold, and this stimulation was surprisingly abolished by 6.25 nM leptin in the incubation medium (Fig. 2C). Thus we find discordant results between leptin inhibition of insulin secretion and leptin-mediated reduction of preproinsulin mRNA levels at different glucose concentrations and in the presence and absence of GLP-1.
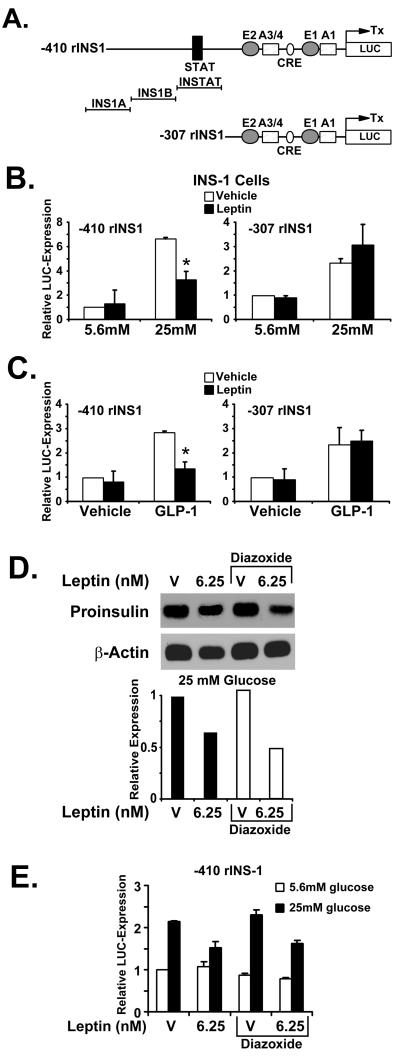
A reporter vector expressing the luciferase gene under the control of 410 bp of the rat insulin I gene promoter transfected into INS-1 cells was typically stimulated 5- to 6-fold by 25 mM glucose over the activity seen at 5.6 mM glucose (Fig. 3B Left). Although no effect of leptin was seen on the promoter activity at 5.6 mM glucose, leptin significantly inhibited reporter gene expression at 25 mM glucose by 50% (Fig. 3B Left). In contrast, the activity of a 5′-deleted reporter gene containing only 307 bp of the rat insulin I promoter was not inhibited by leptin at 25 mM glucose, a concentration that still stimulated promoter activity in this construct 3-fold (Fig. 3B Right). These results indicate that inhibitory effects of leptin on the rat insulin I promoter may be glucose dependent and provide evidence for the existence of leptin-responsive elements within promoter sequences located upstream of −307 bp. Moreover, the findings indicate that leptin-responsive promoter sequences may be different from elements that confer glucose responsiveness to the rat insulin I promoter in INS-1 cells. The glucose-responsive element (E2:A3/4) of the rat insulin I gene has been mapped to bps −223 to −207, which resides within the −307-bp construct (32). However, an overlap of response regions for leptin and glucose cannot be excluded by these results.
Figure 3.
Direct gene regulatory effects of leptin on the rat insulin I promoter in INS-1 cells are independent from KATP channel activation. (A) DNA constructs of the rat insulin I gene promoter used for luciferase reporter gene assays. Major known regulatory elements are indicated. INS1A, INS1B, and INSTAT denote the location of the oligonucleotides used in electrophoretic mobility-shift analysis. (B) Indicated reporter gene constructs were transfected into INS-1 cells and cells treated with leptin (6.25 nM) or vehicle at 5.6 mM and 25 mM glucose. ∗, P < 0.05. (C) Indicated reporter gene constructs were transfected into INS-1 cells and cells treated with leptin (6.25 nM) or vehicle at 11.1 mM glucose in the presence and absence of 10 nM GLP-1. ∗, P < 0.05. (D) Representative Northern blot for rat preproinsulin mRNA in INS-1 cells treated with 6.25 nM leptin or vehicle at 25 mM glucose in the presence or absence of 100 μM diazoxide. The results shown are representative of three independently performed experiments. (E) −410rINS-1 was transfected into INS-1 cells and cells treated with leptin (6.25 nM) or vehicle at 5.6 mM and 25 mM glucose in the presence and absence of 100 μM diazoxide. Means ± SD.
In support of the experiments examining effects of leptin on preproinsulin mRNA levels in INS-1 cells (Fig. 2C), we tested for regulatory effects of leptin on insulin promoter activity at 11.1 mM glucose in the presence or absence of 10 nM GLP-1 (Fig. 3C). We have also tested GLP-1 effects both at the level of preproinsulin mRNA expression and gene transcription at glucose concentrations of 5.6 mM and 25 mM in INS-1 cells (data not shown). We found a smaller insulinotropic effect of GLP-1 at 5.6 mM glucose but no difference between 11.1 mM and 25 mM, indicating that a glucose concentration of 11.1 mM is able to permit full GLP-1 augmentation in INS-1 cells. These findings lead us to choose a glucose concentration of 11.1 mM for the analysis of preproinsulin mRNA (Fig. 2C) and for the transient transfection experiments presented in Fig. 3C. At this glucose concentration, no inhibition of reporter gene expression by leptin was detected in both the −410rINS1 and the −307rINS1 construct. When GLP-1 was added to the medium, however, transcriptional activity of both constructs was typically stimulated between 2- and 3-fold above basal expression, and this induction was significantly inhibited by leptin in the −410rINS1 construct (Fig. 3C Left), but not in the −307rINS1 reporter plasmid (Fig. 3C Right). These results indicate that, similar to the results obtained from the Northern experiments (Fig. 2C), activation of transcription by 10 nM GLP-1 restores the inhibitory actions of leptin on rat insulin I promoter activity at glucose concentrations in which leptin alone fails to reduce insulin promoter activity. The findings further suggest the existence of leptin-responsive DNA sequences located upstream of −307 bp of the rat insulin I promoter, which may be involved in mediating the inhibitory effects of leptin on promoter transactivation in the presence of GLP-1. We conclude from these experiments that GLP-responsive and leptin-responsive DNA elements that reside within the rat insulin I gene promoter are distinct from each other. In addition, leptin does not appear to reduce GLP-1-dependent transcriptional activation of the promoter by interference with GLP-1-responsive elements, because reporter gene expression in −307rINS1 is stimulated by GLP-1 but is resistant to leptin-mediated inhibition of promoter activity (Fig. 3C Right). GLP-1 activates the formation of cAMP in β-cells (31), and the cyclic AMP response element of the rat insulin I promoter resides between bps −183 and −173 (28).
Earlier we demonstrated that leptin inhibits insulin secretion by means of activation of KATP channels (13), and changes in β-cell insulin content by altered secretion patterns are believed to feed back on the regulation of insulin gene expression (33). Thus, we examined whether the inhibition of preproinsulin gene expression by leptin is merely a feedback consequence of leptin inhibition of insulin secretion by means of activation of KATP channels or whether it is an independent effect of leptin in pancreatic β-cells. To test these potential circumstances, INS-1 cells were exposed to leptin for 24 h at 25 mM glucose in the presence or absence of diazoxide (100 μM), an agent that activates KATP channels. The reduction of preproinsulin mRNA expression by leptin detected in the presence of diazoxide is similar to that in the absence of diazoxide (Fig. 3D). To test whether this observation holds true for the direct inhibitory effects of leptin on rat insulin I promoter activity, luciferase expression from the −410rINS1 reporter construct was monitored in response to leptin in INS-1 cells at 5.6 mM and 25 mM glucose in the presence and absence of diazoxide. Analogous to the findings presented in Fig. 3D, inhibition of insulin promoter activity by leptin in INS-1 cells at 25 mM glucose is detected both in the presence and absence of KATP channel activation by diazoxide (Fig. 3E). These findings indicate that inhibition of preproinsulin gene expression by leptin is independent of the activation of KATP channels and suggests that the molecular mechanisms underlying inhibition of insulin secretion and preproinsulin mRNA by leptin may be mediated by different intracellular signaling pathways. The results from the reporter gene assays in INS-1 cells (Fig. 3) provide evidence for the existence of leptin- and glucose-responsive DNA sequences within the rat insulin I gene promoter between bases −307 and −410, although it cannot be excluded from our findings that leptin may also affect more proximal promoter regions.
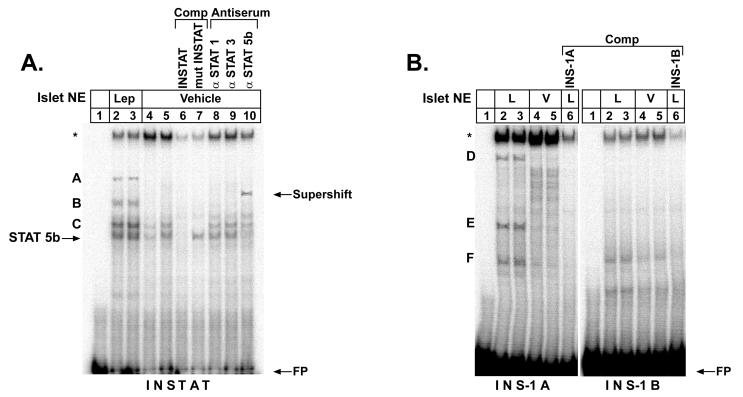
To obtain evidence as to whether the inhibition of rat insulin I promoter activity by leptin may be mediated by means of specific DNA-binding of nuclear proteins in ob/ob islets, electrophoretic mobility-shift analysis was performed on DNA sequences covering the rat insulin I gene promoter between bp −307 and −410 (Fig. 3A). The oligonucleotide INSTAT contains a previously described binding site for isoform 5b of the transcription factor STAT (34). When INSTAT is incubated with nuclear extracts obtained from leptin-treated ob/ob islets, two distinct complexes are observed (A, B) (Fig. 4A, lanes 2 and 3) as compared with vehicle-treated extracts (lanes 4 and 5). The DNA-binding complexes named C and STAT5b are also present in islet extracts treated with vehicle alone (lanes 4 and 5) but appear to increase with leptin treatment (lanes 2 and 3). In competition experiments with a 100-fold excess of unlabeled oligonucleotides, these complexes are abrogated when wild-type INSTAT is used as competitor (lane 6). In contrast, the complex labeled STAT5b remains unchanged in competition with an oligonucleotide in which the STAT binding site is mutated (lane 7), and supershift experiments using specific antisera for the known leptin-associated signal transducers STAT1, STAT3, and STAT5b suggest that this complex contains STAT5b (lanes 8–10).
Figure 4.
Leptin induces formation of distinct DNA binding complexes in extracts of ob/ob pancreatic islets on upstream sequences of the rat insulin I promoter. Pancreatic islets of ob/ob mice were treated with 6.25 nM leptin or vehicle and nuclear extracts analyzed by electrophoretic mobility-shift analysis on rat insulin I promoter sequences. Probes were (A) INSTAT and (B) INS-1A and INS-1B (see Fig. 3A). In A antisera to STAT1, 3, and 5b were used to identify specific DNA-binding proteins. STAT5b antiserum shows an interaction by retardation (supershift) of the mobility of the STAT5b complex. A, B, C, D, E, F, STAT5b, specific complexes; ∗, nonspecific complex; FP, free probe; NE, nuclear extract; L, leptin-treated extract; V, vehicle-treated extract; Comp, ×100 excess of unlabeled competitor; mutINSTAT, competitor with mutated STAT binding site.
Three more complexes (D, E, and F) are formed on the oligonucleotide INS-1A (Fig. 3A) with leptin-treated islet nuclear extracts (Fig. 4B Left, lanes 2 and 3) as compared with vehicle-treated extracts (lanes 4 and 5). These complexes are competed by a 100-fold excess of unlabeled INS-1A, indicating the sequence specificity of these DNA complexes (lane 6). In contrast, no distinct formation of DNA-binding complexes was detected on the oligonucleotide INS-1B. These results provide evidence that leptin induces the formation of specific DNA-binding complexes on upstream sequences in the rat insulin I promoter in pancreatic islets of ob/ob mice. One of these complexes induced by leptin contains STAT5b and is formed on a previously described consensus STAT binding site.
Although leptin signaling by the leptin receptor (OB-RB) is known to activate the JAK-STAT pathway preferentially involving STAT1, STAT3, and STAT5b (8), the previously reported growth hormone-mediated STAT5b binding to the STAT element within the rat insulin I promoter has been proposed to transactivate the promoter (34). Given that the DNA-binding complex containing STAT5b is only one of several complexes that are induced by leptin within the distal promoter of the rat insulin I gene (Fig. 4), we are uncertain whether repression of the rat insulin I promoter by leptin is mediated by means of STAT5b. It is, however, possible that as yet unidentified transcription factors contained in the DNA-binding complexes (labeled A–F in Fig. 4) are involved in the repression.
The doses of leptin used in the in vivo experiment (Fig. 1) are similar to or lower than those initially reported for defining leptin action in the ob/ob mouse model (35). In the in vitro experiments in the cell line INS-1, higher leptin doses were applied, because the “physiological range” of leptin responses may be different in a transformed pancreatic β-cell line, similar to the shifted insulin response to glucose. Whereas the utilized doses of leptin clearly evoked transcriptional effects in the INS-1 cells, we cannot exclude from our experiments that the effects of leptin on preproinsulin gene regulation in vivo may be less pronounced. In particular, differential effects of leptin in pancreatic β-cells at low and high doses have been reported (26).
Notably, in rodent pancreatic β-cells, leptin may exert differential effects on KATP channels and preproinsulin gene regulation in the presence of the insulinotropic hormone GLP-1. Whereas there is evidence that GLP-1 can override short-term inhibition of insulin secretion by leptin mediated through activation of KATP channels (13), such is not observed for the inhibitory regulation of preproinsulin gene expression by leptin (Figs. 2C and 3C). In fact, at normal glucose concentrations preproinsulin gene expression in INS-1 cells is only inhibited by leptin when concomitantly stimulated by GLP-1. This circumstance may indicate that leptin signaling from the adipose tissue as part of an “adipoinsular axis” does not interfere with the well established “enteroinsular axis” (36), which in the short term augments postprandial insulin secretion by the insulinotropic hormone GLP-1. On the other hand, leptin may under certain circumstances also be able to counteract GLP-1-stimulated insulin secretion, as has been demonstrated for the perfused rat pancreas (16) and in mice postprandially (21). At the same time, leptin may inhibit long-term stimulation of preproinsulin gene expression during the fasting state. The inhibitory actions of leptin on preproinsulin gene expression appear to be transmitted through an intracellular signaling pathway that differs from the one affecting the KATP channel and with different sensitivity to ambient glucose concentrations. Whereas short-term inhibition of insulin secretion by leptin has been recently proposed to be mediated through PI 3-kinase-dependent activation of cyclic nucleotide phosphodiesterase 3B (PDE3B) and subsequent suppression of cAMP levels (26), the regulatory effects of leptin on transcription of the preproinsulin gene in pancreatic β-cells may be transmitted directly by means of the JAK-STAT signaling cascade. It is tempting to speculate that leptin serves as an inhibitory control signal provided by adipose tissue to prevent extended stimulation of preproinsulin gene expression in pancreatic β-cells by incretins and glucose and to prevent sustained overproduction of insulin and hyperinsulinemia.
Assuming that mechanisms of leptin actions in human and rodent islets are similar to actions in the hypothalamus, chronic hyperleptinemia may desensitize leptin reception in the pancreatic β-cells in susceptible obese individuals, which may hypothetically lead to increased preproinsulin gene expression, enhanced insulin biosynthesis, and eventually hyperinsulinemia. Whether transcriptional dysregulation of the preproinsulin gene, however, is involved in the development of obesity and adipogenic diabetes mellitus in vivo requires further study.
In conclusion, we demonstrate direct transcriptional inhibition of the rat insulin I gene promoter in pancreatic β-cells by leptin. We propose that this is a molecular mechanism by which leptin modulates pancreatic β-cell function. We further conclude that in pancreatic β-cells, leptin acts in inhibitory fashion at different cellular levels, i.e., the KATP channel (13), cAMP-dependent signal transduction pathways (26), and gene regulation. These actions may integrate β-cell functions in a signaling network that controls appetite, body weight, and glucose metabolism, both in short- and long-term regulatory loops.
Acknowledgments
We thank Dr. S. J. Chan for the plasmid pRat Ins-1, Karen McManus for the insulin and glucose assays, and Townley Budde for help with preparation of the manuscript. J.F.H. is an Investigator with the Howard Hughes Medical Institute. J.S. is supported by the Deutsche Forschungsgemeinschaft (DFG), Grant #Se 787/1–2. The studies were supported in part by United States Public Health Service grants DK30834 and DK30457 (J.F.H.).
ABBREVIATIONS
- OB-R
leptin receptor
- OB-RB
leptin receptor B isoform
- GLP-1
glucagon-like peptide 1
- STAT
signal transducer and activator of transcription
- KATP
ATP-sensitive potassium channel
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Consensus Development Conference on Insulin Resistance. Diabetes Care. 1998;21:310–314. doi: 10.2337/diacare.21.2.310. [DOI] [PubMed] [Google Scholar]
- 2.Ferranini E, Natali A, Bell P, Cavallo-Perin P, Lalic N, Mingrone G. J Clin Invest. 1997;100:1166–1173. doi: 10.1172/JCI119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigal R J, El-Hashimy M, Martin B C, Soeldner J S, Krolewski A S, Warram J H. Diabetes. 1997;46:1025–1029. doi: 10.2337/diab.46.6.1025. [DOI] [PubMed] [Google Scholar]
- 4.Flier J S. J Clin Endocrinol Metab. 1998;83:1407–1413. doi: 10.1210/jcem.83.5.4779. [DOI] [PubMed] [Google Scholar]
- 5.Considine, R. V., Sinha, M. K., Heiman, M. L., Kriauciunas, A., Stephens, T. W., Nyce, M. R., Ohannesian, J. P., Marco, C. C., McKee, L. J. & Bauer, T. L. (1996) N. Engl. J. Med. [DOI] [PubMed]
- 6.Spiegelman B M, Flier J S. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia L A, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards G J, Campfield L A, Clark F T, Deeds J. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 8.Rosenblum C I, Tota M, Cully D, Smith T, Collum R, Qureshi S, Hess J F, Phillips M S, Hey P J, Vongs A. Endocrinology. 1996;137:5178–5181. doi: 10.1210/endo.137.11.8895396. [DOI] [PubMed] [Google Scholar]
- 9.Leibel R L. J Nutr. 1997;127:1908S. doi: 10.1093/jn/127.9.1908S. [DOI] [PubMed] [Google Scholar]
- 10.Emilsson V, Liu Y L, Cawthorne M A, Morton N M, Davenport M. Diabetes. 1997;46:313–316. doi: 10.2337/diab.46.2.313. [DOI] [PubMed] [Google Scholar]
- 11.Kieffer T J, Heller R S, Habener J F. Biochem Biophys Res Commun. 1996;224:522–527. doi: 10.1006/bbrc.1996.1059. [DOI] [PubMed] [Google Scholar]
- 12.Harvey J, McKenna F, Herson P S, Spanswick D, Ashford M L. J Physiol (London) 1997;504:527–535. doi: 10.1111/j.1469-7793.1997.527bd.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieffer T J, Heller R S, Leech C A, Holz G G, Habener J F. Diabetes. 1997;46:1087–1093. doi: 10.2337/diab.46.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spanswick D, Smith M A, Groppi V E, Logan S D, Ashford M L. Nature (London) 1997;390:521–525. doi: 10.1038/37379. [DOI] [PubMed] [Google Scholar]
- 15.Chen N G, Swick A G, Romsos D R. J Clin Invest. 1997;100:1174–1179. doi: 10.1172/JCI119629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehmann H C, Bode H P, Ebert T, Karl A, Gàke B. Horm Metab Res. 1997;29:572–576. doi: 10.1055/s-2007-979103. [DOI] [PubMed] [Google Scholar]
- 17.Fehmann H C, Berghofer P, Brandhorst D, Brandhorst H, Hering B, Bretzel R G, Göke B. Acta Diabetol. 1997;34:249–252. doi: 10.1007/s005920050083. [DOI] [PubMed] [Google Scholar]
- 18.Fehmann H C, Peiser C, Bode H P, Stamm M, Staats P, Hedetoft C, Lang R E, Göke B. Peptides (Tarrytown, NY) 1997;18:1267–1273. doi: 10.1016/s0196-9781(97)00135-6. [DOI] [PubMed] [Google Scholar]
- 19.Ishida K, Murakami T, Mizuno A, Iida M, Kuwajima M, Shima K. Regul Pept. 1997;70:179–182. doi: 10.1016/s0167-0115(97)01002-1. [DOI] [PubMed] [Google Scholar]
- 20.Koyama K, Chen G, Wang M T, Lee Y, Shimabukuro M, Newgard C B, Unger R H. Diabetes. 1997;46:1276–1280. doi: 10.2337/diab.46.8.1276. [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni R N, Wang Z L, Wang R M, Hurley J D, Smith D M, Ghatei M A, Withers D J, Gardiner J V, Bailey C J, Bloom S R. J Clin Invest. 1997;100:2729–2763. doi: 10.1172/JCI119818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ookuma M, Ookuma K, York D A. Diabetes. 1998;47:219–223. doi: 10.2337/diab.47.2.219. [DOI] [PubMed] [Google Scholar]
- 23.Pallet A L, Morton N M, Cawthorne M A, Emilsson V. Biochem Biophys Res Commun. 1997;238:267–270. doi: 10.1006/bbrc.1997.7274. [DOI] [PubMed] [Google Scholar]
- 24.Poitout V, Rouault C, Guerre-Millo M, Briaud I, Reach G. Endocrinology. 1997;139:822–826. doi: 10.1210/endo.139.3.5812. [DOI] [PubMed] [Google Scholar]
- 25.Roduit R, Thorens B. FEBS Lett. 1997;415:179–182. doi: 10.1016/s0014-5793(97)01115-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhao A Z, Bornfeldt K E, Beavo J A. J Clin Invest. 1998;102:869–873. doi: 10.1172/JCI3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seufert J, Weir G C, Habener J F. J Clin Invest. 1998;101:2528–2539. doi: 10.1172/JCI2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu M, Seufert J, Habener J F. J Biol Chem. 1997;272:28349–28359. doi: 10.1074/jbc.272.45.28349. [DOI] [PubMed] [Google Scholar]
- 29.Brasier A R, Tate J E, Habener J F. BioTechniques. 1989;7:1116–1122. [PubMed] [Google Scholar]
- 30.Asfari M, Janjic D, Meda P, Li G, Halban P A, Wollheim C B. Endocrinology. 1992;130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 31.Drucker D J, Philippe J, Mojsov S, Chick W L, Habener J F. Proc Natl Acad Sci USA. 1987;84:3434–3438. doi: 10.1073/pnas.84.10.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.German M S, Moss L G, Rutter W J. J Biol Chem. 1990;265:22063–22066. [PubMed] [Google Scholar]
- 33.Docherty K, Clark A R, Scott V, Knight S W. Proc Nutr Soc. 1991;50:553–558. doi: 10.1079/pns19910068. [DOI] [PubMed] [Google Scholar]
- 34.Galsgaard E D, Gouilleux F, Groner B, Serup P, Nielsen J H, Billestrup N. Mol Endocrinol. 1996;10:652–660. doi: 10.1210/mend.10.6.8776725. [DOI] [PubMed] [Google Scholar]
- 35.Pelleymounter M A, Cullen M J, Baker M B, Hecht R, Winters D, Boone T, Collins F. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 36.Habener J F. Endocrinol Metab Clin North Am. 1993;22:775–794. [PubMed] [Google Scholar]