Abstract
In streptomycete anthracycline biosynthetic gene clusters, small open reading frames are located just upstream of minimal polyketide synthase genes. aknX is such a gene found in the aklavinone-aclacinomycin biosynthetic gene cluster of Streptomyces galilaeus. In order to identify its function, the aknX gene was expressed in Escherichia coli. The cell extract prepared from E. coli cells overexpressing AknX protein exhibited anthrone oxygenase activity, which converted emodinanthrone to anthraquinone emodin. This indicates that AknX and related gene products such as DnrG and SnoaB are involved in the formation of aklanonic acid from its anthrone precursor, as suggested by their homology with TcmH and ActVA6. The AknX protein fused with a His6 tag was efficiently purified to homogeneity by Ni2+ affinity and anion-exchange column chromatography. The native molecular mass of AknX was estimated to be 42 kDa by gel filtration. Thus, native AknX is considered to have a homotrimeric subunit structure. AknX, like TcmH and ActVA6, possesses no apparent prosthetic group for oxygen activation. Site-directed mutagenesis was carried out to identify the key amino acid residue(s) involved in the oxygenation reaction. Of seven AknX mutants expressed, the W67F mutant showed significantly reduced oxygenase activity, suggesting the important role of the W67 residue in the AknX reaction. A possible mechanism for the reaction via peroxy anion intermediate is proposed.
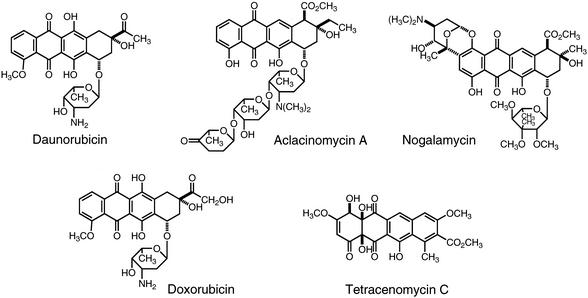
Aclacinomycins, which are anthracycline antibiotics produced by Streptomyces galilaeus, show potent antitumor activity by inhibiting complex formation of DNA and topoisomerase II and thus have lower cardiotoxicity than other anthracyclines that inhibit topoisomerase II by stabilization of cleavable complex (8). Aclacinomycin A, also called aclarubicin, has been clinically used in France, Japan, and other Asian countries for the treatment of carcinoma of the stomach, pulmonary carcinoma, oophoroma, malignant lymphadenoma, and acute leukemia. The aglycone moiety of aclacinomycins is aklavinone, which also serves as a common precursor for aglycones of other anthracyclines such as daunomycinone, pyrromycinone, and ɛ-rhodomycinone, etc. Figure 1 shows the chemical structures of representative anthracycline antibiotics.
FIG. 1.
Chemical structures of anthracycline antibiotics.
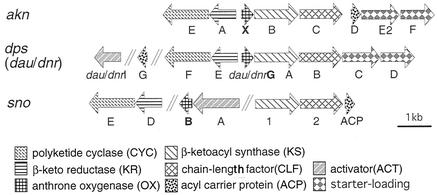
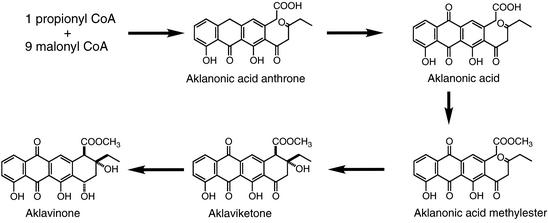
Previously, we cloned the aklavinone biosynthetic gene cluster from S. galilaeus strain 3AR-33, a mutant strain accumulating aklavinone (18). The 3.4-kb BamHI fragment complemented aklavinone production in the mutant strain ANR-58 as well as aclacinomycin production in the strain KE-303 (18). Nucleotide sequence analysis of the fragment showed the presence of open reading frames (ORFs) most typical of bacterial type II polyketide synthase (PKS) genes that code for β-ketoacyl synthase (KS), the so-called chain-length factor, and acyl carrier protein (3). In addition to these minimal PKS genes, we noted the presence of a small ORF, aknX, just upstream of the KS gene aknB. Such genes are not found in typical type II PKS gene clusters like that of actinorhodin (2). However, similar small ORFs exist in known anthracycline biosynthetic gene clusters such as dauG/dnrG in the daunorubicin/doxorubicin biosynthetic gene cluster (6, 20), and snoaB in the nogalamycin biosynthesis gene cluster (21) (Fig. 2). They show high homology with each other and also with tcmH, which was shown to be involved in the oxygenation of naphthacenone to naphthacenequinone in tetracenomycin biosynthesis (15). This similarity suggests that aknX and related dauG/dnrG and snoaB ORFs code for oxygenases responsible for quinone formation in aklavinone biosynthesis. However, their actual functions have not been confirmed. From detailed genetic and biochemical studies of daunorubicin/doxorubicin biosynthesis, the aklavinone biosynthetic scheme in Streptomyces sp. strain C5/Streptomyces peucetius has been established, as shown in Fig. 3. Aklanonic acid anthrone is an assumed direct PKS product and is oxidized to aklanonic acid by the oxygenation reaction. Such oxygenation is also known to be involved in anthraquinone formation in fungi (3). To confirm the role of aknX and related genes in anthracycline biosynthesis, we carried out the expression and functional analysis of the AknX protein.
FIG. 2.
Organization of PKS gene clusters for anthracycline antibiotics. The dau/dps genes are from S. peucetius and Streptomyces sp. strain C5, which produce daunorubicin/doxorubicin, and the sno genes are from S. nogalater, which is a producer of nogalamycin. The akn genes are from S. galilaeus for aklavinone-aclacinomycin biosynthesis.
FIG. 3.
Biosynthesis scheme of aklavinone.
MATERIALS AND METHODS
Strains, plasmids, biochemicals, and chemicals.
Escherichia coli strain XL1-Blue (Stratagene) and NovaBlue (Novagen) were used for transformation and amplification of plasmids. E. coli BL21(DE3)pLysE (Novagen) was used as a host for protein overexpression. Luria-Bertani medium was used for culture of E. coli. The pT7Blue T-vector (Novagen) was used for cloning of PCR products. The expression vectors used were pET22b (Novagen) and pLM 1 (16). DNA restriction and modifying enzymes were from Promega, Roche, Takara Shuzo, and Toyobo. All chemicals and biochemicals were from Sigma and Wako.
Construction of expression plasmids for aknX.
The aknX gene in the 3.4-kb BamHI fragment was originally cloned from chromosomal DNA of S. galilaeus strain 3AR-33 (18). For native AknX expression, plasmid pLM 1 was used in which the T7 gene 10 ribosome binding site and translational leader sequence were designed under the control of the T7 promoter. A sense oligonucleotide primer was designed to introduce an EcoRI site (shown underlined) and codons modified to those preferably used in E. coli (shown in boldface type) as follows: 5′-GCA TGC GAA TTC AGG AGA TAT ACA TAT GAC TGA TCA TGA ACC AGG TAC TGA AGG TGC CGA-3′. An antisense oligonucleotide primer was designed to introduce a HindIII site (shown underlined) at its end as follows: 5′-TCT AGA AAG CTT TCA TAT CTC CTC CCC TTC GGA-3′. The amplified product was subcloned into the pT7Blue T-vector and sequenced to confirm that no unintended mutations had been introduced. The EcoRI-HindIII fragment cleaved from the pT7Blue subclone was reintroduced into EcoRI- and HindIII-digested pLM 1 to construct pLM X-1.
The plasmid pET hisC-X was constructed to express AknX with a His6 tag at its C terminus (AknX His-C). The sense oligonucleotide primer pET-X-F was designed to introduce an NdeI site (shown underlined) and codons modified to those preferably used in E. coli (shown in boldface type) as follows: 5′-GAA TTC CAT ATG ACT GAT CAT GAA CCA GGT ACT GAA GGT GCC GAC GCC GTC ACC-3′. The antisense oligonucleotide primer pET-X-R was designed to replace the stop codon with an XhoI site (shown underlined) as follows: 5′-GCA TGC CTC GAG TCA TAT CTC CTC CCC TTC GGA-3′. The amplified product was subcloned into the pT7Blue T-vector, and the plasmid was named pT7 hisC-X; the insert sequence for this plasmid was confirmed to avoid unintended incorporation of mutations. The NdeI-XhoI fragment of pT7 hisC-X was ligated with NdeI- and XhoI-digested pET22b vector to construct pET hisC-X, which could express AknX His-C.
The plasmid pET hisN-X was also constructed to express AknX with a His6 tag at its N terminus (AknX His-N). A sense oligonucleotide primer was designed to introduce an NdeI site (shown underlined), six consecutive His codons, and codons modified to those preferably used in E. coli (shown in boldface type) as follows: 5′-GAG CAT ATG CAC CAT CAT CAT CAT CAT ACT GAT CAT GAA CCA GGT ACC GAG GGG GCC GAC-3′. An antisense primer was designed to introduce a PstI site (shown underlined) after the termination codon as follows: 5′-GGA CTG CAG TCA TAT CTC CTC CCC TTC GGA-3′. The amplified product was subcloned into pT7Blue T-vector, and the plasmid was named pT7 hisN-X; the insert sequence for this plasmid was confirmed. pT7 hisN-X was digested with PstI and then blunt ended with T4 DNA polymerase. Then, it was further digested with NdeI to obtain an aknX gene fragment with an NdeI end and a blunt end. This fragment was ligated with pET22b which was blunt ended at its EcoRI cleaved site and then digested with NdeI. The resultant plasmid was named pET hisN-X and could express AknX His-N.
Site-directed mutagenesis by PCR.
Site-directed mutations of the aknX gene were introduced by PCR. First PCR was performed to amplify the region from the mutation site to the start codon or stop codon. Then, the first PCR product was used as a primer to amplify the full-length mutated aknX gene. The following primers were used as mutation primers for the first PCR (mutated bases are shown in boldface type): R42K antisense primer, 5′-GAC GAA GCC CGG CTG CTT TGC CAT GAA CTC-3′; H49A antisense primer, 5′-GCA GAG CGT GGC CCG GAC GAA GCC CGG CTG-3′; H49Q antisense primer, 5′-GCA GAG CGT CTG CCG GAC GAA GCC CGG CTG-3′; C52S antisense primer, 5′-CCG TTC CGC GTG CCG TGA GAG CGT GTG GCG-3′; W67F sense primer, 5′-AAC GTC GCC GAG TTT CGG GAC CTC GCC TCG-3′; R74K sense primer, 5′-CTC GCG TCG TTC AAG GCG GCC GTC TCG CAC-3′; and H85A sense primer, 5′-GAC TTC CGG CCG GCA GCC GGC GCG CTG CGC-3′
For R42K, H49A, H49Q, and C52S mutations, the first PCR was performed using the pET-X-F primer and antisense mutation primer with the template aknX gene. The amplified product was purified on agarose gel and used as a primer for single-sided amplification with the template aknX gene. After 10 cycles of amplification, the pET-X-F primer and pET-X-R primer were added to amplify the full-length mutated aknX gene by an additional 25 cycles. For W67F, R74K, and H85A mutations, the first PCR was performed using a sense mutation primer and pET-X-R primer with the template aknX gene. The amplified product was purified on agarose gel and used as a primer for single-sided amplification with the template aknX gene. After 10 cycles of amplification, the pET-X-F primer and pET-X-R primer were added to amplify the full-length mutated aknX gene for an additional 25 cycles. The amplified product was subcloned into the pT7Blue T-vector. The sequence of the entire mutant aknX gene was determined to confirm that the intended mutation was introduced correctly. The cloned mutated aknX gene was cleaved with NdeI and XhoI digestion. Then, the aknX fragment was ligated with NdeI- and XhoI-digested pET22b vector to construct the expression plasmid for mutant AknX.
Overxpression of recombinant AknX.
For expression of AknX and its mutant proteins, expression plasmids were introduced into E. coli BL21(DE3)pLysE. Overnight preculture was inoculated in 10 volumes of Luria-Bertani medium and incubated at 37°C for 2 h (to an optical density at 600 nm of 0.4 to 1). Protein expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactoside (IPTG). After further incubation for 3 h at 37°C, the cells were harvested by centrifugation at 3,200 × g for 5 min. The precipitated cells were stored at −20°C.
Purification of recombinant AknX.
All steps were performed on ice or at 4°C. In a typical purification procedure, 26.5 g of cells was suspended in 79.5 ml of lysis buffer (50 mM sodium phosphate buffer, pH 7.8, containing 0.5 M NaCl). After addition of 0.133 mM phenylmethylsulfonyl fluoride and 21.2 mg of lysozyme, the cell suspension was stored at 4°C for 20 min. Then, 106 mg of sodium deoxycholate was added, and the mixture was incubated at 4°C for 20 min and then at 37°C for 5 min. Following DNase I digestion (70 U/g of cell) at room temperature for 20 min, the cell lysate was passed through a 0.2-μm-pore-size membrane filter to obtain a crude cell extract. For the purification of AknX with a His6 tag, crude extract was applied to a Probond Ni2+ affinity column (Invitrogen), and AknX with the His6 tag was eluted with 200 to 500 mM imidazole. AknX fractions collected were passed through a PD-10 column (Pharmacia) equilibrated with 25 mM Tris-HCl buffer, pH 7.5, for buffer exchange. The AknX fraction was divided into 10 subfractions, and each was applied to a Mono Q HR 5/5 anion-exchange column (Pharmacia) for further purification. AknX protein was eluted with a linear NaCl gradient of 0.15 to 0.5 M in 25 mM Tris-HCl buffer, pH 7.5. About 40 mg of purified AknX protein with the His6 tag was obtained by this two-step purification procedure.
Protein analysis.
Protein concentrations were determined spectrophotometrically by the method of Bradford (1), and bovine serum albumin was used as the standard protein. The concentration of the purified protein was determined by absorption at 280 nm. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using the buffer system of Laemmli (10) in 15% polyacrylamide gel, and protein bands were visualized by staining with Coomassie brilliant blue R-250. The exact molecular masses of purified proteins were determined using a time of flight mass spectrometer (TOF-MS) (KOMPACT MALDI IV; Shimadzu-Kratos). The matrix used was α-cyano-4-hydroxycinnamic acid dissolved in a 1:1 mixture of 0.1% trifluoroacetic acid and ethanol.
Assay of anthrone oxygenase activity.
Anthrone oxygenase activity was measured spectrophotometrically by following the increase of absorbance at 490 nm resulting from the formation of emodin from emodinanthrone at 30°C (3). The reaction mixture consisted of 1.8 ml of 0.5 M potassium phosphate buffer, pH 6.5, and 1 ml of ethylene glycol monomethyl ether containing 0.2 μmol of emodinanthrone. The reaction was started by adding enzyme solution. The reaction rate was calculated using the molecular extinction coefficient difference at 490 nm between emodinanthrone and emodin (Δɛ = 6.35 × 103).
Nucleotide sequence accession number.
The nucleotide sequence of the aklavinone-aclacinomycin biosynthetic gene cluster of S. galilaeus including aknX has been submitted to the DDBJ/GenBank/EMBL database under the accession no. AB008466.
RESULTS
Expression of AknX protein.
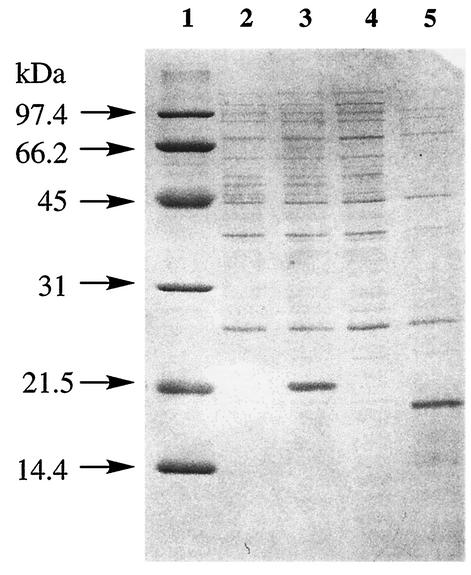
The deduced AknX protein consists of 122 amino acid residues with a molecular mass of 13,663 Da and a pI of 5.2. First, expression of AknX was attempted using the pET vector system (17) without modifying the native aknX coding sequence. However, no detectable expression of AknX protein could be observed by SDS-PAGE analysis. It is known that the expression of Streptomyces gene products is low in heterologous hosts due to the high GC content and that changing their N-terminal codons affects protein production in E. coli (5). Thus, codons of the aknX N-terminal region were changed to those preferably used in E. coli. Two kinds of expression plasmid were constructed: plasmid pLM X-1 for expression of intact AknX protein and plasmid pET hisC-X for expression of AknX with the His6 tag at its C terminus (AknX His-C). E. coli transformants harboring plasmids for AknX expression were cultured and treated with 1 mM IPTG at 37°C for 3 h to induce AknX expression. SDS-PAGE analysis of the resultant cell extracts of these transformants is shown in Fig. 4. Transformants of both plasmids showed high expression of the target protein of 19.4 kDa (pLM X-1) and 21.5 kDa (pET hisC-X), respectively. Due to the addition of a His6 tag at its C terminus, AknX His-C from pET hisC-X appeared to be larger than that from pLM X-1. However, their calculated molecular masses based on their deduced amino acid sequences were 13,663 and 14,728 Da, respectively. As described below, the exact molecular mass of AknX His-C was measured to be 14,728 Da by TOF-MS analysis. Therefore, the abnormal mobility of AknX proteins on SDS-PAGE was not due to erroneous expression of the protein.
FIG. 4.
SDS-PAGE analysis of the AknX expression in E. coli transformants. After electrophoresis, the gel (15% acrylamide) was stained with Coomassie brilliant blue. Lane 1, protein size marker; lane 2, pET-22b; lane 3, pET hisX; lane 4, pLM 1; lane 5, pLM X-1.
Enzyme activity of AknX protein.
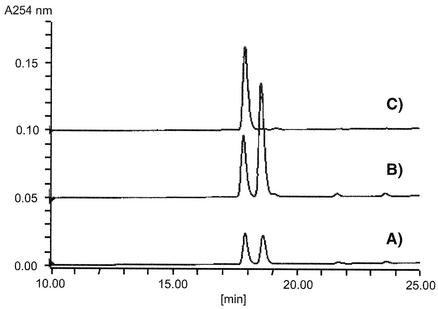
The amino acid sequence of AknX is 35% identical with that of Streptomyces glaucescens TcmH, which was shown to be an oxygenase converting tetracenomycin F1, a naphthacenone, to tetracenomycin D3, the 2,5-naphthacenequinone (15). ActVA6, which is 31% identical to AknX, was expressed in E. coli and showed the same oxygenase activity (9). Thus, the function of the AknX protein was assumed to be as an oxygenase involved in aklavinone biosynthesis, possibly converting aklanonic acid anthrone to aklanonic acid. Since aklanonic acid anthrone was not available and we previously detected emodinanthrone oxygenase activity in S. galilaeus cell extract (3), emodinanthrone was used as an alternative substrate for the AknX oxygenase assay (Fig. 5). Enzyme activity was measured spectrophotometrically by monitoring the increase in absorbance at 490 nm resulting from emodin formation (3). The cell extracts prepared from both E. coli transformants with pLM X-1 and pET hisC-X showed anthrone oxygenase activity (5.39 and 3.35 nmol/min/mg, respectively) that was absent in the host E. coli and transformants with empty vectors. Figure 6 shows the typical high-performance liquid chromatography profile of conversion of emodinanthrone to emodin by the AknX enzyme reaction. The product was confirmed to be emodin as previously described (3).
FIG. 5.
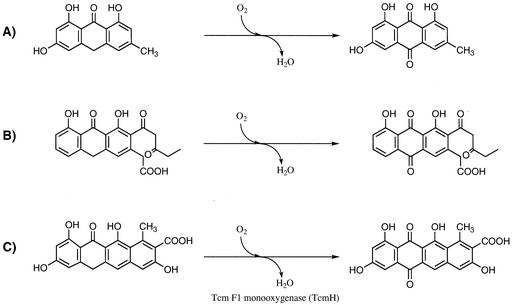
Reaction schemes of emodinanthrone to emodin (A), aklanonic acid anthrone to aklanonic acid (B), and tetracenomycin F1 to tetracenomycin D3 (C).
FIG. 6.
High-performance liquid chromatography analysis of enzyme reaction product. A TSK ODS-120T column (TOSOH) was eluted with gradient of acetonitrile-0.1% acetic acid from 50 to 90% at a flow rate 0.6 ml/min. Elution was monitored at a UV wavelength of 254 nm. (A) Enzyme reaction mixture; (B) enzyme reaction mixture with authentic emodin; (C) emodinanthrone.
Characterization of AknX protein.
The purified AknX His-C appeared as a single band of 21.5 kDa on the SDS-PAGE gel. The precise molecular mass of AknX His-C was found to be 14,728 Da by TOF-MS analysis, which exactly corresponded with the calculated molecular mass of Akn His-C. The native molecular mass of AknX His-C was estimated to be 42 kDa by gel-filtration chromatography. Thus, the AknX protein was assumed to be a homotrimer in solution, and the reason for the abnormal mobility of AknX protein on SDS-PAGE remains unclear. Interestingly, the AknX protein was found to be highly heat stable, did not lose oxygenase activity even after boiling for 5 min, and showed maximum activity at 45°C. However, its enzyme activity was lost by digestion with proteinase K at 56°C for 30 min. Metal chelating agents, such as EDTA or Chelex 100, did not affect the AknX oxygenase activity. Superoxide dismutase did not inhibit the enzyme, thus excluding the participation of superoxide anion in oxygenase reaction.
Expression of mutant AknX.
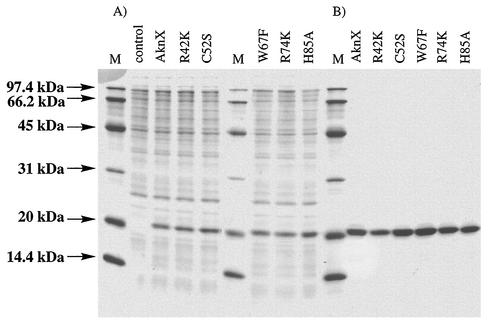
As was reported for related oxygenase enzymes such as TcmH (15) and ActVA6 (9), no apparent prosthetic group was involved in the oxygenation reaction of AknX. In general, the activation of molecular oxygen is required to incorporate oxygen into the substrate molecule (7, 13, 19). Thus, some amino acid residues might have a role in oxygen activation. To identify such key amino acid residues, site-directed mutagenesis was carried out. Of the conserved basic and/or aromatic amino acid residues in AknX, TcmH, and ActVA6, Arg-42, His-49, Trp-67, Arg-74, and His-85 were chosen as target residues. In addition, Cys-52 was included because Shen and Hutchinson commented on the possible involvement of a sulfhydryl group in TcmH monooxygenase (15). Expression plasmids for mutant AknXs were constructed based on PCR mutagenesis as described in Materials and Methods. Mutant AknXs, R42K, H49A, H49Q, C52S, W67F, R74K, and H85A, were expressed with the His6 tag at their C termini in E. coli and purified by Ni2+ affinity column chromatography as performed for AknX His-C expressed by pET hisC-X. The results of SDS-PAGE analysis of mutant AknX expression and purification are shown in Fig. 7. All purified mutant proteins appeared as apparent single bands at 21.5 kDa.
FIG. 7.
SDS-PAGE analysis of mutant AknXs. (A) Cell extracts of control transformant with pET22b and transformants expressing mutant AknXs. (B) Ni2+ affinity purified mutants. Lanes M: molecular mass marker. After electrophoresis, the gel (15% acrylamide) was stained with Coomassie brilliant blue.
Kinetic analysis of AknX and mutants.
All AknX mutants prepared showed emodinanthrone oxygenase activity, and none lost activity completely. To characterize the catalytic properties of the mutant AknXs, kinetic parameters were determined. As shown in Table 1, the mutant W67F showed significantly lower enzyme activity, with less than 0.2% of the Vmax/Km value of wild-type AknX. Interestingly, R74K mutant was found to have a Vmax 4 times greater and a Km value 10 times greater than those of the wild-type AknX. These results were not conclusive but indicated the important role of the Trp-67 residue in the AknX oxygenation reaction.
TABLE 1.
Kinetic analysis of mutated AknXsa
| Mutant | Vmax (nmol/min/mg) | Km (μM) | Vmax/Km |
|---|---|---|---|
| AknX | 73.5 (1.00) | 39.8 (1.0) | 1.85 (1.0) |
| R42K | 76.5 (1.04) | 68.7 (1.7) | 1.12 (0.61) |
| C52S | 61.2 (0.83) | 98.7 (2.5) | 0.62 (0.34) |
| W67F | 15.3 (0.21) | 286 (7.2) | 0.05 (0.03) |
| R74K | 282 (3.84) | 404 (10.2) | 0.70 (0.38) |
| H85A | 61.0 (0.83) | 442 (11.1) | 0.14 (0.08) |
AknX mutants purified by Ni2+ affinity column chromatography were used in kinetic assays in which concentrations of substrate emodinanthrone were varied between 5 and 100 μM. Relative values are shown in parentheses.
Large-scale purification of recombinant AknX.
Large-scale purification of AknX with the His6 tag at its C terminus (AknX His-C) was attempted. However, a minute impurity of lower molecular weight could not be removed even after Ni2+ affinity column and Mono Q anion-exchange column chromatography. This minor protein was considered to be an alternative translation product of aknX started from the GTG codon downstream from the normal start codon ATG. Thus, the new expression plasmid pET hisN-X was constructed to express AknX protein with the His6 tag at its N terminus (AknX His-N). An alternative translation protein, if produced, could be removed by Ni2+ affinity column.
AknX His-N also showed anthrone oxygenase activity and was purified to homogeneity by Ni2+ affinity column and then Mono Q anion-exchange column chromatography as judged by SDS-PAGE (21.5 kDa) and TOF-MS (14,486 Da) analyses. Starting from an 8-liter induction culture of pET hisN-X transformant, 38.9 mg of homogeneous AknX His-N was obtained. The results of the purification are summarized in Table 2.
TABLE 2.
Purification of recombinant AknX His-Na
| Sample | Protein (mg) | Sp act (nmol/min/mg) |
|---|---|---|
| Cell-free extract | 3,075 | 7.7 |
| Ni2+ affinity column | 58.3 | 31.4 |
| Mono Q column | 38.9 | 37.2 |
Purification of recombinant AknX His-N was started from cells collected from 8-liter induction cultures and carried out as described in Materials and Methods.
DISCUSSION
In the aklavinone biosynthetic pathway, aklanonic acid was isolated as the first cyclized intermediate (14) which was considered to be an oxygenation product of its anthrone precursor. However, to the best of our knowledge, no anthrone oxygenase involved in anthraquinone aklanonic acid formation has been reported. In the study of fungal emodinanthrone oxygenase, Fujii et al. reported the detection of emodinanthrone oxygenase activity in S. galilaeus cell extract (3). Subsequently, Shen and Hutchinson reported the oxygenase activity of TcmH involved in tetracenomycin biosynthesis (15). They purified the native oxygenase protein from the cell extract of S. glaucescens and determined that the purified oxygenase was a tcmH gene product. This small protein contains no prosthetic groups such as metal, or heme, etc., which raises the question of how the enzyme activates molecular oxygen for fixation in the substrate. A similar protein was noted in the biosynthetic gene cluster of actinorhodin. This protein, encoded by actVA6, was expressed in E. coli, and its enzyme activity was detected using the TcmH assay system (9). However, its actual catalytic function in actinorhodin biosynthesis has not yet been confirmed.
In genetic studies on anthracycline biosynthesis, similar small ORFs were identified in the biosynthetic gene clusters just upstream from the KS gene in S. peucetius (6), Streptomyces sp. strain C5 (20), Streptomyces nogalater (21), and S. galilaeus (4). Their homology with tcmH suggested that the encoded enzyme might catalyze the oxygenation of aklanonic acid anthrone to form aklanonic acid. However, their catalytic activities have not been identified. In this report, we overexpressed AknX protein and identified it as an anthrone oxygenase.
Shen and Hutchinson proposed that the reaction mechanism of TcmH monooxygenase is a radical process including generation of superoxide anion radical and radical enzyme (15). However, the anthrone oxygenase activity of AknX was not affected by superoxide dismutase. They also commented on the possible involvement of a sulfhydryl group for TcmH monooxygenase activity, although Cys residues are not conserved in other streptomycete oxygenases of this type and the C52S mutant of AknX retained its oxygenase activity.
Kendrew et al. suggested that His-52 may have a role as a key amino acid in ActVA6 enzyme activity (9). They proposed that His-52 acts as a general base catalyzing the dehydration of the putative peroxide intermediate. We expressed mutants of AknX His-49, which corresponds to His-52 of ActVA6. AknX H49Q and H49A showed 40 and 445% of the enzyme activities of AknX His-49, respectively. This implies that His-49 is not the essential residue, at least in the AknX enzyme reaction. Kinetic analysis of mutant AknXs suggested that the Trp-67 residue plays a key role in the AknX oxygenation reaction.
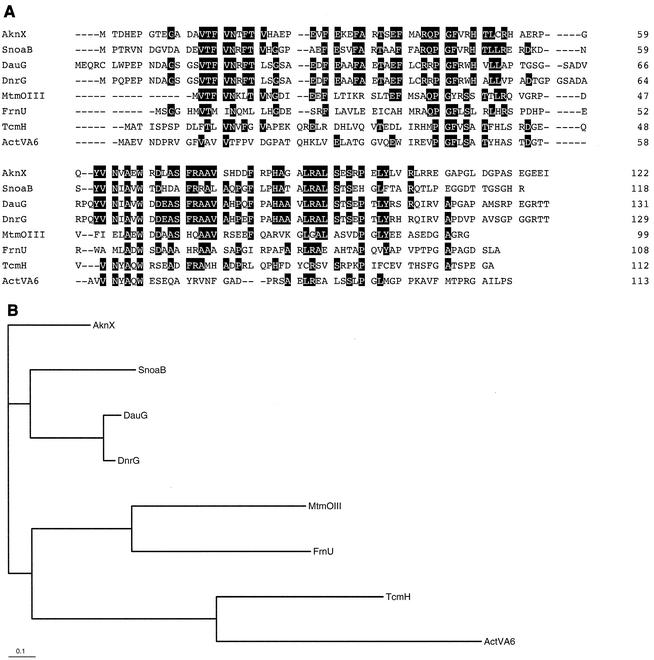
In addition to aknX, tcmH, and actVA6, several genes such as dnrG of S. peucetius (6), dauG of Streptomyces sp. strain C5 (20), snoaB of S. nogalater (21), mtmOIII of Streptomyces argillaceus (11), and frnU of Streptomyces roseofulvus, have been reported to be possibly involved in similar oxygenation reactions. The phylogenetic relationship of their gene products is shown in Fig. 8. The possible aklanonic acid anthrone oxygenases AknX, DnrG, DauG, and SnoaB are closely related in one branch and have a distal relationship with other oxygenases: TcmH, ActVA6, MtmOIII, and FrnU. Their amino acid sequence alignment shows that only four amino acid residues—Pro-44, Gly-45, Ala-65, and Trp-67—in the AknX sequence are fully conserved in these proteins. This also suggested the important role of the Trp-67 residue in the catalytic function of AknX.
FIG. 8.
Phylogenetic tree and amino acid sequence alignment of AknX and related streptomycete oxygenases. (A) The sequences were aligned and homology scores were obtained using the program CLUSTAL W. Highly conserved amino acid residues are in black boxes. (B) The phylogenetic tree was developed with the same program using the neighbor-joining method and drawn with TREE VIEW. The indicated scale represents 0.1 amino acid substitution per site.
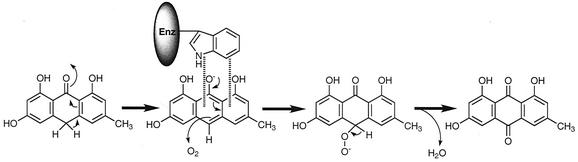
Enzymatic addition of molecular oxygen without cofactor has been observed in luciferin-luciferase reactions of some higher organisms with bioluminescence, for example, the oxidation of renilla-luciferin (colenterazine) by luciferase of Renilla reniformis (an anthozoan coelenterate) (12). A high-energy intermediate, luciferin peroxide, is formed in a nonradical reaction. Provided that the AknX oxygenation reaction is not a radical process without cofactor and that Trp-67 has a key role in the reaction, the following possible mechanism is proposed. The reaction starts with the formation of anthranol anion, which is stabilized by π-π complexation with Trp-67 in a fixed orientation. Then, hydroperoxyanthorne is formed by dioxygen addition and subsequently dehydrated to form anthraquinone (Fig. 9).
FIG. 9.
Proposed mechanism for the AknX-catalyzed oxygenase reaction.
Oxygen activation and the fixation mechanism catalyzed by AknX and related streptomycete oxygenases appear unique compared with known oxygenases with heme, nonheme iron, or other prosthetic groups. Elucidation of the three-dimensional structure of AknX is indispensable to understanding its unique reaction mechanism. Some 600 crystallization conditions have so far been screened to obtain AknX protein crystals. A needle-like crystal and piles of small crystals were formed. However, their shapes and sizes were not suitable for X-ray crystallographic analysis. Further screening of crystallization conditions and optimization is now under way.
Acknowledgments
We thank Mercian Corp. for the kind gift of S. galilaeus strains. We are grateful to M. Sodeoka (Tohoku University, Sendai, Japan) for providing the expression plasmid pLM 1.
This work was in part supported by a grant-in-aid for scientific research (C) (08672411 to I.F.) from The Ministry of Education, Culture, Sports, Science, and Technology, Tokyo, Japan.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez-Moreno, M. A., E. Martinez, L. Boto, D. A. Hopwood, and F. Malpartida. 1992. Nucleotide sequence and deduced functions of a set of co-transcribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J. Biol. Chem. 267:19278-19290. [PubMed] [Google Scholar]
- 3.Fujii, I., Z.-G. Chen, Y. Ebizuka, and U. Sankawa. 1991. Identification of emodinanthrone oxygenase in fungus Aspergillus terreus. Biochem. Int. 25:1043-1049. [PubMed] [Google Scholar]
- 4.Fujii, I., and Y. Ebizuka. 1997. Anthracycline biosynthesis in Streptomyces galilaeus. Chem. Rev. 97:2511-2523. [DOI] [PubMed] [Google Scholar]
- 5.Gramajo, H., J. White, C. R. Hutchinson, and M. J. Bibb. 1991. Overproduction and localization of components of the polyketide synthase of Streptomyces glaucescens involved in the production of the antibiotic tetracenomycin C. J. Bacteriol. 173:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimm, A., K. Madduri, A. Ali, and C. R. Hutchinson. 1994. Characterization of the Streptomyces peucetius ATCC 29050 genes encoding doxorubicin polyketide synthase. Gene 151:1-10. [DOI] [PubMed] [Google Scholar]
- 7.Hayaishi, O. 1974. Molecular mechanisms of oxygen activation. Academic Press, New York, N.Y.
- 8.Holm, B., P. B. Jensen, M. Sehested, and H. Hansen. 1994. In vivo inhibition of etoposide-mediated apoptosis, toxicity, and antitumor effect by the topoisomerase II-uncoupling anthracycline aclarubicin. Cancer Chemother. Pharmacol. 34:503-508. [DOI] [PubMed] [Google Scholar]
- 9.Kendrew, S. G., D. A. Hopwood, and E. N. G. Marsh. 1997. Identification of a monooxygenase from Streptomyces coelicolor A3(2) involved in biosynthesis of actinorhodin: purification and characterization of the recombinant enzyme. J. Bacteriol. 179:4305-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lombo, F., G. Blanco, E. Fernandez, C. Mendez, and J. A. Salas. 1996. Characterization of Streptomyces argillaceus genes encoding a polyketide synthase involved in the biosynthesis of the antitumor mithramycin. Gene 172:87-91. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz, W. W., R. O. McCann, M. Longiaru, and M. J. Cormier. 1991. Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc. Natl. Acad. Sci. USA 88:4438-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oritz de Montellano, P. R. 1986. Cytochrome P-450: structure, mechanism and biochemistry. Plenum Press, New York, N.Y.
- 14.Schlegel, B., C. Stengel, G. Schumann, H. Prauser, and K. Eckardt. 1987. Aklanonic acid-producing mutants of Streptomyces galilaeus and Streptomyces peucetius var. caesius. J. Basic Microbiol. 27:107-111. [DOI] [PubMed] [Google Scholar]
- 15.Shen, B., and C. R. Hutchinson. 1993. Tetracenomycin F1 monooxygenase: oxidation of a naphthacenone to a naphtahcenequinone in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry 32:6656-6663. [DOI] [PubMed] [Google Scholar]
- 16.Sodeoka, M., C. J. Larson, L. Chen, K. P. LeClair, and G. L. Verdine. 1993. A multifunctional plasmid for protein expression by ECPCR: overproduction of the p50 subunit of NF-κB. Bioorg. Med. Chem. Lett. 3:1089-1094. [Google Scholar]
- 17.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dudendorff. 1990. Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 18.Tsukamoto, N., I. Fujii, Y. Ebizuka, and U. Sankawa. 1992. Cloning of aklavinone biosynthesis genes from Streptomyces galilaeus. J. Antibiot. 45:1286-1294. [DOI] [PubMed] [Google Scholar]
- 19.Walsh, C. 1979. Enzymatic reaction mechanisms. Freeman & Co., San Francisco, Calif.
- 20.Ye, J., M. L. Dickens, R. Plater, Y. Li, J. Lawrence, and W. R. Strohl. 1994. Isolation and sequence analysis of polyketide synthase genes from the daunomycin-producing Streptomyces sp. strain C5. J. Bacteriol. 176:6270-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yhilihonko, K., J. Tuikkanen, S. Jussila, L. Cong, and P. Mäntsälä. 1996. A gene cluster involved in nogalamycin biosynthesis from Streptomyces nogalater: sequence analysis and complementation of early-block mutations in the anthracycline pathway. Mol. Gen. Genet. 251:113-120. [DOI] [PubMed] [Google Scholar]