Abstract
The 3′ ends of the genes for the C-terminal region of C5a peptidase from 15 Streptococcus pyogenes isolates were analyzed by PCR. Amplicons were found to differ in size. DNA sequence analysis revealed that the differences between PCR fragment sizes accorded with the number of R repeats in the C5a peptidase gene.
Streptococcus pyogenes (group A streptococci [GAS]) is the cause of a number of contagious human diseases, including pharyngitis, skin infections, and scarlet fever, as well as generalized infections like necrotizing fasciitis and toxic shock syndrome. It is well documented that GAS, together with group B streptococci (GBS) and group G streptococci, express on their surfaces streptococcal C5a peptidase (SCP) (1, 2, 3, 6). This enzyme is related to serine proteases and specifically cleaves the C5a complement component (3, 7), resulting in the inactivation of C5a and the destruction of its ability to act as an anaphilatoxin and a strong chemoattractant (11, 12, 13, 14, 16). Thus, SCP is considered an important virulence factor of pathogenic streptococci. However, virulent gene heterogeneity, which usually reflects the degree of host response, has so far not been demonstrated for SCP genes from different GAS strains (scpA). Moreover, scpA has been reported to be highly homologous to SCP genes from GBS (scpB) (4, 5); the genes were found to differ only by 51 bp. This region was characterized with so-called R repeats that encode the C-terminal part of SCP (3, 4).
In order to investigate the degree of scpA heterogeneity, several GAS isolates belonging to different M types and ribotypes were selected for the present study. SCP genes from GAS were analyzed by PCR and sequence analysis.
Bacterial strains and DNA techniques.
GAS strains were isolated from patients. GAS strain SF370 serotype M1 and GBS strain 090R serotype Ia were obtained from the collection of the University of Oklahoma. Sera from the PHLS Respiratory and Systemic Infection Laboratory (Colindale, United Kingdom) were used to perform serotyping.
Chromosomal DNAs from GAS and GBS strains were extracted with phenol-chloroform and ethanol precipitation according to standard methods (15). Ribotyping of the strains was carried out by digestion with PvuII and EcoRI (P and E ribotypes) and by using the rrs gene, labeled with digoxigenin, as a 16S rRNA probe (Roche, Mannheim, Germany). The sizes of DNA fragments were estimated with the computer program SEQAID.
Chromosomal DNA was used as a template for amplification by PCR (9, 20). The 5′-ACAATGGAAGGCTCTACTGTTC-3′ (forward) and 5′-ACCTGGTGTTTGACCTGAACTA-3′ (reverse) primers corresponded to the 3′ end of the SCP gene.
Sequencing was accomplished by the sequencing facility of the Microbiology Department of the University of Oklahoma. Computer analysis of DNA and amino acid sequences was performed with the Genetics Computer Group sequence analysis package (program manual for the Wisconsin Package, version 8). The analysis of the amino acid sequence for the region rich in proline, glutamic acid, serine, and threonine (PEST-positive region) was conducted with the PEST FIND program (PC/Gene).
Amplification of SCP genes.
The 3′ ends of scpA genes from 15 GAS strains and that of scpB from one GBS (090R) strain were amplified by PCR. All GAS examined revealed four different amplicon sizes that corresponded to the C-terminal partof SCP. The amplicon size of scpB was found to be 255 bp, as previously reported (8). The amplicon sizes of scpA genes ranged from 255 to 396 bp (Table 1). The amplicon sizes of scpA genes from strains of similar M types and different E ribotypes (strains M1, M6, and M11) were different. On the other hand, the amplicon sizes of scpA genes for strains of similar M types belonging to similar P and E ribotypes were identical (strains M83 and M89). The scpA amplicon sizes of 306 and 357 bp were found to be the most common. One strain (M68) among all GAS strains examined produced the 396-bp amplicon fragment. The amplified region of scpA from strain M5 was the smallest of all and equal in length to the 255-bp scpB fragment.
TABLE 1.
Sizes of SCP genes amplified by PCR and PEST analysis of the C-terminal parts of their products
| Straina (isolate no.) | 16S rRNA gene profileb
|
Size of amplicon fragment (bp) | No. of PEST-positive sequences with a PEST score of:
|
||
|---|---|---|---|---|---|
| PvuII | EcoRI | 9.91c | 4.75d | ||
| 090R | NA | NA | 255 | 2 | 0 |
| M1 (SF370) | NA | NA | 357 | 2 | 1 |
| M1 (11) | P3 | E3 | 306 | ||
| M5 (8) | P5 | E4 | 255 | 0 | 0 |
| M6 (4) | P1 | E1 | 357 | ||
| M6 (18) | P1 | E2 | 306 | 1 | 1 |
| M11 (7) | P1 | E5 | 357 | ||
| M11 (19) | P1 | E1 | 306 | ||
| M22 (12) | P3 | E3 | 357 | ||
| M64 (13) | P3 | E3 | 306 | ||
| M68 (24) | P1 | E2 | 396 | 0 | 0 |
| M83 (14) | P3 | E3 | 357 | 1 | 2 |
| M83 (20) | P3 | E3 | 357 | ||
| M87 (15) | P3 | E3 | 357 | ||
| M89 (2) | P2 | E2 | 357 | ||
| M89 (10) | P2 | E2 | 357 | ||
All strains are GAS with the exception of strain 090R, which is a GBS.
NA, not analyzed.
Amino acid sequence, PEQDGSGQTPD.
Amino acid sequence, PEQDGSGQAPD.
DNA sequencing and sequence analysis.
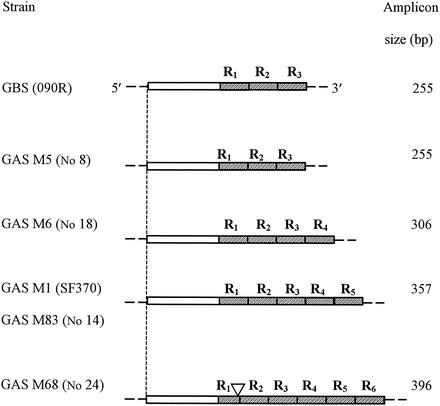
Amplified fragments of scpAs from GAS of types M1 (isolate SF370), M5 (isolate 8), M6 (isolate 18), M68 (isolate 24), and M83 (isolate 14) (scpA1, scpA5, scpA6, scpA68, and scpA83, respectively) were cloned in Escherichia coli (DHIB) with subsequent sequence analysis. DNA sequence analysis revealed the highest degree of homology between scpA83 and scpA1 (97%) and scpA5 and scpB (90%), which accorded with their similarity in amplicon size. A comparison of analyzed fragments of scpBs and scpAs from five GAS strains is schematically presented in Fig. 1. As shown in Fig. 1, all scpA genes examined differed from each other by 51-bp sequences, which resulted in various numbers of R repeats corresponding to 17 amino acids. Only one R repeat in scpA68 contained a 12-bp deletion.
FIG. 1.
Comparison of the 3′ ends of C5a peptidase genes. DNA sequence analysis revealed that SCP genes from GBS strain 090R and GAS strains M1 (isolate SF370), M5 (isolate 8), M6 (isolate 18), M68 (isolate 24), and M83 (isolate 14) differed from each other in their numbers of 51-bp R repeats. Symbols: shaded box, R repeats; ▿, 12-bp deletion.
It was previously reported that the proline-rich C terminus of SCP carried several PEST-positive regions (4). The PEST regions were found to be sensitive to proteolysis, which led to rapid destruction of the proteins (17, 18, 19). The data from a deduced amino acid analysis of the C-terminal part of SCPs from GAS strains revealed significant differences in PEST scores (Table 1). SCPs from different isolates either did not comprise any PEST-positive regions (strains M5 and M68) or comprised several regions with a significant PEST score. Every 17-amino-acid repeat sequence except the last one contained a 6-residue motif (PDKKPE). It has been reported that the lysine pairs (labeled “KK”) in the sequence of some proteins might play a role in proteolysis (10). The 6-residue sequence in the proline-rich region of SCP may also serve as a target for proteolysis.
Heterogeneity demonstrated in the scpA region encoding the C terminus of SCP can be explained by selective pressure from the host to delete the protease-sensitive regions from the surface-expressed proteins. This may be important when the bacteria (like GBS) have a tendency for long-lasting colonization in the protease-rich milieu. Another explanation might reflect the fact that the cleavage sites for eukaryotic proteases can be important for the secretion of SCP from the cell surface.
Acknowledgments
We are grateful to J. Ferretti for valuable help in DNA sequencing. We also thank A. Dmitriev for helpful discussions.
This work was supported by Public Health Service grant AI19304, NIH grant TW00188, and RFFI grant 00-04-49360a.
REFERENCES
- 1.Bohnsack, J. F., K. W. Mollison, A. M. Buko, J. C. Ashworth, and R. H. Hill. 1991. Group B streptococci inactivate C5a by enzymatic cleavage at the carboxy terminus. Biochem. J. 273:635-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohnsack, J. F., X. Zhou, P. A. Williams, P. P. Cleary, C. J. Parker, and H. R. Hill. 1991. Purification of the proteinase from group B streptococci that inactivates human C5a. Biochim. Biophys. Acta 1079:222-228. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. C., and P. P. Cleary. 1990. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J. Biol. Chem. 265:3161-3167. [PubMed] [Google Scholar]
- 4.Chmouryguina, I., A. Suvorov, P. Ferrieri, and P. P. Cleary. 1996. Conservation of the C5a peptidase genes in group A and B streptococci. Infect. Immun. 64:2387-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleary, P. P., J. Handley, A. N. Suvorov, A. Podbielski, and P. Ferrieri. 1992. Similarity between the group B and A streptococcal C5a peptidase genes. Infect. Immun. 60:4239-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary, P. P., J. Peterson, C. Chen, and C. Nelson. 1991. Virulent human strains of group G streptococci express a C5a peptidase enzyme similar to that produced by group A streptococci. Infect. Immun. 59:2305-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary, P. P., U. Prahbu, J. B. Dale, D. E. Wexler, and J. Handley. 1992. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect. Immun. 60:5219-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dmitriev, A., L. Tkáèiková, A. Suvorov, M. Kantíková, I. Mikula, and A. Totolian. 1999. Comparative genetic study of group B streptococcal strains of human and bovine origin. Folia Microbiol. 44:449-453. [DOI] [PubMed] [Google Scholar]
- 9.Erlich, H. A. 1991. Recent advances in the polymerase chain reaction. Science 252:1643-1651. [DOI] [PubMed] [Google Scholar]
- 10.Gomes, A. V., and J. A. Barnes. 1995. Pest sequences in EF-hand calcium-binding proteins. Biochem. Mol. Biol. Int. 37:853-860. [PubMed] [Google Scholar]
- 11.Hill, H. R., J. F. Bohnsack, E. Z. Morris, N. H. Augustine, C. J. Parker, P. P. Cleary, and J. T. Wu. 1988. Group B streptococci inhibit the chemotactic activity of the fifth component of complement. J. Immunol. 141:3551-3556. [PubMed] [Google Scholar]
- 12.Ji, Y., B. Carlson, A. Kondagunta, and P. P. Cleary. 1997. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect. Immun. 65:2080-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji, Y., L. McLandsborough, A. Kondagunta, and P. P. Cleary. 1996. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect. Immun. 64:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji, Y., N. Schnitzler, E. DeMaster, and P. Cleary. 1998. Impact of M49, Mrp, Enn, and C5a peptidase proteins on colonization of the mouse oral mucosa by Streptococcus pyogenes. Infect. Immun. 66:5399-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.O'Connor, S. P., and P. Cleary. 1987. In vivo Streptococcus pyogenes C5a peptidase activity: analysis using transposon and nitrosoguanidine-induced mutants. J. Infect. Dis. 156: 495-504. [DOI] [PubMed] [Google Scholar]
- 17.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 18.Rechsteiner, M. 1990. PEST sequences are signals for rapid intracellular proteolysis. Semin. Cell Biol. 1:433-440. [PubMed] [Google Scholar]
- 19.Roger, S., R. Wells, and M. Rechsteiner. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234:364-369. [DOI] [PubMed] [Google Scholar]
- 20.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]