Abstract
Transposases encoded by various transposable DNA elements and retroviral integrases belong to a family of proteins with three conserved acidic amino acids, D, D, and E, constituting the D-D-E motif that represents the active center of the proteins. IS1, one of the smallest transposable elements in bacteria, encodes a transposase which has been thought not to belong to the family of proteins with the D-D-E motif. In this study, we found several IS1 family elements that were widely distributed not only in eubacteria but also in archaebacteria. The alignment of the transposase amino acid sequences from these IS1 family elements showed that out of 14 acidic amino acids present in IS1 transposase, three (D, D, and E) were conserved in corresponding positions in the transposases encoded by all the elements. Comparison of the IS1 transposase with other proteins with the D-D-E motif revealed that the polypeptide segments surrounding each of the three acidic amino acids were similar. Furthermore, the deduced secondary structures of the transposases encoded by IS1 family elements were similar to one another and to those of proteins with the D-D-E motif. These results strongly suggest that IS1 transposase has the D-D-E motif and thus belongs to the family of proteins with the D-D-E motif. In fact, mutant IS1 transposases with an amino acid substitution for each of the three acidic amino acids possibly constituting the D-D-E motif were not able to promote transposition of IS1, supporting this hypothesis. The D-D-E motif identified in IS1 transposase differs from those in the other proteins in that the polypeptide segment between the second D and third E in IS1 transposase is the shortest, 24 amino acids in length. Because of this difference, the presence of the D-D-E motif in IS1 transposase has not been discovered for some time.
IS1 is an insertion element first identified in Escherichia coli (for a review, see reference 28). IS1 is only 768 bp in length and has imperfect terminal inverted repeats (IR) of about 30 bp (19, 30). IS1 transposes to a new site and generates a duplication of a target sequence of 9 bp (in most cases) or 8 bp (in some cases) (13, 14, 17, 29). IS1 has two open reading frames (ORFs), insA and B′-insB, which are required for transposing itself (18, 25, 26). insB is in the −1 frame with respect to insA. The −1 frameshifting occurs during translation at the AAAAAAC (A6C) sequence in the overlapping region between insA and B′-insB. The resulting transframe protein, InsA-B′-B, is IS1 transposase (10, 37).
IS1 generates the InsA protein from insA unless translational frameshifting occurs. The InsA protein, which has the ability to bind to terminal IR sequences (43, 52, 54), inhibits transposition of IS1 (24, 53). IS1 with a 1-bp insertion in the A6C sequence, causing the two frames insA and B′-insB to be in the same frame, produces transposase without frameshifting and can thus transpose at a very high frequency (37, 39). These facts suggest that the InsB segment in IS1 transposase forms a catalytic domain. Many elements with structural features similar to those seen in IS1 have been identified in various bacteria and their plasmids (for a review, see reference 28). Most of them are highly homologous to IS1 and to one another (more than 90% sequence identity), whereas a few, such as IS1(NuXi) from Shigella dysenteriae, have low homology, about 55% (31).
Transposases encoded by IS3 family elements and retroviral integrases have been found to have three acidic amino acid residues, D, D, and E, constituting a motif called the D-D-E motif, at positions where a polypeptide segment 35 amino acids long is usually present between the second D and third E residues (11, 23). The D-D-E motif has also been found in transposases encoded by IS630, Tn7, Tn552, and others and in integrases encoded by retrotransposons (7, 34). Transposases encoded by IS10 and phage Mu also have the D-D-E motif, but a longer polypeptide segment of 130 or 55 amino acids is present between the second D and the E (4, 5, 49). The polypeptide segment with the D-D-E motif forms a catalytic domain in which the three acidic amino acid residues have an essential role in capturing Mg2+ and other divalent cations (4). Tertiary structures of the catalytic core domains of human immunodeficiency virus type 1 (HIV-1) integrase and phage Mu transposase (MuA) and full-length Tn5 transposase have been determined by X-ray crystallography (6, 9, 35). These proteins characteristically have three β sheets in tandem in the segment with the D-D-E motif, such that the first D residue is present in the first β sheet.
The transposase encoded by IS1 has been thought not to belong to the family of proteins with the D-D-E motif but to the phage λ integrase (Int) family (44) because IS1 transposase has the H-R-Y motif, which is conserved in all the Int family proteins (1, 3). In this study, we report that several IS1 family elements with rather low homology to IS1 are present not only in eubacteria but also in archaebacteria and that the transposases encoded by these IS1 family elements have three acidic amino acid residues (D, D, and E) that are conserved in corresponding positions. We show that these three acidic amino acid residues constitute a functional D-D-E motif with the shortest polypeptide segment, only 24 amino acids in length, between the second D and the E residues of all those in the family of proteins with the D-D-E motif.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used were Escherichia coli K-12 derivatives JM109 (50), JE6638 (polA1) (National Institute of Genetics collection), and NM554 (33). pNS17 is the pUC-based ampicillin resistance (Apr) plasmid carrying mini-IS1 (46). Plasmid pSEK80 (40) is the R100-based spectinomycin resistance (Spr) plasmid used as the target for mini-IS1 transposition. pKEN200G is the p15A-based kanamycin resistance (Kmr) plasmid that carries the IS1 transposase gene, which is expressed from the promoter region of the araBAD operon of Salmonella enterica serovar Typhimurium (47). In pKEN200G, the A6C sequence was changed to GA2GA3C, so this transposase gene produces wild-type transposase but not the InsA protein. The pSIN plasmids used were constructed from pKEN200G as described below. The plasmid DNA used for cloning and nucleotide sequencing was prepared with the Concert rapid plasmid miniprep system (Gibco-BRL). The LiCl method (16) was used for rapid plasmid DNA preparation and identification.
Media.
Culture media used were L broth, L-rich broth, φ-medium (51), and SOC medium (8). L-agar plates contained 1.5% (wt/vol) agar (Wako) in L broth. When necessary, ampicillin, chloramphenicol, spectinomycin, kanamycin, or arabinose was added to the media.
Enzymes and synthetic oligonucleotide primers.
Restriction endonucleases AatII, BamHI, BglII, BslI, BstEI, MluI, SalI, Tsp509I, and XhoI (New England Biolabs or Takara), T4 DNA ligase, and DNA polymerase I (Klenow fragment) (Takara) were used as recommended by the suppliers. Synthetic oligonucleotide primers used are listed in Table 1. Oligonucleotides were obtained from Sawady Technology. Most of these primers were used for PCR, which was performed as described previously (42).
TABLE 1.
Oligonucleotide primers used in this studya
| Primer | Sequence (5′ → 3′) | Positionb |
|---|---|---|
| P1 | tcatgaggatccctAgTTAcTGgTAGTGTTTTAT | 755-736 |
| P2 | GTTTTACGCGTATGACAGGCTCCGGAAGACGGTTGTTGCGCACGTATTCGGTGAACGCACT ATGGCGACGCTGGGGCGTCTTATGAGCCTGCTGTCACCCTTTGACGTGGTGATATGGATG ACGGcTGGCTGG | 426-558 |
| P3 | GGCAGTGACGTCATCGTCTGCGCGGAAATGGcCGAACAGTG | 349-389 |
| P4 | GTTTTACGCGTATGACAGGCTCCGGAAGACGGTTGTTGCGCACGTATTCGGTGAACGCACTA TGGCGACGCTGGGGCGTCTTATGAGCCTGCTGTCACCCTTTGcCGTGGTGATAT | 426-541 |
| P5 | CAGTCGGTAACCTCGCGCATACAGCCGGGCAGTGcCGTCATCG | 322-374 |
| P6 | GTTTTACGCGTATGcCAGGCTCCG | 426-449 |
| P7 | GTTTTACGCGTATGACAGGCTCCGGAAGACGGTTGTTGCGCACGTATTCGGTGCACGcACTAT GGC | 426-491 |
| P8 | gggggctcgagTGAGGTGCTCCAATGGCTTCTGTTTCTATCAGCTGTCCCTCCTGTTCAGCTACTGcC GGGGTG | 44-106 |
| P9 | CGCAGACGATGACGTCACTGCCCGGCTG | 370-345 |
| P10 | CCGGGCAGTGACGTCATCGTCT | 346-367 |
| P11 | ggggatccCTTATTATTGATAGTGTTTTATGTTCAGATAATGCCCGATGACTTTGTCATGCAGCgC CACCG | 754-695 |
| P12 | ggggatccCTTATTGATAGTGTTTTATGTTCAGATAATGCCCGATGACTTTGgCATGCAGC | 754-702 |
| P13 | CTGTATGcATCCCGCCTGAA | 562-581 |
| P14 | GCGGGATgCATACAGCGGCCAGCCA | 576-556 |
| P15 | GCGAATTGcGCGGCATAACC | 618-637 |
| P16 | TGCCGCgCAATTCGCTGCGT | 632-617 |
| P17 | GGCAGTGACGTCATCGTCTGCGCGGcAATGGA | 349-380 |
| P18 | GGCAGTGACGTCATCGTCTGCGCGGAAATGGACGCACAGTGG | 349-390 |
| P19 | GCGCGAGGTTACCGACTGCGGCCTGAGTTTcTTcAAATGGCGGAAAATCGTGTTGAGGCCAA CGCCCATAATGCGGGCGGTTGCCCGGCATCCAACGCCATTCATGGCCATAGCAaTGAT | 339-221 |
| P20 | gggggctcgagTGAGGTGCTCCAaTGGCT | 44-61 |
| P21 | tcatgaggatccctAgTTAcTGgTAGTGTTTTATGTTCAGATAATGCCCGATGACTTTGTtATGC | 755-706 |
| P22 | tcatgaggatccctAgTTAcTGgTAGTGTTTTATGTTCAGATAATCCCGATGACTTTcTCATGC | 755-706 |
| P23 | GCAGTGACGTCATCGTCTGCGCGGAAATGaACGAA | 350-384 |
| P24 | GCAGTGACGTCATCGTCTGCGCGGAAATGGACcAACAG | 350-387 |
| P25 | TGTTTTACGCGTATaACAGGCTCCGGAAGAC | 425-455 |
| P26 | CTGGCCGCTGTATGAATCCC | 555-574 |
| P27 | GTGCAGCTTTCCCTTCAGGCGGGATTCATACAGCGGCCAGCCATtCGTCA | 594-545 |
| P28 | GTGCAGCTTTCCCTTCAGGCGGGATTgATACA | 594-563 |
| P29 | AGATTCAGGTTATGCCGCTCAA | 644-623 |
| P31 | GCAGTGACGTCATCGTCTGCGCGGAAATGGAaGAACAG | 350-387 |
| P32 | GCAGTGACGTCATCGTCTGCGCGGAAATGGACGAtCAGTG | 350-389 |
| P33 | TGTTTTACGCGTATGAaAGGCTCCGGAAGAC | 425-455 |
| P34 | GTGCAGCTTTCCCTTCAGGCGGGATTCATACAGCGGCCAGCCtTCCGTCA | 594-545 |
| P35 | CGCAGCGAATTGAtCGGCATAACC | 614-637 |
Primers were used for construction of pSIN plasmids (see Materials and Methods). Lowercase letters indicate additional nucleotide sequences with a restriction site or nucleotides altered to introduce mutations.
Positions are indicated by coordinates for IS1.
Computer analysis.
The nucleotide sequence homology search in the DDBJ/GenBank/EMBL databases was performed with software programs FASTA (32) and Blast (2). The secondary structures of proteins were analyzed with the software program PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) (20, 27). A phylogenetic tree was constructed with the neighbor-joining method with the Genetyx software program (Software Development).
Construction of pSIN plasmids.
The pSIN plasmids carry the mutant transposase gene that produces mutant transposase with an amino acid substitution (see Fig. 6 in the Results section). pSIN1 was constructed by replacing the MluI-BglII fragment of the IS1 transposase gene in pKEN200G with the MluI-BamHI fragment, which was obtained by PCR with BstBI-digested pKEN200G DNA as the template and with primers P1 and P2, followed by digestion with MluI and BamHI. Five pSIN plasmids (pSIN3, pSIN7, pSIN8, pSIN27, and pSIN36) were constructed as described above with the MluI-BamHI fragment derived from the PCR product amplified with primers P1 and P4, P1 and P6, P1 and P7, P1 and P25, and P1 and P33, respectively.
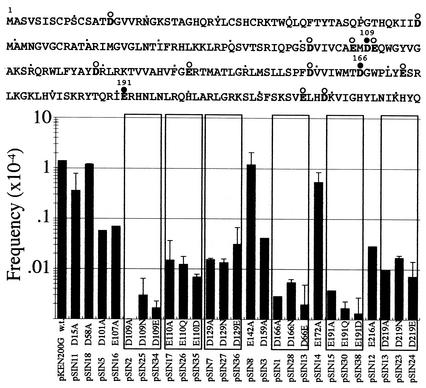
FIG. 6.
Mini-IS1 transposition mediated by mutant transposases. The amino acid sequence of IS1 transposase is shown at the top. In this sequence, boldface letters indicate acidic amino acids. Solid and open circles above the sequence indicate acidic amino acids constituting the D-D-E motif and other acidic amino acids, respectively. Frequencies of mini-IS1 transposition mediated by wild-type (w.t.) and mutant transposases are shown by the graph at the bottom. pSIN plasmids that produced mutant transposases with amino acid substitutions are shown. Results from the analysis of mutant transposases with substitutions for the same amino acid residue are boxed.
pSIN2 was constructed by replacing the AatII-BamHI fragment with the AatII-BamHI fragment which was obtained by PCR with primers P1 and P3, followed by digestion with AatII and BamHI. Similarly, 11 pSIN plasmids (pSIN5, pSIN12, pSIN13, pSIN16, pSIN17, pSIN23, pSIN24, pSIN25, pSIN26, pSIN34, and pSIN35) were constructed with the AatII-BamHI fragment from the PCR product amplified with primers P1 and P5, P10 and P11, P10 and P12, P1 and P17, P1 and P18, P10 and P21, P10 and P22, P1 and P23, P1 and P24, P1 and P31, and P1 and P32, respectively.
pSIN11 was constructed by replacing the AatII-SalI fragment with the AatII-XhoI fragment derived from the PCR product amplified with primers P8 and P9. pSIN18 was constructed by replacing the BstEII-SalI fragment with the BstEII-XhoI fragment from the PCR product amplified with primers P19 and P20.
pSIN14 was constructed as follows. Two fragments were first obtained by PCR with primers P1 and P13 or P10 and P14 and treated with DNA polymerase I. The two fragments were used as templates for PCR with primers P10 and P1 to derive the AatII-BamHI fragment. pSIN14 was then constructed by replacing the AatII-BglII fragment with the AatII-BamHI fragment. Similarly, pSIN15 was constructed by using three pairs of primers, P1 and P15, P10 and P16, and P1 and P10.
pSIN29 was constructed as follows. One fragment was obtained by PCR with primers P1 and P26 and treated with BslI and BamHI, whereas the other was obtained by PCR with primers P10 and P27 and treated with MluI and BslI. pSIN29 was constructed by replacing the MluI-BglII fragment with the BslI-BamHI and BslI-MluI fragments. Similarly, pSIN37 was constructed with two fragments from the two PCR products amplified with two pairs of primers, P1 and P26, and P10 and P34.
pSIN30 (or pSIN37) was constructed by replacing the AatII-BglII fragment with the Tsp509I-BamHI and Tsp509I-AatII fragments, which were obtained by PCR with two pairs of primers, P1 and P28 (or P1 and P29) and P10 and P29 (or P10 and P35), respectively.
All mutations introduced into the transposase gene were confirmed by DNA sequencing.
DNA sequencing.
DNA sequencing was carried out by the dideoxynucleotide chain termination method with an ABI Dye terminator cycle sequencing kit (Applied Biosystems) with AmpliTaq DNA polymerase FS (Perkin-Elmer) and an oligodeoxyribonucleotide primer, pKENseq/K (5′-ATTGGTTGTAACACTGG-3′) or pKENseq/B (5′-GGCCGACGAAATCACTC-3′). The sequencing reaction was performed with Catalyst A800 (Perkin-Elmer), and the reaction products were analyzed with the ABI 377 DNA sequencing system (Applied Biosystems).
IS1 transposition assay.
Three plasmids, pNS17, pSEK80, and pKEN200G (or a pSIN plasmid), were introduced by transformation into E. coli strain NM554 in φ-medium. Cells were grown in 5 ml of L-rich broth at 37°C overnight. The cell culture (75 μl) was added to 5 ml of fresh L-rich broth and incubated by shaking at 37°C. When the optical density was 0.4, 100 μl of 10% arabinose was added to the cell culture. After incubation of the culture at 37°C for 16 h, plasmid DNAs were extracted from cells with a Quantum Prep plasmid miniprep kit (Bio-Rad). The plasmid DNA prepared was dissolved in 10 μl of water, and 1 μl of this solution was used for electroporation into cells of JE6638 (polA1) with a Gene Pulser (Bio-Rad). The cells were incubated in 1 ml of SOC medium at 37°C for 1 h and plated on L-agar plates containing 100 μg of spectinomycin and 150 μg of chloramphenicol per ml and also on L-agar plates containing 100 μg of spectinomycin per ml. After incubation of the plates at 37°C overnight, the number of colonies was counted. Transposition frequency was calculated by dividing the number of Spr Cmr transformants by the number of Spr transformants.
RESULTS
Identification and characterization of IS1 family elements.
A computer-aided homology search of the databases was done with the software program FASTA with the amino acid sequence encoded by the insB frame of IS1 or its family element IS1(NuXi) as the query. We found several IS elements with significant homology (up to 48% similarity) not only in eubacteria, but also in archaebacteria: ISEhe5 from Erwinia herbicola (15), ISY120 from Synechocystis sp. strain PCC6803 (21), and ISC1173 from Sulfolobus solfataricus (45). With the homologous sequence found in ISC1173 as the query, we found two kinds of repeated sequences (796 and 1,173 bp in length) in the Sulfolobus tokodaii chromosome (22) and another (740 bp in length) in the Methanosarcina acetivorans chromosome (13) to encode proteins with significant homology to the query sequence. These sequences were named ISSt796, ISSt1173, and ISMac740 because they appeared to have terminal inverted repeats and to generate duplication of a target site sequence, like ISC1173, as described below. All these IS elements were assumed to be new members belonging to the IS1 family.
ISEhe5, ISY120, ISSt796, and ISMac740 were 787, 802, 796, and 740 bp in length, respectively, as large as IS1 and IS1(NuXi), which are 768 and 766 bp, respectively (Fig. 1). These IS elements had terminal IR sequences 19 to 23 bp in length, as large as the IRs of 23 bp in both IS1 and IS1(NuXi) (Fig. 1). ISC1173 and ISSt1173 were 1,174 and 1,173 bp in length, respectively, which is larger than those of the other elements (Fig. 1). These two elements have IRs 44 and 50 bp in length, respectively, larger than those of the other elements (Fig. 1).
FIG. 1.
Structures of IS1 family elements and terminal IR sequences. In the structures shown on the left panel, left- and roght-terminal IRs, IRL and IRR, are indicated by solid triangles. Horizontal thick arrows indicate ORFs. In the elements having two ORFs, the positions of a run of adenines present in the overlapping region between the two ORFs are indicated by broken vertical lines. Numbers are coordinates given to each element, in which the nucleotide at the end of IRL is defined as 1. Names of host species are given in parentheses. Nucleotide sequences of the terminal regions containing IRL and IRR are shown on the right panel in the direction 5′ to 3′. Boldface letters indicate IR sequences, and asterisks show nucleotides that are identical between two IR sequences. ISC1173 and ISSt1173 have long IR sequences of 44 and 50 bp, but the entire sequences of these are not shown.
IS1 and IS1(NuXi) duplicate a target sequence of 9 or 8 bp upon transposition, and thus, direct repeats of the target sequence appear in the regions flanking each element. A 9-bp sequence was present as a direct repeat in the regions flanking ISY120, whereas an 8-bp sequence was present in the regions flanking ISC1173, ISSt1173, and ISMac740. A 9- or 8-bp sequence was present as a direct repeat in the regions flanking ISSt796. These findings suggest that the direct repeats found in the IS1 family elements are the target sequences duplicated upon transposition. However, no direct repeat sequences were found in the regions flanking ISEhe5. This suggests that an IS-mediated genomic rearrangement, probably a deletion, has occurred at an end of ISEhe5 with the other sequence as the target, resulting in the loss of direct repeats in the flanking regions.
Presence of three acidic amino acids conserved in transposases encoded by IS1 family elements.
ISEhe5, ISY120, and ISMac740 were found to have two ORFs which appeared to correspond to insA and insB of IS1 and IS1(NuXi) (Fig. 1). In the overlapping region between the two ORFs in ISEhe5, ISY120, and ISMac740, a run of adenines were present, as in the region between insA and insB of IS1 and IS1(NuXi) (Fig. 1). This suggests that ISEhe5, ISY120, and ISMac740 also produce their transposases by translational frameshifting, as do IS1 and IS1(NuXi). Interestingly, however, ISSt796, ISC1173, and ISSt1173 were found to have only one ORF (Fig. 1).
The amino acid sequences of the transposases encoded by the IS1 family elements were aligned (Fig. 2). Note here that the transposases encoded by IS1(NuXi), ISEhe5, ISY120, and ISMac740 with two ORFs were defined as transframe proteins produced by −1 translational frameshifting occurring at the run of adenines present in the overlapping region between the two ORFs, similar to the transposase encoded by IS1. The alignment revealed that several amino acid residues were conserved in all the transposases in positions corresponding to those in IS1 transposase (Fig. 2). This shows that all the elements examined are closely related to one another and supports the above assumption that they are members of the IS1 family.
FIG. 2.
Alignment of transposases encoded by IS1 family elements. The amino acid sequences of transposases encoded by eight elements (IS1, IS1[NuXi], ISEhe5, ISY120, ISMac740, ISSt796, ISC1173, and ISSt1173) are aligned. Numbers are coordinates given to the amino acid sequence of IS1 transposase. Amino acid residues constituting the D-D-E motif are shadowed. The other amino acid residues conserved in all the elements or in seven of eight elements are shown in boldface. Asterisks indicate amino acid residues that are thought to constitute the H-R-Y motif (44). The sequence encoded by insA in each transposase is underlined.
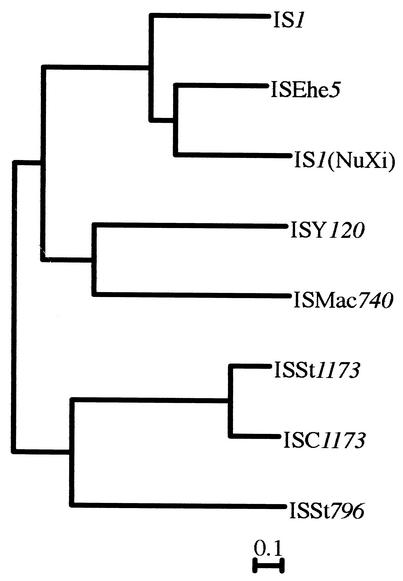
It was particularly interesting that out of 14 acidic amino acid residues present in IS1 transposase, two aspartic acid residues at amino acid positions 109 (D109) and 166 (D166) and one glutamic acid residue at position 191 (E191) were present in all the transposases encoded by the other IS1 family elements (Fig. 2). However, three amino acid residues, histidine at position 200 (H200), arginine at position 203 (R203), and tyrosine at position 231 (Y231), which are thought to constitute the H-R-Y motif in the IS1 transposase, were not conserved in corresponding positions in the transposases encoded by IS1 family elements (see Fig. 2). A phylogenetic tree constructed on the basis of the amino acid sequences of the transposases encoded by all the IS1 family elements revealed that these elements were apparently divided into two groups, of which one included only those from the Sulfolobus strains (Fig. 3).
FIG. 3.
Phylogenetic tree of IS1 family elements. The tree was constructed by the neighbor-joining method based on the amino acid sequences of transposases encoded by IS1 family elements.
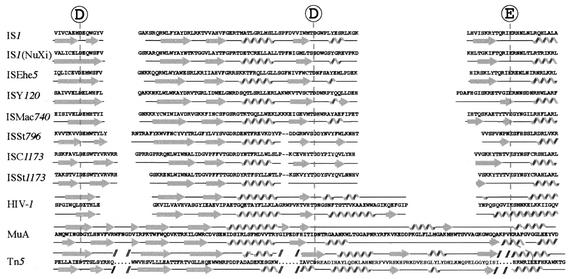
The three acidic amino acids, D, D, and E, conserved in the transposases encoded by all the IS1 family elements are assumed to constitute the D-D-E motif. The amino acid sequence of the polypeptide segment including this motif-like sequence in the IS1 transposase was therefore compared with those of proteins with the D-D-E motif, such as transposases encoded by other ISs and transposons and integrases encoded by retroviruses and retrotransposons (Fig. 4). The alignment revealed that some amino acid residues in IS1 transposase were conserved around each of the three acidic amino acids constituting the D-D-E motif (Fig. 4). This supports the assumption above that the three acidic amino acids (D, D, and E) conserved in the transposases encoded by all the IS1 family elements constitute the D-D-E motif.
FIG. 4.
Alignment of transposases and integrases. Polypeptide segments including D, D, and E and constituting the D-D-E motif in transposases encoded by IS1 family elements, other ISs or transposons (IS3, IS630, Tn3, Tn5, Tn7, Tn552, and Tc1), phage Mu transposase (MuA), integrases of retroviruses (HIV-1, Rous sarcoma virus [RSV], and Moloney murine leukemia virus [MoMLV]), and retrotransposons (Ty3-2, Gypsy, and Copia) are shown. Acidic amino acids constituting the D-D-E motif are shadowed, and other conserved amino acids are shaded. Amino acids frequently observed at each of several positions are shown at the top.
Secondary-structure analysis of transposases encoded by IS1 family elements.
To further support the above assumption, the secondary structure of the polypeptide segment with the D-D-E motif-like sequence in each of the transposases encoded by IS1 family elements was analyzed with the software program PSIPRED. In the four elements IS1(NuXi), ISEhe5, ISY120, and ISMac740, the deduced secondary structures of the polypeptide segments were very similar and had the characteristic three β sheets in tandem, with the first D residue present in the first of the three β sheets (Fig. 5). In the other IS1 family elements, IS1, ISSt796, ISC1173, and ISSt1173, the deduced secondary structures of the segments with the D-D-E motif-like sequence were very similar to one another and to those from the above four elements (IS1[NuXi], ISEhe5, ISY120) and ISMac740 except that the first of the three characteristic β sheets seen in IS1(NuXi), ISEhe5, and ISY120 was not present in IS1, ISSt796, ISC1173, or ISSt1173; instead, two short β sheets were present so as to flank a sequence of 2 to 4 amino acids including the first D residue (Fig. 5).
FIG. 5.
Comparison of secondary structures of transposases and an integrase. Secondary structures of the polypeptide segments with the D-D-E motif in transposases encoded by IS1 family elements were predicted by PSIPRED and are shown under the polypeptide sequence of each element. Secondary structures based on the tertiary structures of transposases encoded by phage Mu and Tn5 and HIV-1 integrase (PDB codes: 1BCM, 1F3I:M, and 1BHL, respectively) are shown above the polypeptide sequence of each element. α-Helices are indicated by ribbons, whereas β sheets are indicated by arrows. Other features are indicated by straight solid lines. Positions of acidic amino acids constituting the D-D-E motif are indicated by vertical broken lines.
The tertiary structures of HIV-1 integrase, phage Mu transposase (MuA), and Tn5 transposase have been determined by X-ray crystallography. Based on their tertiary structures, the secondary structures of the polypeptide segments with the D-D-E motif in these proteins are shown schematically in Fig. 5. Although the lengths of the regional segments between the first D and second D or between the second D and the E differ from one another, the overall secondary structures appear to be similar, particularly in the segments containing three β sheets (Fig. 5). Note that the three β sheets are arranged in tandem, and the first D residue is present in the first of the three β sheets, as it is in the four IS1 family elements IS1(NuXi), ISEhe5, ISY120, and ISMac740 (Fig. 5).
With the software program PSIPRED, the secondary structures of the polypeptide segments with the D-D-E motif in HIV-1 integrase, phage Mu transposase (MuA), and Tn5 transposase were deduced, as shown schematically in Fig. 5. The deduced secondary structures were generally similar to those based on their tertiary structures except that the segments with the D-D-E motif in the Mu and Tn5 transposases do not have the first β sheet but instead have two short β sheets flanking a sequence of 3 or 4 amino acids including the first D residue (Fig. 5). This difference may be derived from the parameters used for predicting the secondary structure.
It should be noted here that the secondary structures deduced for the integrase and transposases are similar to those of transposases from the four IS1 family elements IS1, ISSt796, ISC1173, and ISSt1173 (see Fig. 5). This suggests that the regional segment including the first D actually forms one β sheet even in transposases encoded by the four IS1 family elements. This hypothesis should be considered when discussing the difference between the secondary structure based on the tertiary structure and that deduced by PSIPRED in the phage Mu and Tn5 transposases. The similarity in the secondary structures also supports the suggestion that transposases encoded by the IS1 family elements have the D-D-E motif.
IS1 transposition mediated by mutant transposases.
We have developed a genetic assay system for the transposition of IS1. pNS17 is an ampicillin resistance (Apr) plasmid carrying mini-IS1, which has the chloramphenicol resistance (Cmr) gene replacing the transposase-coding region in IS1 (46). Mini-IS1 in this plasmid can transpose to the spectinomycin resistance (Spr) plasmid pSEK80 harbored within the same cell of E. coli strain NM554 when transposase is supplied from the kanamycin resistance (Kmr) plasmid pKEN200G, which produces IS1 transposase without translational frameshifting due to the presence of the sequence GA2GA3C in place of the A6C sequence (46). The IS1 transposase gene in pKEN200G can be expressed from a promoter induced by arabinose.
Therefore, for the actual experiment, NM554 cells harboring the three plasmids were grown overnight in broth containing arabinose. Plasmid DNA extracted from the cells was introduced by electroporation into E. coli strain JE6638(polA1), in which pNS17 carrying mini-IS1 cannot replicate, and thus, cells harboring target plasmids with mini-IS1 can be selected as Spr Cmr transformants (see Materials and Methods). In this system, the Spr Cmr transformants were obtained at a frequency of 1.4 × 10−4 per target plasmid (Fig. 6).
To see whether the three acidic amino acid residues (D109, D166, and E191) in IS1 transposase constitute a functional D-D-E motif, we constructed pSIN plasmids, derivatives of pKEN200G, producing mutant transposases with an amino acid substitution for each of the three acidic amino acids and measured the transposition frequency of mini-IS1 as described above. Mutant transposases with an alanine substitution (D109A, D166A, or E191A) mediated transposition of mini-IS1 at a greatly reduced frequency, 1/1,000th or less of that in wild-type transposase (Fig. 6). Mutant transposases with glutamic acid (or aspartic acid) instead of aspartic acid (or glutamic acid) (D109E, D166E, or E191D) or with asparagine (or glutamine) instead of aspartic acid (or glutamic acid) (D109N, D166N, or E191Q) also mediated transposition of mini-IS1 at a greatly reduced frequency, similar to that of those with an alanine substitution (Fig. 6). These results show that the three acidic amino acids, D109, D166, and E191, are important for transposition of IS1 and thus constitute a D-D-E motif.
We also constructed pSIN plasmids producing mutant transposases with an alanine residue substituted for each of the 11 acidic amino acid residues other than those in the D-D-E motif in the IS1 transposase (see Fig. 6) and measured the frequency of transposition of mini-IS1, as described above. Each of eight mutant transposases with a substitution (D15A, D58A, D101A, E107A, E142A, D159A, E172A, or D216A) mediated transposition of mini-IS1 at almost the same frequency as that in wild-type transposase or at a slightly lower frequency (Fig. 6). Each of the remaining three mutant transposases with a substitution (E110A, D129A, or D219A) mediated transposition of mini-IS1 at a reduced frequency, about 1/100th of that in wild-type transposase (Fig. 6).
We then constructed plasmids encoding mutant transposases with different amino acid substitutions for each of these three amino acid residues (E110Q, E110D, D129N, D129E, D219N, and D219E) and measured the frequency of mini-IS1 transposition. Each of the mutant transposases mediated transposition of mini-IS1 as efficiently as the mutant transposases with E110A, D129A, or D219A (Fig. 6). These results suggest that the 11 acidic amino acid residues are functionally less important than those in the D-D-E motif.
DISCUSSION
In this study, we have shown that IS1 family elements with homology to one another are widely distributed not only in eubacteria but also in archaebacteria. The phylogenetic tree constructed on the basis of the amino acid sequences of the transposases encoded by these elements showed that they are divided into two groups. Interestingly, the IS elements in one group have two ORFs, whereas those in the other group have only one ORF. This shows that the IS elements from each of the two groups are also structurally related to one another. These findings suggest that IS1 family elements diverged into two groups very early in evolutionary time.
It is assumed that the high frequency of transposition of an IS element causes deleterious effects on host cells. Some IS elements, such as IS1 and IS3, have two ORFs, and the product from one of the two ORFs acts as the negative regulator, inhibiting transposition of each element (24, 40, 53). These IS elements produce transposases as a transframe protein by translational frameshifting between the two ORFs (10, 37, 38, 41). This is thought to be another negative regulatory mechanism for the transposition of IS elements, because the frameshifting occurs at low efficiency, resulting in the production of a limiting amount of transposase (28). The IS1 family elements with only one ORF may therefore have a different regulatory mechanism(s) from those involved in the transposition of IS1 and IS3.
In this study, we have shown that the three acidic amino acids D109, D166, and E191 in IS1 transposase are conserved in corresponding positions in the transposases encoded by all the IS1 family elements. We also showed that the polypeptide segments including the three acidic amino acids are similar in primary structure as well as in secondary structure to those of the family of proteins with the D-D-E motif. The mutant IS1 transposases with an amino acid substitution for each of the three acidic amino acids lost the ability to promote transposition of IS1 almost completely. A few transposition products from the IS1 mutants were generated, probably due to the presence of six copies of IS1 in the chromosome of E. coli K-12 (48), in which even mutants with a deletion within IS1 can transpose at a very low frequency by the action of transposase produced from these IS1 copies (25).
In fact, the IS1 mutants producing transposases with an amino acid substitution for each of the three acidic amino acids did not transpose at all in E. coli strain ECOR46 (unpublished results), which is known to have no IS1 (36). These findings show that the three acidic amino acids are critically important for transposase to mediate transposition of IS1 and thus appear to constitute a D-D-E motif. This indicates that the transposase encoded by IS1, one of the smallest transposable elements in bacteria, also belongs to this family of proteins, including most transposases and integrases encoded by other transposable elements. The D-D-E motif identified in IS1 transposase, however, differs from those in the other elements in that the length of the polypeptide segment between the second D and the E is the shortest, 24 amino acids in length. Because of this difference, the presence of the D-D-E motif in IS1 transposase was not clarified for a long time, although IS1 was one of the first elements discovered in bacteria.
Previously, IS1 transposase was thought to belong to the Int family with the H-R-Y motif (44). All Int family proteins have histidine, arginine, and tyrosine residues constituting the H-R-Y motif in corresponding positions (1, 3). However, these amino acid residues in IS1 transposase (H200, R203, and Y231 [44]) are not found in corresponding positions in transposases encoded by the other IS1 family elements. This suggests that these three amino acid residues do not constitute the active center of IS1 transposase. In fact, the mutant IS1 transposases with amino acid substitution H200R or Y231A were able to promote transposition of mini-IS1 (data not shown), supporting this hypothesis.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Abremski, K. E., and R. H. Hoess. 1992. Evidence for a second conserved arginine residue in the integrase family of recombination protein. Protein Eng. 5:87-91. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Argos, P., A. Landy, K. Abremski, J. B. Egan, E. Haggard-Ljungquist, R. H. Hoess, M. L. Kahn, B. Kariolis, S. V. L. Narayana, L. S. Pierson III, N. Sternberg, and J. M. Leong. 1986. The integrase family of site-specific recombinase: regional similarities and global diversity. EMBO J. 5:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, T. A., and L. Luo. 1994. Identification of residues in the Mu transposase essential for catalysis. Proc. Natl. Acad. Sci. USA 91:6654-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolland, S., and N. Kleckner. 1996. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell 84:223-233. [DOI] [PubMed] [Google Scholar]
- 6.Davies, D. R., I. Y. Goryshin, W. S. Reznikoff, and I. Rayment. 2000. Three-dimensional Tn5 synaptic complex transposition intermediate. Science 289:77-85. [DOI] [PubMed] [Google Scholar]
- 7.Doak, T. G., F. P. Doerder, C. L. Jahn, and G. Herrick. 1994. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc. Natl. Acad. Sci. USA 91:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dower, W. J., J. M. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 10.Escoubas, J. M., M. F. Prère, O. Fayet, I. Salvignol, D. Galas, D. Zerbib, and M. Chandler. 1991. Translational control of transposition activity of the bacterial insertion sequence IS1. EMBO J. 10:705-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fayet, O., P. Ramond, P. Polard, M. F. Prère, and M. Chandler. 1990. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol. Microbiol. 4:1771-1777. [DOI] [PubMed] [Google Scholar]
- 12.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galas, D. J., M. P. Calos, and J. H. Miller. 1980. Sequence analysis of Tn9 insertions in the lacZ gene. J. Mol. Biol. 143:19-41. [DOI] [PubMed] [Google Scholar]
- 14.Grindley, N. D. F. 1978. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell 13:419-426. [DOI] [PubMed] [Google Scholar]
- 15.Guo, M., S. Manulis, H. Mor, and I. Barash. 2002. The presence of diverse IS elements and an avrPphD homologue that acts as a virulence factor on the pathogenicity plasmid of Erwinia herbicola pv. gypsophilae. Mol. Plant-Microbe Interact. 15:709-716. [DOI] [PubMed] [Google Scholar]
- 16.He, M., A. Wilde, and M. A. Kaderbhai. 1990. A simple single-step procedure for small-scale preparation of Escherichia coli plasmid. Nucleic Acids Res. 18:1660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iida, S., R. Hiestand-Nauer, and W. Arber. 1985. Transposable element IS1 intrinsically generates target duplications of variable length. Proc. Natl. Acad. Sci. USA 82:839-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakowec, M., P. Prentki, M. Chandler, and D. J. Galas. 1988. Mutational analysis of the open reading frames in the transposable element IS1. Genetics 120:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnsrud, L. 1979. DNA sequence of the transposable element IS1. Mol. Gen. Genet. 169:213-218. [DOI] [PubMed] [Google Scholar]
- 20.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 22.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-No, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagi, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 23.Kulkosky, J., K. S. Jones, R. A. Katz, J. P. G. Mack, and A. M. Skalka. 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machida, C., and Y. Machida. 1989. Regulation of IS1 transposition by the insA gene product. J. Mol. Biol. 208:567-574. [DOI] [PubMed] [Google Scholar]
- 25.Machida, Y., C. Machida, and E. Ohtsubo. 1984. Insertion element IS1 encodes two structural genes required for its transposition. J. Mol. Biol. 177:229-245. [DOI] [PubMed] [Google Scholar]
- 26.Machida, Y., C. Machida, H. Ohtsubo, and E. Ohtsubo. 1982. Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. Proc. Natl. Acad. Sci. USA 79:277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 28.Ohtsubo, E., and Y. Sekine. 1996. Bacterial insertion sequences. Curr. Top. Microbiol. Immunol. 204:1-26. [DOI] [PubMed] [Google Scholar]
- 29.Ohtsubo, E., M. Zenilman, and H. Ohtsubo. 1980. Plasmids containing insertion elements are potential transposons. Proc. Natl. Acad. Sci. USA 77:750-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtsubo, H., and E. Ohtsubo. 1978. Nucleotide sequence of an insertion element, IS1. Proc. Natl. Acad. Sci. USA 75:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtsubo, H., K. Nyman, W. Doroszkiewicz, and E. Ohtsubo. 1981. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature 292:640-643. [DOI] [PubMed] [Google Scholar]
- 32.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raleigh, E. A., N. E. Murray, H. Revel, R. M. Blumenthal, D. Westaway, A. D., P. W. Rigby, J. Elhai, and D. Hanahan. 1988. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 16:1563-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rezsöhazy, R., B. Hallet, J. Delcour, and J. Mahillon. 1993. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 9:1283-1295. [DOI] [PubMed] [Google Scholar]
- 35.Rice, P., and K. Mizuuchi. 1995. Structure of the bacteriophage Mu transposase core: a common structure motif for DNA transposition and retroviral integration. Cell 82:209-220. [DOI] [PubMed] [Google Scholar]
- 36.Sawyer, S. A., D. E. Dykhuizen, R. F. DuBose, L. Green, T. Mutangadura-Mhlanga, D. F. Wolczyk, and D. L. Hartl. 1987. Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics 115:51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekine, Y., and E. Ohtsubo. 1989. Frameshifting is required for expression of IS1 transposase. Proc. Natl. Acad. Sci. USA 86:4609-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekine, Y., and E. Ohtsubo. 1991. Translational frameshifting in IS elements and other genetic systems, p. 243-261. In M. Kimura and N. Takahata (ed.), New aspects of the genetics of molecular evolution. Japan Science Society Press, Tokyo, Japan.
- 39.Sekine, Y., H. Nagasawa, and E. Ohtsubo. 1992. Identification of the site of translational frameshifting required for production of the transposase encoded by insertion sequence IS1. Mol. Gen. Genet. 235:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekine, Y., K. Izumi, T. Mizuno, and E. Ohtsubo. 1997. Inhibition of transpositional recombination by OrfA and OrfB proteins encoded by insertion sequence IS3. Genes Cells 2:547-557. [DOI] [PubMed] [Google Scholar]
- 41.Sekine, Y., N. Eisaki, and E. Ohtsubo. 1994. Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 235:1406-1420. [DOI] [PubMed] [Google Scholar]
- 42.Sekine, Y., N. Eisaki, K. Kobayashi, and E. Ohtsubo. 1997. Isolation and characterization of IS1 circles. Gene 191:183-190. [DOI] [PubMed] [Google Scholar]
- 43.Sekino, N., Y. Sekine, and E. Ohtsubo. 1995. IS1-encoded proteins, InsA and the InsA-B′-InsB transframe protein (transposase): functions deduced from their DNA-binding ability. Adv. Biophys. 31:209-222. [DOI] [PubMed] [Google Scholar]
- 44.Serre, M. C., C. Turlan, M. L. Bortolin, and M. Chandler. 1995. Mutagenesis of the IS1 transposase: importance of a His-Arg-Tyr triad for activity. J. Bacteriol. 177:5070-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C.-Y. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. de Moors, G. Erauso, C. Fletcher, P. M. K. Gordon, I. H. Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiga, Y., Y. Sekine, Y. Kano, and E. Ohtsubo. 2001. Involvement of H-NS in transpositional recombination mediated by IS1. J. Bacteriol. 183:2476-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiga, Y., Y. Sekine, and E. Ohtsubo. 1999. Transposition of IS1 circles. Genes Cells 4:551-561. [DOI] [PubMed] [Google Scholar]
- 48.Umeda, M., and E. Ohtsubo. 1991. Four types of IS1 with differences in nucleotide sequence reside in the Escherichia coli K-12 chromosome. Gene 98:1-5. [DOI] [PubMed] [Google Scholar]
- 49.Williams, T. L., E. L. Jackson, A. Carritte, T. A. Baker. 1999. Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev. 13:2725-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC10 vectors. Gene 33:103-109. [DOI] [PubMed] [Google Scholar]
- 51.Yoshioka, Y., H. Ohtsubo, and E. Ohtsubo. 1987. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J. Bacteriol. 169:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zerbib, D., M. Jakowec, P. Prentki, D. J. Galas, and M. Chandler. 1987. Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. EMBO J. 6:3163-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zerbib, D., P. Polard, J. M. Escoubas, D. Galas, and M. Chandler. 1990. The regulatory role of the IS1-encoded InsA protein in transposition. Mol. Microbiol. 4:471-477. [DOI] [PubMed] [Google Scholar]
- 54.Zerbib, D., P. Prentki, P. Gamas, E. Freund, D. J. Galas, and M. Chandler. 1990. Functional organization of the ends of IS1: specific binding site for an IS1-encoded protein. Mol. Microbiol. 4:1477-1486. [PubMed] [Google Scholar]