Abstract
Methylobacterium extorquens AM1, a serine cycle facultative methylotroph, accumulates poly-β-hydroxybutyrate (PHB) as a carbon and energy reserve material during growth on both multicarbon- and single-carbon substrates. Recently, the identification and mutation of the genes involved in the biosynthesis and degradation of PHB have been described for this bacterium, demonstrating that two of the genes of the PHB cycle (phaA and phaB) are also involved in C1 and C2 metabolism, as part of a novel pathway for glyoxylate regeneration in the serine cycle (N. Korotkova and M. E. Lidstrom, J. Bacteriol. 183:1038-1046, 2001; N. Korotkova, L. Chistoserdova, V. Kuksa, and M. E. Lidstrom, J. Bacteriol. 184:1750-1758, 2002). In this work, three new genes involved in PHB biosynthesis in this bacterium have been investigated via mutation and phenotypic analysis: gap11, gap20, and phaR. We demonstrate that gap11 and gap20 encode two major granule-associated proteins (phasins) and that mutants with mutations in these genes are defective in PHB production and also in growth on C2 compounds, while they show wild-type growth characteristics on C1 or multicarbon compounds. The phaR mutant shows defects in both PHB accumulation and growth characteristics when grown on C1 compounds and has defects in PHB accumulation but grows normally on C3 and C4 compounds, while both PHB accumulation and growth rate are at wild-type levels during growth on C2 compounds. Our results suggest that this phenotype is due to altered fluxes of acetyl coenzyme A (CoA), a major intermediate in C1, C2, and heterotrophic metabolism in M. extorquens AM1, as well as the entry metabolite for the PHB cycle. Therefore, it seems likely that PhaR acts to control acetyl-CoA flux to PHB in this methylotrophic bacterium.
Poly-β-hydroxybutyrate (PHB) and other polyhydroxyalkanoates (PHAs) are accumulated by many bacteria as an energy and carbon reserve material (1). While the enzymology and the genetics of PHA biosynthesis have been extensively studied for a number of organisms and are now well understood, less is known about the regulation of this process. A series of recent studies have been undertaken to unravel the mechanisms by which PHA biosynthesis is controlled in a number of bacteria with different modes of metabolism, and the emerging patterns seem to be complex. So far, two major types of PHA accumulation regulators have been investigated: (i) phasins, the proteins that bind to PHA granules and promote PHA synthesis, and (ii) regulators having an effect ranging from regulating phasin biosynthesis to more complex, pleiotropic effects. These are known under two different names, PhaR and AniA (11, 12, 28, 29, 46). In addition, two different types of transcriptional regulators have been reported elsewhere to be involved in PHB production in Azotobacter vinelandii (4, 45).
So far, phasins appear to be present in all PHA-synthesizing bacteria, and even though they generally are not conserved in sequence and seem to be species specific, they are believed to fulfill the same function, binding to PHA granules and promoting PHA synthesis, in a manner still poorly understood (19). It has been shown elsewhere that phasins stabilize the PHA granules, prevent them from coalescing, and control the granule size (15, 18, 19, 34). In accordance with one hypothesis, phasins must bind directly to PHA and possibly to the PHA-cycling enzymes, PHA synthase and/or PHA depolymerase (15, 18), while in accordance with a second hypothesis, they may bind to the granules themselves and prevent binding of proteins not related to PHA metabolism (18, 19). The second type of regulator, now identified in many bacteria and designated PhaR, was first identified in close proximity to a gene encoding a phasin in Paracoccus denitrificans and implicated in its negative regulation (28, 29). Negative regulation of a phasin by a PhaR homolog was also reported for Ralstonia eutropha; however, in the latter case, PhaR also influenced PHB production in a phasin-independent fashion, which remains unknown (46). In Sinorhizobium meliloti, a homolog of PhaR was called AniA, because of its expression during anaerobic growth. An even more complex function was suggested for this protein, in partitioning of carbon flow in cells, affecting not only PHB production but also production of extracellular polymeric substances and nitrogen fixation (36). AniA was also investigated in another rhizobial species, Rhizobium etli, and suggested to play a role not only in PHB production but also as a global regulator of protein expression and of carbon and energy fluxes, based on the pleiotropic effect of mutation in AniA on metabolism of this bacterium (11, 12).
Methylobacterium extorquens AM1 accumulates PHB and forms PHB granules like other PHB-producing bacteria (13, 14). PHB content in M. extorquens AM1 cells varies depending on growth substrate (23). Genes and enzymes participating in the biosynthesis and degradation of PHB have recently been identified in M. extorquens AM1, and their roles in PHB cycling have been studied via mutation (23). Mutants with mutations in the first two enzymes for PHB synthesis from acetyl coenzyme A (CoA), β-ketothiolase (PhaA) and NADPH-linked acetoacetyl-CoA reductase (PhaB), revealed not only a PHB deficiency but also a deficiency in growth on C1 and C2 compounds. It was determined that this growth deficiency was due to the participation of these genes in the novel pathway for regeneration of glyoxylate from acetyl-CoA in the serine cycle (23, 24). Thus, in this methylotrophic bacterium, the PHB biosynthesis pathway overlaps with the central metabolic pathways for C1 and C2 assimilation. Such a central placement of the PHB cycle in the metabolic network of M. extorquens AM1 would suggest specific mechanisms for regulating synthesis and degradation of PHB depending on growth substrate-specific metabolic modes. In this work we identify the phaR gene in M. extorquens AM1 and investigate the effect of mutation in this gene on growth and PHB production on different substrates. We also identify two proteins associated with PHB granules in this organism, Gap11 and Gap20.
MATERIALS AND METHODS
Bacterial strains, growth conditions, plasmids, and DNA manipulations.
Escherichia coli strains DH5α (Gibco-BRL, Rockville, Md.), S17-1 (40), and BL21(DE3) (Novagen, Inc., Madison, Wis.) used in the study were grown in Luria-Bertani medium in the presence of appropriate antibiotics as described by Maniatis et al. (30). M. extorquens AM1 was grown in the minimal medium described previously (16). Succinate (20 mM), pyruvate (20 mM), methanol (100 mM), or ethylamine (20 mM) was used as a substrate. The antibiotic concentrations used for M. extorquens AM1 were as follows: tetracycline, 10 μg ml−1; kanamycin, 100 μg ml−1; and rifamycin, 50 μg ml−1. Growth responses of the phaR mutant were tested in methanol-containing medium supplemented with acetate (0.1 to 0.2 mM). For methanol induction, strains were grown on succinate to mid-logarithmic phase, pelleted, and incubated in methanol (100 mM)-containing medium overnight with shaking at 30°C. The following cloning vectors were used: pUC19 (Amersham Pharmacia Biotech, Inc. Piscataway, N.J.) for cloning and subcloning, pAYC61 (5) as a suicide vector, pRK2013 (10) as a helper plasmid, pCR2.1 (Invitrogen, Carlsbad, Calif.) for cloning of PCR products, pET21d(+) (Novagen, Inc.) as an expression vector, and pCM130 (31) as a promoter probe vector. Plasmid isolation, E. coli transformation, restriction enzyme digestion, and ligation were carried out as described by Maniatis et al. (30). The chromosomal DNA of M. extorquens AM1 was isolated by the procedure of Saito and Miura (38).
Gene amplification and cloning.
Data from the M. extorquens AM1 genome project (http://vixen.microbiol.washington.edu/) were used for designing primers specific for putative genes of M. extorquens AM1 encoding PhaR, Gap11, and Gap20 (primer sequences may be provided on request). The identity of the amplified DNA fragments was confirmed by sequencing from both strands performed by the Department of Biochemistry, University of Washington Sequencing Facility.
Mutant generation.
Insertion mutations in phaR, gap11, and gap20 were generated in vitro with the Kmr gene cartridge as described earlier (5-8). The sites used for inserting the Kmr gene cartridge were as follows: the BsgI site in the first half of phaR, the PstI site in the putative ribosome-binding region of gap11, and the HincII site in the first half of gap20. Triparental or biparental matings between E. coli carrying the respective donor plasmids and M. extorquens AM1 were performed overnight on nutrient agar at 30°C. Cells were then washed with sterile medium and plated on selective medium at appropriate dilutions. In triparental matings, pRK2013 (10) was used as a helper plasmid. Rifamycin was used for E. coli counterselection. In all cases, mutants were selected on succinate plates in the presence of kanamycin, and Kmr colonies were checked for their resistance to tetracycline. Tcs colonies were chosen as potential double-crossover recombinants, while Tcr colonies were assumed to be single-crossover recombinants. The identity of the double-crossover mutants was confirmed by diagnostic PCR.
Computer analysis.
Sequence comparisons and analysis were performed with BLAST and search tools available on the M. extorquens AM1 genome website (http://vixen.microbiol.washington.edu/) and also tools and programs available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Motif searches were performed with two different programs for helix-turn-helix motif prediction available at http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_hth.html and at http://www.es.embnet.org/Services/MolBio/hth.html.
Enzyme assays.
Activities of β-ketothiolase and acetoacetyl-CoA reductase were determined as described previously (39), and the activity of catechol 2,3-dioxygenase was assayed as described by Kataeva and Golovleva (21) in M. extorquens AM1 crude extracts obtained by passing cells through a French pressure cell at 1.2 × 108 Pa, followed by centrifugation for 10 min at approximately 15,000 × g. All measurements were done at room temperature (26°C) in a total volume of 1 ml. Spectrophotometric methods (20, 43) were used for protein determination. All assays were done in triplicate, and results agreed within ±15%.
Determination of distribution of 14C from [2-14C]acetate between CO2 and biomass.
Succinate-grown cells were harvested by centrifugation (5 min, 6,000 × g), washed, and resuspended in a medium containing methanol (100 mM). Cells were incubated with shaking at 30°C for 1 h for induction, after which [2-14C]acetate (0.1 mM, 0.2 μCi per reaction vial) was added and samples were taken at 10-min intervals. The percentage of total radioactivity in cell biomass versus CO2 was determined and calculated as described previously (24).
PHB analysis.
PHB concentration in bacterial cells was determined by a gas chromatographic method (2). A model GC-14A capillary gas chromatograph (Shimadzu, Kyoto, Japan) with an AT-WAX capillary column (0.53 mm by 10 m; 1.2-μm film thickness; Alltech, Deerfield, Ill.) and a flame ionization detector was used. The flow rate of helium carrier gas was 2.7 ml/min. The initial column temperature of 60°C was held for 2 min, and then the temperature was increased by 5°C/min up to 160°C.
PHB granule isolation.
PHB granules were isolated on a sucrose gradient by the method of Föllner and colleagues (13). The pelleted granules were resuspended in gel loading buffer. After 5 min of incubation at 100°C, the granule-associated proteins (GAPs) were separated by gel electrophoresis (sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) according to the method of Laemmli (25) with 8 to 12% gradient acrylamide gels (Bio-Rad Laboratories, Hercules, Calif.).
xylE fusion construction.
Putative promoter regions for phaA (650 bp), phaB (450 bp), phaC (740 bp), phaR (650 bp), gap11 (450 bp), and gap20 (620 bp) were amplified by PCR and cloned into the pCR2.1 vector. These fragments covered the entire intergenic regions, contained the 5′ termini of the genes whose promoters were being tested, and also overlapped with the neighboring genes by their 5′ or 3′ termini. The resulting fragments were subsequently cut out with appropriate restriction enzymes and inserted into the promoter probe vector pCM130 (31), upstream of a promoterless xylE gene.
Electrophoretic mobility shift assays.
An 0.65-kb PCR product containing the entire phaR gene and its putative ribosome-binding site was cloned into pCR2.1, redigested with XbaI-BamHI, and cloned into pET21d(+) in the orientation allowing transcription from the T7 promoter. The resulting plasmid was transferred into the expression strain E. coli BL21(DE3). The resulting E. coli strain was grown in Luria-Bertani medium supplemented with 100 μg of ampicillin/ml at 37°C with shaking at 350 rpm to an optical density of 0.4 to 0.6 at 600 nm. Expression of PhaR was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), followed by a 4-h incubation. Cells were collected by centrifugation, resuspended in 3 ml of 50 mM sodium phosphate (pH 7.5)-200 mM KCl, and disrupted with a French pressure cell at 1.2 × 108 Pa, followed by centrifugation for 20 min at approximately 15,000 × g. The supernatant was used for gel shift assays.
Target DNA fragments of about 400 to 500 bp were amplified by PCR and end labeled by phosphorylation with T4 DNA kinase (Promega, Madison, Wis.). The cell extract containing PhaR (8 to 10 μg) was incubated with the labeled DNA for 20 min at room temperature in the following gel shift binding buffer (Promega): 5 mM Tris HCl (pH 7.5)-50 mM NaCl-1 mM MgCl2-0.5 mM dithiothreitol-0.5 mM EDTA-4% glycerol-0.05 μg of poly(dI-dC)/ml. Cell extract of E. coli containing no PhaR was used as a control. After incubation, the mixtures were loaded on a Novex 6% retardation gel (Invitrogen) or a 6% nondenaturing acrylamide gel in 0.5× Tris-borate-EDTA and electrophoresed at 300 V. Gels were subsequently dried and exposed to X-ray film (Kodak).
Nucleotide sequence accession numbers.
The nucleotide sequences of fragments containing phaR, gap11, and gap20 have been deposited with GenBank under accession no. AF287907, AF442748, and AF442749, respectively.
RESULTS
Identification of phaR, gap11, and gap20.
In a previous study, phaA and phaB were identified as encoding, respectively, β-ketothiolase and NADPH-linked acetoacetyl-CoA reductase on a single 6.1-kb fragment of the M. extorquens AM1 chromosome (23). orf3 was found upstream of phaA (23). The potential polypeptide translated from this gene showed high identity (up to 56%) with proteins translated from regions containing genes for PHB biosynthesis in a number of proteobacteria (3, 11, 12, 36). For rhizobia, this gene has been designated aniA and implicated in a complex regulation of carbon flow in these bacteria (12, 36). Orf3 also showed lower identities with PhaR proteins in P. denitrificans and R. eutropha, which have been implicated elsewhere in negative regulation of GAPs in these organisms (28, 29, 46). Therefore, we selected orf3 for further analysis, as a potential regulator of PHB synthesis, and redesignated it phaR.
GAPs, also called phasins, have been implicated elsewhere in regulating PHB accumulation (18, 22, 37, 46). To identify genes potentially encoding GAPs in M. extorquens AM1, we used the N-terminal amino acid sequences for two GAPs, of 11 and 20 kDa (GA11 and GA20, respectively), previously identified for a similar strain, M. extorquens MB371 (14), as queries to search the genomic database of M. extorquens AM1 (http://vixen.microbiol.washington.edu/). We identified the two respective genes and designated them gap11 and gap20. gap11 and gap20 are not linked on the chromosome to each other or to other genes known to be involved in PHB biosynthesis in M. extorquens AM1. The molecular masses deduced for the polypeptides translated from gap11 and gap20 (12.8 and 19.1 kDa, respectively) were in agreement with the molecular masses experimentally determined for GA20 and GA11 from M. extorquens MB371, and the N-terminal amino acid sequences translated from gap11 and gap20 were identical to the experimentally determined sequences of GA11 and GA20 (13, 14). Gap20 and Gap11 showed high identity to each other (42%), and they revealed similarity with GAPs from Zoogloea ramigera (17) and unknown proteins from PHB-synthesizing α-proteobacteria Rhodopseudomonas palustris, Mesorhizobium loti MAFF303099, and S. meliloti (http://jgi.doe.gov/JGI_microbial/html/index.html).
Gap11, Gap20, and PhaR were inactivated by site-directed mutagenesis to test their role in PHB accumulation and general metabolism of M. extorquens AM1.
Mutations in Gap11 and Gap20 affect PHB accumulation and growth of M. extorquens AM1.
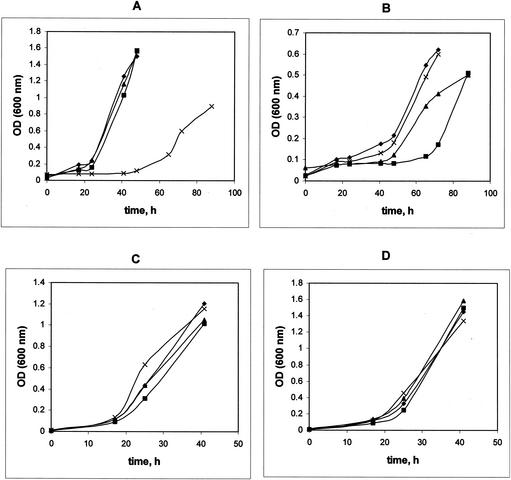
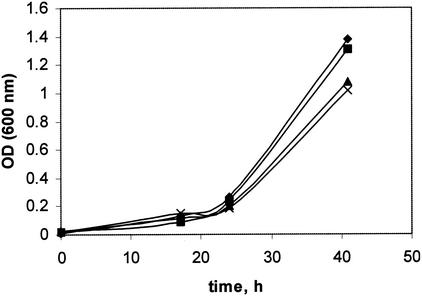
Mutations in Gap11 and Gap20 were generated as described in Materials and Methods, and the resulting mutants were characterized with respect to PHB accumulation, PHB granule content, and growth characteristics. Four types of growth substrates were tested: a C1 substrate (methanol), a C2 substrate (ethylamine), a C3 substrate (pyruvate), and a C4 substrate (succinate). Mutation in Gap11 or Gap20 had no effect on growth of M. extorquens AM1 on the C1, C3, or C4 substrate, while in both cases growth on the C2 substrate was altered (Fig. 1). PHB accumulation was also affected in the mutants, and different patterns were observed depending on the growth substrate (Table 1). Lower PHB levels were determined for both mutants grown on methanol, and the effect was more dramatic in the Gap20 mutant. PHB accumulation dropped by about one-half in cells of both mutants grown on ethylamine, while it remained at wild-type level in cells grown on pyruvate. The effect on PHB accumulation during succinate growth was different for the two mutants. The Gap20 mutant accumulated a reduced amount of PHB compared to the wild-type strain, while in the Gap11 mutant PHB accumulation increased about twofold. These data suggest that the two GAPs in M. extorquens AM1 must have distinct functions in controlling PHB accumulation and these functions must be different in different metabolic circumstances.
FIG. 1.
Growth of M. extorquens AM1 and gap20, gap11, and phaR mutants on methanol (A), ethylamine (B), succinate (C), and pyruvate (D). Symbols: diamond, M. extorquens AM1; square, gap11 mutant; triangle, gap20 mutant; cross, phaR mutant. OD, optical density.
TABLE 1.
PHB content in mutant strains and wild-type M. extorquens AM1 grown on C1, C2, and multicarbon substrates
| Strain | PHB (% of dry biomass [wt/wt])
|
|||
|---|---|---|---|---|
| Methanol | Ethylamine | Pyruvate | Succinate | |
| M. extorquens AM1 | 22-25 | 32-30 | 36-38 | 17-20 |
| gap11 mutant | 9-12 | 11-13 | 34-36 | 30-40 |
| gap20 mutant | 3-5 | 16-18 | 35-37 | 4-6 |
| phaR mutant | 0-0.2 | 26-28 | 1-2 | 0-1 |
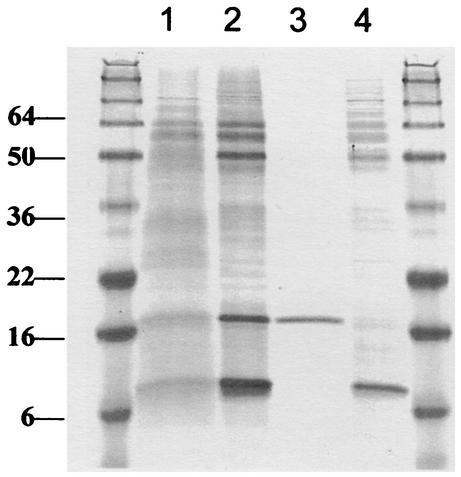
To prove that Gap11 and Gap20 are associated with PHB granules in M. extorquens AM1 and to assess and compare their levels, we analyzed PHB granules from wild-type M. extorquens AM1 and from the Gap11 and Gap20 mutants. Granules were isolated from methanol- and succinate-grown cultures and subjected to electrophoresis in denaturing conditions. Two major proteins are present in wild-type granules, with molecular masses of approximately 11 and 20 kDa (Fig. 2). In the Gap11 mutant, only the 20-kDa band is present, and in the Gap20 mutant, only the 11-kDa band is seen, confirming that the products of gap11 and gap20 are the major GAPs in M. extorquens AM1. As seen in Fig. 2, Gap11 and Gap 20 are present at different levels in the granules, with Gap11 being much more abundant than Gap20. These data were also confirmed by measuring expression levels for gap11 and gap20, employing transcriptional fusions to a reporter gene, xylE, encoding catechol 2,3-dioxygenase (XylE). As seen from Table 2, the difference in XylE activities in M. extorquens AM1 carrying the Pgap11:xylE and Pgap11:xylE fusions is about 60-fold. These data suggest that gap11 and gap20, in addition to being involved in controlling PHB levels, are themselves subjects of regulation. An obvious candidate as a regulator was the phaR gene.
FIG. 2.
SDS-PAGE of the proteins associated with PHB granules isolated from wild-type M. extorquens AM1 and gap11 and gap20 mutants. Lanes 1 and 2, wild type grown on methanol and succinate, respectively; lane 3, gap11 mutant grown on succinate; lane 4, gap 20 mutant grown on succinate. Size markers (molecular masses in kilodaltons) are shown in the flanking lanes.
TABLE 2.
Activities of catechol dioxygenase (milliunits) in wild-type M. extorquens AM1 and the phaR mutant carrying transcriptional fusions, grown on succinate or grown on succinate and induced with methanola
| Fusion | Wild type
|
phaR mutant
|
||
|---|---|---|---|---|
| Succinate | Methanol | Succinate | Methanol | |
| phaA:xylE | 75 | 135 | 50 | 400 |
| phaB:xylE | 75 | 80 | 50 | 350 |
| phaC:xylE | 175 | 330 | 165 | 700 |
| phaR:xylE | 135 | 385 | 150 | 410 |
| gap11:xylE | 1,990 | 1,760 | 1,885 | 1,715 |
| gap20:xylE | 30 | 20 | 25 | 30 |
Background activity of XylE expressed without a promoter is about 1 mU.
Mutation in phaR affects PHB accumulation and growth of M. extorquens AM1.
PhaR has been implicated previously in playing a role in negative regulation of GAPs (phasins) in some bacteria (i.e., P. denitrificans and R. eutropha), while for others (rhizobia) a PhaR homolog, AniA, was shown to play a role in global regulation of carbon flow, and mutation in this gene caused highly pleiotropic effects (11, 12, 36). To test the role of PhaR in M. extorquens AM1, we generated a null mutation in phaR and investigated the mutant phenotype with respect to growth characteristics on various substrates, PHB accumulation during growth on various substrates, and expression of genes involved in PHB biosynthesis. Unlike mutations in the GAPs, the mutation in PhaR had a dramatic effect on growth of M. extorquens AM1 on the C1 compound (Fig. 1A), while growth on the C2, C3, and C4 compounds was not affected (Fig. 1B to D). PHB accumulation was also dramatically affected in the PhaR mutant when grown on the C1, C3, and C4 compounds. However, PHB accumulation on the C2 compound was at wild-type levels (Table 1). These data suggest that PhaR must be involved in controlling PHB biosynthesis during growth on C1, C3, and C4 compounds, but not on C2 compounds. The growth defect on methanol is not due to the decreased PHB synthesis per se, as the GAP mutants both had greatly reduced PHB synthesis but normal growth on methanol. In addition, a mutant has been previously described that is unable to synthesize PHB but which grows normally on methanol (23).
Investigation of PhaR as a transcriptional regulator in M. extorquens AM1.
For P. denitrificans, PhaR was shown to specifically bind to chromosomal regions upstream of phaP encoding a phasin and to phaR and to negatively regulate expression of these genes (29). However, the expression results showed that in M. extorquens AM1 PhaR is transcribed at a high level, comparable to that of major metabolic enzymes (see below), an unusual attribute of a transcriptional regulator. It is worth noting that PhaR polypeptides from M. extorquens AM1 and P. denitrificans share only about 30% identical amino acid residues, while identities with the rhizobial AniA polypeptides are much higher (about 56%). In this work, we used a number of approaches to test if PhaR in M. extorquens AM1 might be a transcriptional regulator.
We analyzed the amino acid sequence translated from phaR for the presence of DNA-binding motifs by using two different programs devoted to helix-turn-helix motif detection (see Materials and Methods). The searches resulted in low scores, predicting that the probability of PhaR being able to bind DNA is low. As controls, we performed the same searches with proteins known to bind DNA, for example, LysR-type regulators from rhizobia, and obtained high scores with the same programs.
To test transcription levels for key genes in PHB biosynthesis, we cloned promoter regions for phaA, phaB, and phaC, encoding the enzymes of PHB biosynthesis, and also promoter regions for gap11, gap20, and phaR upstream of the promoterless xylE gene, and tested the activity of XylE in the wild-type background and in the phaR-minus background (Table 2). Transcription from gap11, gap20, and phaR promoters was at a similar level in wild-type and phaR backgrounds, indicating that phaR has no role in transcriptional regulation of phasins in M. extorquens AM1, or in its own transcriptional regulation. Transcription from phaA, phaB, and phaC promoters was increased two- to fourfold in the phaR background in the methanol-induced cultures, while it was not affected in the succinate-grown cultures. These data were supported by testing the activities of PhaA (β-ketothiolase) and PhaB (acetoacetyl-CoA reductase) in the phaR mutant, and they were somewhat increased after methanol induction (PhaA, 0.52 U; PhaB, 0.05U), but not on succinate (PhaA, 0.71 U; PhaB, 0.03U), compared to the wild type (0.46 and 0.03 versus 1.18 and 0.4 U, respectively).
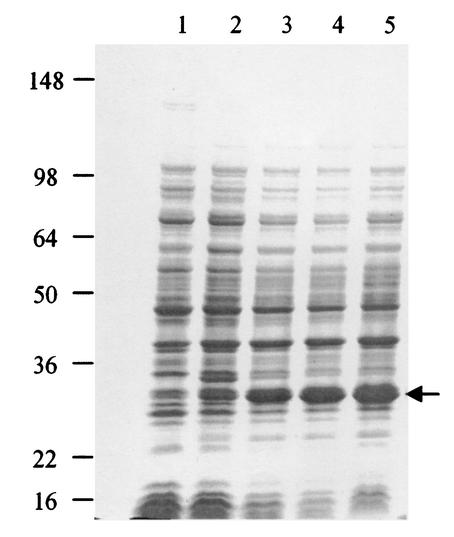
We further tested the ability of PhaR to bind DNA by overexpressing phaR in E. coli (Fig. 3) and performing gel retardation assays with the resulting cell extracts. All the promoter regions tested for promoter activity and listed in Table 2 were employed. However, no retardation was observed with any of the DNA fragments tested (data not shown). These data suggest that, although transcription of some genes is altered in the phaR mutant, PhaR may not act directly as a transcriptional regulator.
FIG. 3.
Expression of PhaR in E. coli BL21(DE3). Proteins were separated by SDS-PAGE. Lane 1, total proteins of E. coli carrying the plasmid with phaR before induction by IPTG; lanes 2 to 5, total cellular proteins of E. coli carrying the plasmid with phaR after 1, 2, 3, and 4 h of induction by IPTG, respectively. The arrow denotes the PhaR band. Numbers at left are molecular masses in kilodaltons.
Role of PhaR in partitioning the flow of acetyl-CoA.
The evidence showing that PhaR is not a transcriptional regulator of phasins suggested that it might play a role more similar to that of AniA. A number of results in the literature point toward AniA, a homolog of PhaR playing a role in directing carbon flow in rhizobial species (12, 36). However, the key metabolite in this regulation or the targets of such a regulation still remain elusive
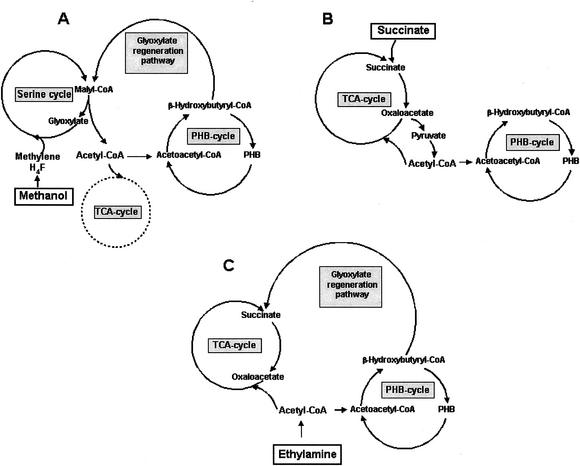
In M. extorquens AM1, the phenotype of the PhaR mutant points towards PhaR having a role in partitioning the flow of acetyl-CoA into the PHB cycle. Figure 4 shows the different sources and fates of acetyl-CoA in M. extorquens AM1 growing on C1, C2, and heterotrophic substrates. Based on the schemes shown in Fig. 4 and the presence of significant PhaA and PhaB activity in the phaR mutant, the defect in PHB accumulation during growth on C1 and C3-C4 compounds in the phaR mutant could be explained if the acetyl-CoA pool is not sufficient for PHB biosynthesis in the absence of PhaR. In that case, the specific growth defect on methanol could be due to low availability of acetyl-CoA for the glyoxylate regeneration cycle, an essential pathway for growth on C1 compounds (8, 23, 24). The long lag and eventual recovery of growth (Fig. 1A) could reflect the time that it takes to build up an effective acetyl-CoA pool to drive C1 metabolism. During growth on C2 compounds, all of the carbon and energy flux passes through acetyl-CoA. This larger, direct flux may bypass the PhaR defect, thereby allowing normal PHB production during growth on C2 compounds. Several lines of evidence were pursued to test this hypothesis.
FIG. 4.
Schematic representation of major pathways producing and consuming acetyl-CoA in M. extorquens AM1 during growth on different substrates. (A) Methanol. The main source of acetyl-CoA is the malyl-CoA lyase reaction (6, 42). The main fate of acetyl-CoA is its conversion to β-hydroxybutyryl-CoA, which is partitioned roughly equally between PHB synthesis and the glyoxylate regeneration pathway (23, 24). Very little acetyl-CoA is oxidized via the TCA cycle during methylotrophic growth. (B) Succinate. The main source of acetyl-CoA is the pyruvate dehydrogenase reaction. The main fate of acetyl-CoA is oxidation through the TCA cycle, with a lesser flux into PHB. (C) Ethylamine. Acetyl-CoA is made directly from the C2 substrate, and acetyl-CoA is partitioned between the TCA cycle and β-hydroxybutyryl-CoA. A significant flux occurs into both PHB and the glyoxylate regeneration cycle, which generates the C4 acceptors for acetyl-CoA oxidation via the TCA cycle (23, 24, 41).
A prediction of this hypothesis is that the PhaR mutant should grow normally on methanol in the presence of supplementing acetate. Figure 5 shows that the phaR mutant does grow normally on methanol in the presence of 0.1 to 0.2 mM acetate, supporting the idea that the PhaR mutant is defective in acetyl-CoA production and/or partitioning. M. extorquens AM1 does not grow on acetate as a sole source of carbon and energy, and so this growth is not due to utilization of the acetate by itself. We further demonstrated that the addition of higher concentrations of acetate (10 to 50 mM) not only complements growth of the phaR mutant on methanol but also restores wild-type levels of PHB production on this substrate, while PHB production on C3 and C4 compounds remained unchanged (data not shown).
FIG. 5.
Growth of M. extorquens AM1 and gap20, gap11, and phaR mutants on methanol in the presence of 0.2 mM acetate. Symbols: diamond, M. extorquens AM1; square, gap11 mutant; triangle, gap20 mutant; cross, phaR mutant. OD, optical density.
The partitioning of acetyl-CoA was also tested directly by measuring 14CO2 production from 2-14C-labeled acetate in whole cells. As shown in Fig. 4, acetyl-CoA has three major fates in M. extorquens AM1, depending on the growth substrate: oxidation by the tricarboxylic acid (TCA) cycle, incorporation into PHB, or conversion to C4 compounds by the glyoxylate regeneration pathway. It has been shown that the C-1 atom of acetyl-CoA is converted to CO2 in the glyoxylate regeneration pathway but the C-2 atom is not (24). The C-2 atom, however, is converted to CO2 in the TCA cycle. Therefore, the production of 14CO2 from 2-14C-labeled acetate is a measure of TCA cycle activity. Incorporation of [2-14C]acetate into CO2 and cells increased linearly with time in M. extorquens AM1 and the PhaR mutant during incubation in the presence of methanol (data not shown). Under these conditions, in M. extorquens AM1 the proportion of the total [2-14C]acetate utilized that was detected in CO2 was less than 0.5%, confirming that the TCA cycle functions at very low activity in the wild-type strain during methylotrophic metabolism (42). In the PhaR mutant, the proportion of 14CO2 production from the total 2-14C-labeled acetate utilized was about 11%, indicating that in this mutant the TCA cycle activity is greatly increased compared to that in the wild type. These results demonstrate the redistribution of acetyl-CoA partitioning in this mutant, with a significant flux being redirected to the TCA cycle.
DISCUSSION
M. extorquens AM1 accumulates PHB during growth on all substrates tested, and the levels of PHB accumulation seem to be determined by the nature of the substrate, suggesting substrate-specific regulatory mechanisms (23). In this work we investigated three genes potentially involved in controlling PHB accumulation in M. extorquens AM1, via mutation and phenotypic analysis: gap11 and gap20, encoding GAPs, and phaR, encoding a putative regulator.
Phasins have been implicated previously in playing an important role in PHB granule formation, as well as in other aspects of PHB accumulation and degradation (15, 18). Several phasins have been isolated and characterized before, from R. eutropha (44), Rhodococcus ruber (34, 35), P. denitrificans (28), Chromatium vinosum (27), and Pseudomonas oleovorans (22, 37). In this work we have identified gap11 and gap20, encoding the two phasins in M. extorquens AM1. The amino acid sequences translated from gap 11 and gap20 reveal low or no identity to phasins characterized from non-Methylobacterium strains, while they show a significant degree of identity to each other. Despite the sequence similarity, however, the two phasins differ in size, are expressed at different levels, and apparently fulfill different roles in PHB synthesis regulation, as mutants with mutations in the two phasins have different phenotypes. Further work will be necessary to determine the mechanism of action of these two phasins.
In this study we concentrated on investigating the role of PhaR in M. extorquens AM1. While in P. denitrificans PhaR was demonstrated elsewhere to be involved in controlling phasin levels as well as its own expression (29), our results indicate that PhaR does not regulate phasins in M. extorquens AM1 at the level of transcription. Instead, our evidence suggests that PhaR is involved in controlling metabolite flows, a role suggested elsewhere for its homolog, AniA in rhizobia (12, 36). The mutation in phaR virtually eliminates PHB production in M. extorquens AM1 during growth on methanol, succinate, and pyruvate and also results in significant growth reduction on methanol. However, a wild-type growth phenotype on methanol and wild-type PHB accumulation on this substrate were restored by the addition of acetate, pointing toward the acetyl-CoA pool as a potential regulation target. This hypothesis is supported by the ability of the phaR mutant to grow and synthesize PHB at wild-type levels on ethylamine, when the supply of acetyl-CoA should not be limited. Finally, our results demonstrating that PhaR mutants redirect the flow of acetyl-CoA from PHB precursors to the TCA cycle indicate a role in regulating acetyl-CoA flux. These results may explain the increase in transcription and enzyme activity observed for the PHB-synthesizing enzymes in the PhaR mutant. This increase was initially puzzling, as it is the opposite trend that would be expected to result in low PHB production. However, if PhaR controls acetyl-CoA partitioning in the cell, the metabolic state of the PhaR mutant would be expected to be very different from that of the wild type and could result in significantly altered cellular signals. This in turn could alter regulation of other enzymes in the cell, including the PHB synthesis enzymes, but would not result in increased PHB synthesis if the acetyl-CoA flux was directed elsewhere. In all PHB-synthesizing bacteria the TCA cycle and the PHB cycle compete for acetyl-CoA (9, 26, 32, 33, 39). Most bacteria accumulate PHB only under conditions when carbon is abundant but oxygen, nitrogen, or sulfate is limited (9, 26, 33, 39). Under these conditions cells possess high ratios of NAD(P)H/NAD(P) that are shown to inhibit citrate synthase and isocitrate dehydrogenase, effectively decreasing the rate of acetyl-CoA metabolism via the TCA cycle. Reduced consumption via the TCA cycle redirects the pool of acetyl-CoA into the PHB cycle, with reoxidation of NAD(P)H. In M. extorquens AM1, the overlap of the PHB biosynthesis pathway with the central metabolic pathways for C1 and C2 assimilation (23, 24) suggests a continuous acetyl-CoA flux via the PHB-synthesizing enzymes β-ketothiolase and acetoacetyl-CoA reductase during C1 and C2 growth. In Methylobacterium rhodesianum, a methylotrophic bacterium very similar to M. extorquens AM1, the affinity of β-ketothiolase (PhaA) for acetyl-CoA was shown previously to be about 20-fold lower than the affinity of citrate synthase (32). However, the flux into the TCA cycle in this bacterium is not significant compared to the flux into both the PHB cycle and the glyoxylate regeneration cycle during growth on methanol. These results imply the presence of specific mechanisms for regulating fluxes of acetyl-CoA in serine cycle methylotrophs, to support the glyoxylate regeneration pathway during methylotrophic growth. Our results show that PhaR is a key player in regulating acetyl-CoA fluxes in M. extorquens AM1.
Although the mechanism by which PhaR acts is not yet known, the evidence so far is not supportive of a direct role in transcriptional regulation. It is possible that PhaR regulates either the acetyl-CoA-producing reactions (malyl-CoA lyase and pyruvate dehydrogenase) or the acetyl-CoA-consuming reactions (β-ketothiolase and citrate synthase), perhaps by altering enzyme activity in vivo. Future work to confirm this hypothesis will require experiments with purified enzymes and PhaR. However, the results presented here suggest that PhaR is a central regulator of carbon flow in this serine cycle methylotroph and assists in redirecting carbon through alternative pathways as the cell switches between different metabolic modes.
Acknowledgments
We thank K. Korotkov for assisting with the preparation of PHB granules, M. Kalyuzhnaya for useful discussions, and H. Rothfuss for help with graphics.
This work was supported by a grant from the NIH (GM58933).
REFERENCES
- 1.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braunegg, G., B. Sonnleitner, and R. M. Lafferty. 1978. A rapid method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur. J. Appl. Microbiol. Biotechnol. 6:29-37. [Google Scholar]
- 3.Cabrera, J. E., G. Panzetta-Dutari, J. L. Pruneda, and S. Genti-Raimondi. 1997. A new Comamonas testosteroni steroid-inducible gene: cloning and sequence analysis. J. Steroid Biochem. Mol. Biol. 63:91-98. [DOI] [PubMed] [Google Scholar]
- 4.Castaneda, M., J. Gusman, S. Moreno, and G. Espin. 2000. The GacS sensor kinase regulates alginate and poly-β-hydroxybutyrate production in Azotobacter vinelandii. J. Bacteriol. 182:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistoserdov, A. Y., L. V. Chistoserdova, W. S. McIntire, and M. E. Lidstrom. 1994. Genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J. Bacteriol. 176:4052-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chistoserdova, L., and M. E. Lidstrom. 1997. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology 143:1729-1736. [DOI] [PubMed] [Google Scholar]
- 7.Chistoserdova, L. V., and M. E. Lidstrom. 1992. Cloning, mutagenesis, and physiological effect of a hydroxypyruvate reductase gene from Methylobacterium extorquens AM1. J. Bacteriol. 174:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chistoserdova, L. V., and M. E. Lidstrom. 1996. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquens AM1. Microbiology 142:1459-1468. [DOI] [PubMed] [Google Scholar]
- 9.Chohan, S. N., and L. Copeland. 1998. Acetoacetyl coenzyme A reductase and polyhydroxybutyrate synthesis in Rhizobium (Cicer) sp. Strain CC 1192. Appl. Environ. Microbiol. 64:2859-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ditta, G., T. Schmidhauser, F. Yakobson, P. Lu, X. Liang, D. Finlay, D. Guiney, and D. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, M. F., G. Araiza, S. Encarnacion, M. del Carmen Vargas, and J. Mora. 2002. Effect of aniA (carbon flux regulator) and phaC (poly-β-hydroxybutyrate synthase) mutations on pyruvate metabolism in Rhizobium etli. J. Bacteriol. 184:2296-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Encarnacion, S., M. del Carmen Vargas, M. F. Dunn, A. Davalos, G. Mendoza, Y. Mora, and J. Mora. 2002. AniA regulates reserve polymer accumulation and global protein expression in Rhizobium etli. J. Bacteriol. 184:2287-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Föllner, C. G., W. Babel, and A. Steinbüchel. 1995. Isolation and purification of granule-associated proteins relevant for poly(3-hydroxybutyric acid) biosynthesis from methylotrophic bacteria relying on the serine pathway. Can. J. Microbiol. 41(Suppl. 1):124-130. [Google Scholar]
- 14.Föllner, C. G., M. Madkour, F. Mayer, W. Babel, and A. Steinbüchel. 1997. Analysis of the PHA granule-associated proteins GA20 and GA11 in Methylobacterium extorquens and Methylobacterium rhodesianum. J. Basic Microbiol. 37:11-21. [DOI] [PubMed] [Google Scholar]
- 15.Hanley, S. Z., D. J. Pappin, D. Rahman, A. J. White, K. M. Elborough, and A. R. Slabas. 1999. Re-evaluation of the primary structure of Ralstonia eutropha phasin and implications for polyhydroxyalkanoic acid granule binding. FEBS Lett. 447:99-105. [DOI] [PubMed] [Google Scholar]
- 16.Harder, W., M. Attwood, and J. R. Quayle. 1973. Methanol assimilation by Hyphomicrobium spp. J. Gen. Microbiol. 78:155-163. [Google Scholar]
- 17.Inagawa, Y., Y. Inoue, M. Shiraki, and T. Saito. 2002. Identification and characterization of poly-3-hydroxybutyrate granule-associated proteins, PGA12 and PGA16 in Zoogloea ramigera I-16-M. Int. J. Biol. Macromol. 30:55-61. [DOI] [PubMed] [Google Scholar]
- 18.Jossek, R., R. Reichelt, and A. Steinbuchel. 1998. In vitro biosynthesis of poly(3-hydroxybutyric acid) by using purified poly(hydroxyalkanoic acid) synthase of Chromatium vinosum. Appl. Microbiol. Biotechnol. 49:258-266. [DOI] [PubMed] [Google Scholar]
- 19.Jurasek, L., and R. H. Marchessault. 2002. The role of phasins in the morphogenesis of poly(3-hydroxybutyrate) granules. Biomacromolecules 3:256-261. [DOI] [PubMed] [Google Scholar]
- 20.Kalb, V. F., and R. W. Bernlohr. 1977. A new spectrophotometric assay for protein in cell extracts. Anal. Biochem. 82:362-371. [DOI] [PubMed] [Google Scholar]
- 21.Kataeva, I. A., and L. A. Golovleva. 1990. Catechol 2,3-dioxygenase from Pseudomonas aeruginosa 2x. Methods Enzymol. 188:115-121. [DOI] [PubMed] [Google Scholar]
- 22.Klinke, S., G. de Roo, B. Witholt, and B. Kessler. 2000. Role of phaD in accumulation of medium-chain-length poly(3-hydroxyalkanoates) in Pseudomonas oleovorans. Appl. Environ. Microbiol. 66:3705-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korotkova, N., and M. E. Lidstrom. 2001. A connection between poly-β-hydroxybutyrate biosynthesis and growth on C1 and C2 compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 183:1038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korotkova, N., L. Chistoserdova, V. Kuksa, and M. E. Lidstrom. 2002. The glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 184:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lee, Y., M. K. Kim, H. N. Chang, and Y. H. Park. 1995. Regulation of poly-β-hydroxybutyrate biosynthesis by nicotinamide nucleotide in Alcaligenes eutrophus. FEMS Microbiol. Lett. 131:35-39. [Google Scholar]
- 27.Liebergesell, M., B. Schmidt, and A. Steinbüchel. 1992. Isolation and identification of granule-associated proteins relevant for poly(3-hydroxyalkanoic acid) biosynthesis in Chromatium vinosum D. FEMS Microbiol. Lett. 99:227-232. [DOI] [PubMed] [Google Scholar]
- 28.Maehara, A., S. Ueda, H. Nakano, and T. Yamane. 1999. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J. Bacteriol. 181:2914-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maehara, A., S. Taguchi, T. Nishiyama, Y. Takagi, T. Yamane, and Y. Doi. 2002. A repressor protein, PhaR, regulates polyhydroxyalkanoate (PHA) synthesis via its direct interaction with PHA. J. Bacteriol. 184:3992-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 32.Mothes, G., I. S. Rivera, and W. Babel. 1997. Competition between β-ketothiolase and citrate synthase during poly (β-hydroxybutyrate) synthesis in Methylobacterium rhodesianum. Arch. Microbiol. 166:405-410. [DOI] [PubMed] [Google Scholar]
- 33.Park, J. S., and Y. H. Lee. 1996. Metabolic characteristics of isocitrate dehydrogenase leaky mutant of Alcaligenes eutrophus and its utilization of poly-β-hydroxybutyrate production. J. Ferment. Bioeng. 81:197-205. [Google Scholar]
- 34.Pieper-Fürst, U., M. H. Madkour, F. Mayer, and A. Steinbüchel. 1994. Purification and characterization of 14-kilodalton protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J. Bacteriol. 176:4328-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieper-Fürst, U., M. H. Madkour, F. Mayer, and A. Steinbüchel. 1995. Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J. Bacteriol. 177:2513-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Povolo, S., and S. Casella. 2000. A critical role for aniA in energy-carbon flux and symbiotic nitrogen fixation in Sinorhizobium meliloti. Arch. Microbiol. 174:42-49. [DOI] [PubMed] [Google Scholar]
- 37.Prieto, M. A., B. Bühler, K. Jung, B. Witholt, and B. Kessler. 1999. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GRo1 involved in the regulatory expression system for pha genes. J. Bacteriol. 181:858-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito, H., and K.-I. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 39.Senior, P. J., and E. A. Dawes. 1973. The regulation of poly-3-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem. J. 134:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon, R., U. Priefer, and A. Puhler. 1983. Vector plasmids for in vivo manipulations of Gram-negative bacteria, p. 98-106. In A. Puhler (ed.), Molecular genetics of the bacteria-plant interactions. Springer-Verlag, Berlin, Germany.
- 41.Taylor, I. J., and C. Anthony. 1976. Acetyl-CoA production and utilization during growth of the facultative methylotroph Pseudomonas AM1 on ethanol, malonate and 3-hydroxybutyrate. J. Gen. Microbiol. 95:134-143. [DOI] [PubMed] [Google Scholar]
- 42.Van Dien, S. J., and M. E. Lidstrom. 2002. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C(3) and C(4) metabolism. Biotechnol. Bioeng. 78:296-312. [DOI] [PubMed] [Google Scholar]
- 43.Whitaker, J. R., and P. E. Granum. 1980. An absolute method for protein determination based on the difference in absorbance at 235 and 280 nm. Anal. Biochem. 109:156-159. [DOI] [PubMed] [Google Scholar]
- 44.Wieczorec, R., A. Pries, A. Steinbüchel, and F. Mayer. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, G., A. J. G. Moir, G. Sawers, S. Hill, and R. K. Poole. 2001. Biosynthesis of poly-β-hydroxybutyrate (PHB) is controlled by CydR (Fnr) in the obligate aerobe Azotobacter vinelandii. FEMS Microbiol. Lett. 194:215-220. [DOI] [PubMed] [Google Scholar]
- 46.York, G. M., J. Stubbe, and A. J. Sinskey. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]