Abstract
We analyzed expression of a putative oligopeptide permease (Opp) of Borrelia burgdorferi. Unlike the opp operons of other bacteria for which there is a single substrate binding protein, B. burgdorferi codes for three substrate binding proteins (OppA-I to -III) in its opp operon and an additional two homologs on plasmids (OppA-IV and -V). Instead of a single promoter region regulating transcription of the entire operon, as seen in other bacterial opp operons, it appears that among oppA-I, -II, and -III, as well as oppA-IV and -V, each has a potential upstream promoter region. We tested the function of these putative promoter sequences by fusion to a promoterless β-galactosidase reporter gene in pCB182. Each of the promoter regions was found to be active. The level of activity in the reporter constructs closely paralleled the level of expression of each gene in in vitro-grown B. burgdorferi. Changes in carbon and nitrogen availability differentially affected individual promoters, but no changes in promoter activity were seen when Escherichia coli bacteria (with the promoter constructs) were grown in various concentrations of phosphate and leucine and changes in pH. Expression of specific oppA genes with B. burgdorferi varied significantly between its mouse and fed and unfed tick hosts. Differences in regulation of opp gene expression suggest a potential role in environmental response by the organism.
Borrelia burgdorferi is the causative organism of Lyme disease. One of the striking observations that has been made about B. burgdorferi in the postgenomic era is that of its apparent lack of synthetic machinery. B. burgdorferi does not appear to possess genes for the synthesis of amino acids, fatty acids, and other essential elements (5). As a result, the organism is dependent upon its environment to supply these essential nutrients. Other bacteria typically possess multiple peptide transport systems with different specificities to facilitate the utilization of peptides as a source for amino acids (17). The B. burgdorferi genome appears to code for a single peptide transport system only. This peptide transport system shows a high degree of similarity to both the oligopeptide permease (Opp) system and the dipeptide permease (Dpp) system of Escherichia coli and Salmonella enterica serovar Typhimurium. All of these systems encode a peptide binding protein (OppA or DppA), two transmembrane proteins (Opp or DppB and -C) and two ATP binding proteins (Opp or DppD and -F). The B. burgdorferi opp operon, however, differs in that it encodes two additional OppA-like proteins (OppA-II and OppA-III). While the B. burgdorferi opp operon is chromosomally encoded, at least two other OppA orthologues have been identified on borrelial plasmids (oppA-IV on cp26 and oppA-V on lp-54). In previous studies using a complementation system with the E. coli integral membrane proteins OppB and OppC, the ability of the borrelial OppA orthologues to bind and facilitate transport of peptides was confirmed (10).
The natural life cycle of B. burgdorferi requires it to adapt to wide variations in environmental conditions as it moves between its tick and mammalian hosts. Peptide transporters have been shown to play an important role in a diverse array of environmentally relevant functions, including nutrient acquisition, chemotaxis, quorum sensing, and conjugation (1, 8, 9, 11). The specificity in these functions typically derives from the specificity of the substrate binding protein. Whereas E. coli DppA appears to have fairly narrow substrate specificity, OppA in other bacteria has been shown to have very broad substrate specificity (6, 14-16, 20, 21). In comparison to E. coli OppA, the B. burgdorferi OppA proteins appear to have overlapping but narrower specificities, allowing for the potential of separate roles for each OppA protein (10). We were interested in the mechanisms of regulation and expression of the B. burgdorferi OppA proteins and whether they could be differentially expressed under changing environmental conditions. In this study, we examine the activities of the promoter regions of the B. burgdorferi opp genes and assess the impact of different environmental factors on gene expression.
MATERIALS AND METHODS
Bacterial strains and media.
Borrelia burgdorferi, strain N40, was grown at 37°C or 25°C in Barbour-Stoenner-Kelly (BSK) H medium (Sigma, St. Louis, Mo.) as previously described (7). E. coli bacteria of strain Top10 (Invitrogen) were employed as host cells for the expression of β-galactosidase genes from reporter plasmids.
Plasmid constructions.
Five oppA upstream regions were individually cloned from B. burgdorferi B31 DNA through PCR using specific primers with BamHI and BglII restriction sites. The primers, URA1F (5′-TCGGATCCTAAAACCAGCACCCT-3′) and URA1R (5′-TCAGATCTTGGGATTTTCCTTTT-3′), URA2F (5′-TCGGATCCATAATAAAATTAAGT-3′) and URA2R (5′-TCAGATCTTAATTTTTTATACCT-3′), URA3F (5′-TCGGATCCATAATATAATAAACT-3′) and URA3R (5′-TCAGATCTTATTAACCTTTCCCC-3′), URA4F (5′-TCGGATCCTATTCCTCTCCTTTGA-3′) and URA4R (5′-TCAGATCTTACAAGCATCCTTACA-3′) and URA5F (5′-TCGGATCCTTTGGAGGCGATTTT-3′) and URA5R (5′-TCAGATCTATAGGAATACTGGAA-3′), were used for cloning the upstream regions of oppA-I, -II, -III, IV, and -V, respectively. The sequences underlined represent the restriction sites for BamHI or BglII. PCR was performed with the following cycling parameters: 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final extension cycle of 72°C for 7 min. The DNA fragments obtained from PCR were ligated into pCR2.1 vector (Invitrogen) for further amplification. After digestion by BamHI and BglII, the DNA fragments were recovered using a Qiaquick gel extraction kit (Qiagen) and ligated to the promoter probe vector pCB182 or pCB192 (18) at BamHI and BglII sites. The recombinant plasmids were transformed into Top10 by electroporation (Electroporator 2510; Eppendorf). A similar strategy was used for construction of deletions in oppA upstream regions to identify the DNA fragment with promoter function. All DNA sequences inserted into the promoter-probe vectors were confirmed by DNA sequencing. Plasmid DNA was purified using Qiaprep mini spin columns (Qiagen). All restriction enzymes and primers were purchased from Invitrogen unless otherwise stated.
β-Galactosidase assays.
Bacterial cultures were grown to mid-logarithmic phase under various environmental and nutritional conditions. Growth curves for the bacteria were established experimentally prior to the experiments to ensure that bacteria were collected at similar stages of growth. Bacteria in logarithmic phase were centrifuged, washed and resuspended in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol [pH 7.0]), and incubated on ice (12). After absorbance was recorded at a wavelength of 600 nm, 10 μl of 1% sodium dodecyl sulfate and 20 μl of chloroform were added in each 0.8-ml sample. The samples were vortexed for 15 s and then placed at 30°C for 15 min. o-Nitrophenyl-β-d-galactopyranoside (ONPG) (160 μl of 4 mg/ml, dissolved in 0.1 M sodium phosphate buffer [pH 7.0]) was added and mixed vigorously for 10 s. The samples were incubated at 30°C for 15 to 30 min. 1 M Sodium carbonate (400 μl) was added to quench the reaction. The samples were clarified by centrifugation. β-Galactosidase activity was monitored by measuring the optical density at 420 nm (OD420) and OD550. Enzyme activity was calculated based on the following equation: U = 1,000 × [OD420 − (1.75 × OD550)]/[time × vol × OD600]. One unit of enzyme activity was defined as the ability to hydrolyze 1 μmol of ONPG per min (12). Specific activity was expressed as units per absorbance unit of the culture at a wavelength of 600 nm.
RNase protection assay.
An RNase protection assay (RPA) was employed as a tool to quantitatively detect the target RNA transcribed by each oppA gene in B. burgdorferi. To generate linear DNA templates with the core T7 promoter sequence for the preparation of RNA probes, the oligonucleotide primers listed in Table 1 were separately used to specifically amplify oppA gene fragments from each cloned oppA gene by PCR. Polymerase chain reactions were conducted under the same conditions as described above. The desired DNA fragments, after being amplified from each oppA gene, were separated by 1.5% agarose gel electrophoresis and extracted using a Qiaquick gel extraction kit according to the manufacturer's instructions.
TABLE 1.
Sequences of primers used in this study
| Primer | Sequence | Location on oppA gene |
|---|---|---|
| A1F | 5′-ATGAAATATATAAAAA-3′ | oppAI (1-16 bp) |
| T7A1R | 5′-TAATACGACTCACTATAGGGTTGCTACCGTAAAGG-3′ | oppAI (141-156 bp) |
| A2F | 5′-ATGAAATTACAAAGGTC-3′ | oppAII (1-17 bp) |
| T7A2R | 5′-TAATACGACTCACTATAGGGCGACATTACCTCTGCT-3′ | oppAII (135-152 bp) |
| A3F | 5′-ATGAGCTTTAATAAAACTA-3′ | oppAIII (1-19 bp) |
| T7A3R | 5′-TAATACGACTCACTATAGGTATTGTCTCATCTACCA-3′ | oppAIII (160-177 bp) |
| A4F | 5′-GTATTATTTCTCAATTTAAT-3′ | oppAIV (31-50 bp) |
| T7A4R | 5′-TAATACGACTCACTATAGGGTCCTGGGATCTCCATCT-3′ | oppAIV (197-215 bp) |
| A5F | 5′-ATGATAATAAAAAAAAG-3′ | oppAV (1-17 bp) |
| T7A5R | 5′-TAATACGACTCACTATAGGGCCTATCGCTGCAAAA-3′ | oppAV (136-151 bp) |
To create a specific RNA probe, the linear PCR product containing the core T7 promoter sequence was used as the template DNA. Biotin-labeled RNA probes were individually synthesized with a MAXIscript T7 kit (Ambion), separated on urea-6% polyacrylamide gel, and then eluted into 350 μl of DEPC (diethylpyrocarbonate)-treated H2O at 37°C overnight.
RNA was extracted from B. burgdorferi, using Trizol (Invitrogen) according to the manufacturer's instructions. The amount of RNA probe required to completely hybridize with the target RNA molecules in total RNA and the optimal hybridization temperatures were determined experimentally. Threefold molar excesses of each probe over target mRNA were used to ensure that every target RNA transcript was hybridized. Hybridized RNA was digested with RNase A-RNase T1 at 37°C for 30 min and inactivated according to the manufacturer's instructions (MAXIscript; Ambion). The RNA samples were separated on a 6% polyacrylamide gel containing urea and transferred to a positively charged nylon membrane (Osmonics). Biotinylated RNA probes protected from RNase digestion by hybridizing to cRNA were detected chemiluminescently, using a BrightStar BioDetect nonisotopic detection kit (Ambion). Quantitation was performed by densitometric scanning (KS1D apparatus; Kodak) of exposed X-ray film.
Preparation of tick RNA.
Ixodes dammini ticks were obtained from a laboratory colony derived from an Ipswich, Mass., population that has been determined to be free of inherited spirochetal infection. Outbred C3H mice are infected by nymphs infected by strain N40 (wild type), which is maintained in alternating tick-mouse-tick passages. Larvae are allowed to feed to repletion 3 weeks after the infected nymphs engorge. Upon repletion, engorged larvae are collected and permitted to molt to the nymphal stage at 21°C and 95% relative humidity. Nymphal ticks were fed on uninfected mice for 60 h prior to removal.
Fed and unfed nymphal ticks were pooled into groups of 5 to 10 ticks, suspended in Trizol (Invitrogen), and homogenized for 30 s using a rotor-stator homogenizer. RNA was purified according to the manufacturer's instructions.
Preparation of mouse RNA.
C3H/HeN mice (Jackson Laboratories) were infected subcutaneously with 104 B. burgdorferi (cN40) bacteria. Mice were sacrificed at 2 weeks postinfection, and hearts were snap frozen in RNAlater (Ambion) until use. RNA was prepared using Trizol according to the manufacturer's instructions.
Reverse transcriptase PCR (RT PCR).
Total RNA from B. burgdorferi ticks or mouse tissue was heated to 95°C for 10 min and then chilled. Samples were then treated with RNase-free DNase I (Ambion) at 37°C for 15 min. First-strand cDNA synthesis was performed, using SuperScript (Invitrogen) with random hexamer primers or gene-specific primers, according to the manufacturer's instructions. The generated cDNA was used as a template for real-time PCR amplification (ABI 7700; Applied Biosystems), using SYBR green fluorescent dye (SYBR Green Master Mix; Applied Biosystems) and specific primers for each oppA gene. Cycling parameters were 50°C for 5 min, 95°C for 10 min, followed by 40 cycles of 95°C for 30s, and 55°C for 1 min. Specific primers RTA1F (5′-CTTTAACTAAAGTAGTTTTAAAGGGAAGTTCAGAT-3′) and RTA1R (5′-ACATTTGGTAATTTCCAGTTCTTCTGCTTCCTAGGAA-3′) for oppA-I, RTA2F (5′-GGCTCAAAGTACGTTGAAATGGTTAAATCGGTA-3′) and RTA2R (5′-TAATAGTCGCTTCTTAATTTTAGATTTTTGATTAG-3′) for oppA-II, RTA3F (5′-TGTTCTTACCAACAGCAGAAATACTGGCA-3′) and RTA3R (5′-TAATTCAGAAAGATAATAAACCTCTGATACA-3′) for oppA-III, RTA4F (5′-GTCACAGATAATACCATTACAGCTTA-3′) and RTA4R (5′-TAGAAGAAGTTATATTAAGAAATACA-3′) for oppA-IV, and RTA5F (5′-AAATATTTTAAAAGGTCAATATGAGATCTCAGTAAGGTC-3′) and RTA5R (5′-GTTTCCCGCAACACTGTATATTGGAATTATT-3′) for oppA-V were designed with the program Primer3, which was developed by Steve Rozen and Helen J. Skaletsky. All primer pairs were tested for specificity using templates for each of the other oppA genes. None of the primer pairs showed any cross-reactivity with other opp genes and all showed only a single band with B. burgdorferi DNA. Sequences of the products were confirmed to correspond with the predicted sequence by DNA sequencing. Calculations of relative expression of the gene of interest were normalized to recA gene expression with the ΔΔCt method, in which the amount of target (normalized to an endogenous reference and relative to a calibrator) is given by 2− ΔΔCt, where Ct is the cycle number of the detection threshold. Calculations of absolute copy number per sample were calculated from standard curves generated with each individual primer set.
RESULTS
Analysis of B. burgdorferi opp promoters.
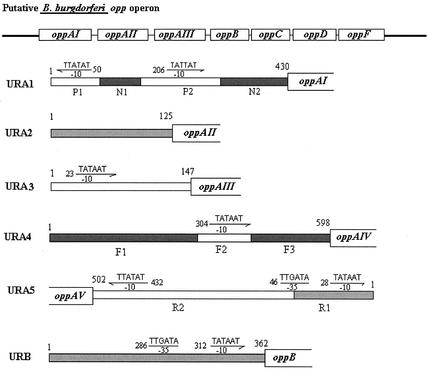
The best-understood OPPs are those of E. coli and S. enterica serovar Typhimurium. Opp proteins in the opp operons of these bacteria appear to be under the regulation of a single promoter region which precedes the oppA gene, with little to no separation between the coding sequences. In contrast, the B. burgdorferi opp operon contains intergenic regions of sufficient size to potentially allow separate regulation of the three chromosomally encoded oppA genes and another potential promoter region upstream of the oppB, oppC, oppD, and oppF coding sequences. In addition, both plasmid-encoded OppA orthologues have upstream sequences that could potentially serve as promoter regions. In the chromosomally encoded opp operon of B. burgdorferi, three oppA genes (oppA-I, oppA-II, and oppA-III) are individually separated by substantial intergenic regions (oppA-I to oppA-II, 123 bp; oppA-II to oppA-III, 145 bp; and oppA-III to oppB, 361 bp), whereas the oppB, oppC, oppD, and oppF genes are separated by only a few base pairs (4 to 13 bp). A total of 430 bp separates oppA-I from the putative glycerol-3-P-O-acyltransferase gene that is transcribed in the reverse direction. There is a 598-bp region separating oppA-IV from an unknown reading frame on the plasmid cp26 and a 501-bp region separating oppA-V from a lipoprotein on the plasmid lp-54.
The B. burgdorferi genome encodes sequences for the classical, σ70-type promoter as well as those for alternative sigma factors such as rpoS (σ38) and ntrA (σ54). An analysis of the upstream sequences of the opp genes for significant identity to the consensus σ70 binding sequences revealed classical −35 and −10 binding sequences upstream of oppB. However, we were unable to find both −35 and −10 σ70-type sequences upstream of any oppA coding region. Putative −10 sequences without corresponding −35 sequences were found for several of the sequences upstream of individual oppA coding regions and are shown in Fig. 1. A search for binding sequences of alternative sigma factors did not reveal any sequences with >60% identity to a σ38- or σ54-dependent promoter region.
FIG. 1.
Schematic representations of opp operon and upstream regions of five oppA genes and oppB. DNA sequences symbolize putative −10 and −35 regions of σ70 promoter; arrows indicate the orientations of promoters. Arrows facing to the right indicate sequences on the sense strand; arrows pointing to the left indicate sequences in the reverse orientation. The numbers represent the nucleotide positions in each upstream region. P1, N1, P2, and N2 represent DNA fragments of the oppA-I upstream region, F1, F2, and F3 represent those of oppA-IV upstream region, and R1 and R2 represent those of oppA-V upstream region.
Cloning of B. burgdorferi oppA promoters into reporter plasmids.
To confirm the promoter function of the upstream sequences of the B. burgdorferi opp genes, we cloned the sequences into the promoter-less β-galactosidase reporter plasmids pCB182 (oppA-I, oppA-II, oppA-III, oppA-IV, and oppB) and pCB192 (oppA-V). pCB182 and pCB192 differ only in the orientations of the multicloning sites upstream of the β-galactosidase gene. In addition, for oppA-I, oppA-IV, and oppA-V, which have large upstream intergenic regions of between 430 and 598 bp, we examined the effects of serial deletions of the upstream regions on promoter function to better localize the active sites. β-Galactosidase activity from strains of E. coli transformed with the various recombinant plasmids was measured to establish the promoter activity for each upstream fragment. Results are shown in Table 2. There are marked differences in the promoter activities of the different upstream sequences, with the highest activity levels seen for oppA-I and oppA-III and minimal activity seen for oppA-II and oppA-IV.
TABLE 2.
β-Galactosidase activity for B. burgdorferi opp genes
| Name of fragment(region)a | Specific activity (U) |
|---|---|
| URA1 (1-430 bp) | |
| P1 (1-87 bp) | 331 ± 54 |
| P1N1 (1-156 bp) | 3.3 ± 0.6 |
| P1N1P2 (1-299 bp) | 249 ± 44 |
| P1N1P2N2 (1-430 bp) | 1.0 ± 0.2 |
| N1P2N2 (71-430 bp) | 87 ± 13 |
| P2N2 (156-430 bp) | 156 ± 19 |
| N2 (275-430 bp) | 11 ± 1.2 |
| P2 (156-299 bp) | 125 ± 19 |
| URA2 (1-125 bp) | 12 ± 1.2 |
| URA3 (1-147 bp) | 128 ± 7.1 |
| URA4 (1-598 bp) | |
| F1F2F3 (1-598 bp) | 1.6 ± 0.7 |
| F1 (1-290 bp) | 0.3 ± 0.5 |
| F2 (290-400 bp) | 8.7 ± 2.2 |
| F2F3 (290-598 bp) | 2.4 ± 0.5 |
| URA5 (1-501 bp) | |
| R1R2 (1-501 bp) | 29 ± 5.3 |
| R1 (1-92 bp) | 71 ± 2.6 |
| R2 (93-501 bp) | 22 ± 1.5 |
| URB (1-362 bp) | 56 ± 8.5 |
URA1 to -5, upstream regions of oppAI to -V; URB, upstream region of oppB.
Analysis of the serial deletions of the putative oppA-I promoter region revealed two separate sequences, designated P1 and P2, with high activity. Both sequences contain a putative −10 region TATATT for P1 and a TATTAT for P2, which are located at nucleotide positions 50 and 206, respectively (Fig. 1). The P1 fragment presumably functions as a promoter to control the transcription of a putative glycerol-3-P-O-acyltransferase gene upstream of oppA-I which is oriented in the reverse direction. Cloning of P1 into pCB192 confirmed that P1 possesses a strong biorientational promoter function. As P1N1 displays 100-fold-lower activity than that for P1 alone, it appears that P1 promoter function is unlikely to participate in regulating oppA-I gene transcription. This speculation is further supported by the finding that specific activity for N1P1 (bp 156 to 1) (data not shown) is sixfold higher than that for P1N1 (bp 1 to 156). Furthermore, P1N1P2 displays activity 75-fold higher than that for P1N1 but only twofold higher than that for P2, indicating that activity of P1N1P2 results primarily from the function of P2. The lack of significant difference (less than twofold) between the specific activities of N1P2N2 and P2N2 suggests that N1 fragments do not interfere with the function of P2.
Of note, the entire upstream region (P1N1P2N2) of oppA-I gene displayed markedly reduced activity compared with that of the truncated sequences. We believe this to be an artifact of the construction and not due to any inhibitory effects of the P1 region. As we will show in subsequent sections in which expression of oppA-I in B. burgdorferi is examined, the activities of the P2 sequences are more reflective of the promoter activity in B. burgdorferi than those revealed by our results using the entire intergenic region.
Although the sequence search in the nonsense chain of the oppA-II upstream region did not reveal any areas with identity to σ-binding promoter sequences, it does appear that the intergenic region between oppA-I and oppA-II has at least weak, independent promoter function (12 units), suggesting that there may be individual regulation of this gene. In contrast to the oppA-II promoter region, the oppA-III upstream region displayed a high specific activity level (128 units), suggesting that this fragment functions as a strong promoter. A perfect −10 σ70-like promoter sequence (TATAAT) was found to be located at nucleotide position 23.
Specific activity for the oppA-IV upstream region located on plasmid cp26 showed the weakest activity (1.6 units). Serial deletions of the upstream sequence revealed negligible activity (0.3 unit) for the F1 fragment (1 to 299 bp), suggesting that the F1 fragment is not involved in promoter function for the oppA-IV gene. The F2F3 fragment (299 to 598 bp) had 2.4 units of specific activity, which is similar to the level of activity found for the intact oppA-IV upstream region (1.6 units). Sequence analysis revealed a putative −10 region sequence (TATAAT) in the F2 fragment (290 to 400 bp). Insertion of the F2 fragment into promoter-probe vector pCB182 gave F2 8.7 units of specific activity.
The promoter activity level of the oppA-V upstream region (1 to 501 bp) was intermediate compared with those of the other promoter regions (18-fold higher than that of oppA-IV and fivefold lower than that of oppA-I). Both the R1 fragment (1 to 92 bp) and the R2 fragment (93 to 501 bp) displayed promoter activity. The activity of the R2 fragment was comparable to the activity for the entire upstream sequence. Sequence analysis did reveal a putative −10 σ70-like binding sequence (TATATT) located at position 432 (Fig. 1), which is close to the initiation codon of oppA-V gene.
Promoter activities for oppA-1 (P1N2), oppA-2 (URA2), oppA-3 (URA3), oppA-4 (URA4), and oppA-5 (URA5) differed significantly from each other, with P < 0.05 by Kruskal-Wallis nonparametric analyses.
Expression of Opp genes in in vitro-grown B. burgdorferi.
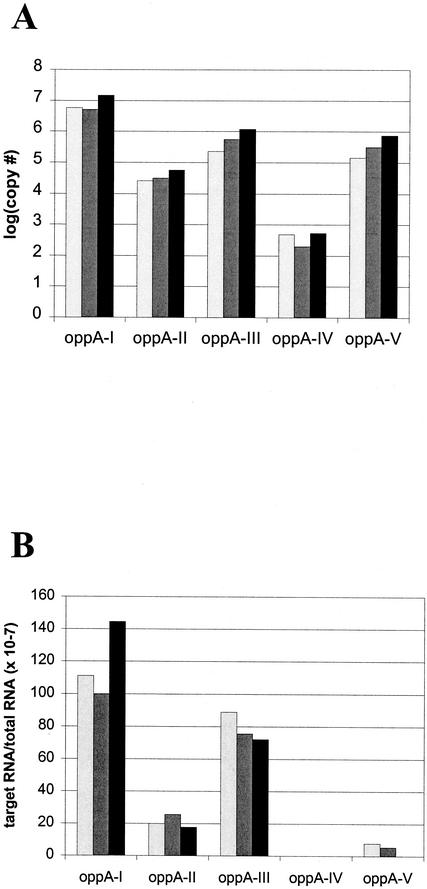
To confirm the results of our reporter assays, we examined expression of the oppA genes directly in B. burgdorferi. Quantitative RT PCR was used to determine the amount of mRNA for each oppA gene. The results are shown in Fig. 2A. Similar to what we found with the promoter activity in our reporter constructs, the amount of oppA-I transcripts in B. burgdorferi grown in BSK medium at 37°C was the largest, followed by those of oppA-III and oppA-V and, finally, oppA-II, in that order. It appeared that there was very little transcription of oppA-IV (over 4 logs fewer copies than that for oppA-I). This data was confirmed using RPAs, which showed similar trends (Fig. 2B).
FIG. 2.
Expression of oppA mRNA transcripts in cultured B. burgdorferi. Expression of the oppA genes was measured by RT PCR (A) and by RPA (B). For RT PCR, RNA was isolated from B. burgdorferi grown in BSK H medium at 37°C. cDNA was generated from total RNA, using gene-specific primers. For each sample, quantitative RT PCR for oppA-I to -V was performed. Determination of copy numbers was calculated by comparison to individual standard curves generated for each primer set. Panel A shows the results of three separate experiments. For RPA, total RNA was hybridized to biotinylated probes specific for each oppA gene and then digested with RNase A-RNase T1. Samples were subjected to electrophoresis in a polyacrylamide gel and transferred to nylon membranes. Detection of biotinylated probes was done using chemiluminescence, and quantitation was performed by scanning densitometry. Data shown are individual results from three separate experiments.
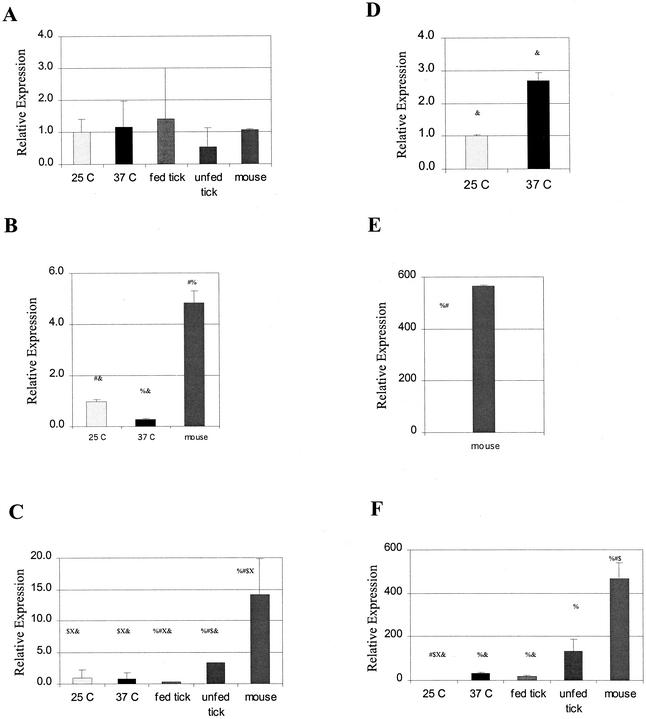
To further establish that our reporter constructs reflect promoter activity in B. burgdorferi, we next sought to compare the effects of changes in temperature on promoter activity in the reporter constructs and on oppA gene expression in B. burgdorferi. Temperature change has been shown to be an important signal for expression of other B. burgdorferi genes and is an important environmental variable in the life cycle of B. burgdorferi as it moves from its tick to mammalian hosts. Bono et al. (3) have previously shown that only oppA-V expression is significantly affected by temperature changes. Using real-time quantitative PCR technology, we confirmed these findings (Fig. 3). There were only small changes (less than fivefold) in the levels of mRNA transcripts for oppA-I, oppA-II, oppA-III, and oppA-IV between organisms grown at 25°C and those grown at 37°C. oppA-V showed a large increase (32-fold) in mRNA transcripts at 37°C versus 25°C.
FIG. 3.
Expression of oppA mRNA transcripts under various environmental conditions. Total RNA was isolated from in vitro-grown organisms at 25°C and 37°C or from fed and unfed Ixodes ticks. cDNA to total RNA was generated using random hexamers. Expression of mRNA for each individual oppA was examined by quantitative real-time RT PCR. Expression levels of oppA-I (A), oppA-II (B), oppA-III (C), oppA-IV (D), and oppA-V (E and F) are shown relative to that of expression of the gene in in vitro-grown organisms at 25°C, which was assigned a relative value of 1.0. Data shown are averages from two to three experiments performed in duplicate. Error bars represent standard errors of the means. Statistical significance was calculated using the Kruskal-Wallis method. %, significant (P < 0.05) differences in expression level compared with samples grown at 25°C; #, significant differences in expression level compared with samples grown at 37°C; $, significant differences in gene expression level in fed ticks; X, significant differences in gene expression level in unfed ticks; &, significant differences from expression levels in mouse tissue. Note that the scale changes for each graph. Data for oppA-IV are split into two graphs (panels E and F) due to the wide range of relative expression.
We next examined the effects of temperature on our promoter constructs in E. coli (Table 3). Bacteria containing the reporter plasmids for the upstream regions of each of the five oppA genes and the region upstream of oppB were incubated at 25°C and 37°C and grown to mid-logarithmic phase. Prior to performing the experiments, bacterial growth curves for each strain at each temperature were established to ensure that samples were taken at the same growth stage. There was no significant difference in plasmid copy numbers in E. coli grown at the two temperatures, as measured by real-time PCR using total DNA as the template and primers for the plasmid and a host chromosomal gene. All of the reporter constructs showed statistically significant increases in activity at 37°C versus 25°C. For the oppA-I, oppA-II, oppA-III, and oppA-IV promoters, the increase was in the range of 3.1- to 3.6-fold. Of the oppA promoters, only the reporter plasmid for the oppA-V promoter region showed an increase in β-galactosidase activity greater than 5-fold (5.5-fold) when bacteria were grown at 37°C versus 25°C.
TABLE 3.
Effect of temperature on E. coli promoter constructs
| Region (DNA fragment)a | β-Galactosidase activityb
|
Ratio (37°C/25°C) | P value | |
|---|---|---|---|---|
| 25°C | 37°C | |||
| URB | 15 ± 1.4 | 66.4 ± 3.6 | 4.4 | < 0.05 |
| URA1 (P1N1N2) | 50.4 ± 4.1 | 155 ± 23.4 | 3.1 | < 0.05 |
| URA2 | 3.9 ± 0.6 | 13.1 ± 0.6 | 3.4 | < 0.05 |
| URA3 | 39 ± 3.2 | 133 ± 6.2 | 3.4 | < 0.05 |
| URA4 (F1F2F3) | 0.5 ± 0.2 | 1.8 ± 0.7 | 3.6 | < 0.05 |
| URA5 (R1R2) | 6.3 ± 0.3 | 34.4 ± 7.3 | 5.5 | < 0.05 |
URB, upstream region of oppB; URA1 to -5, upstream regions of oppAI to -V.
Miller units.
Effects of nutritional changes in the environment on activity of oppA promoters.
Having confirmed that our reporter constructs in E. coli appear to reflect promoter activity in B. burgdorferi, we next sought to use these constructs to test the effects of various environmental changes on promoter activity for the oppA and oppBCDF genes. Because B. burgdorferi does not grow in defined media, many of these manipulations would be difficult or impossible to carry out directly in B. burgdorferi. Since the Opp systems of other bacteria play a major role in nutritional uptake of oligopeptides and may be regulated as part of a “feast or famine” response, we sought to determine whether specific changes in nutrient availability could change opp gene expression.
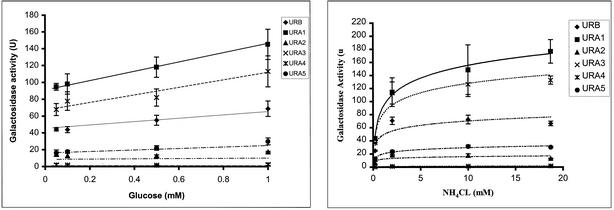
We first examined the effects of variations in the carbon and nitrogen availability in the culture medium on oppA promoter activity. The bacterial cells containing the various reporter plasmids were grown at 37°C in minimal medium, with a wide range of concentrations of d-glucose or NH4Cl. Again, bacterial growth curves in each medium were monitored to ensure that measurements were taken at comparable growth phases. All bacterial cells used in β-galactosidase assays were grown to the mid-logarithm phase.
The promoter activity for the upstream regions of oppA-II and oppA-IV remained fairly stable over a wide range of glucose concentrations (0.05 to 1 mM). In contrast, promoter activity for oppA-I, oppA-III, and oppA-V showed steady increases with increasing glucose concentration; however, none of these increases were greater than twofold (Fig. 4A).
FIG. 4.
Effects of glucose and NH4Cl concentrations on opp promoter activity. Bacteria containing plasmid reporter constructs for each of the oppA promoter regions were grown at 37°C in minimal medium supplemented with 100 μg of ampicillin/ml, 0.2 mM l-leucine, and different glucose (A) and NH4Cl (B) concentrations. Specific activity (U) was calculated from three experiments performed in duplicate. Error bars represent standard errors of the means. URA1, upstream region of oppA-I; URA2, upstream region of oppA-II; URA3, upstream region of oppA-III; URA4, upstream region of oppA-IV; URA5, upstream region of oppA-V; URAB, upstream region of oppB.
Promoter activities for the oppA-I and oppB upstream regions showed increases of approximately threefold, with increases in NH4Cl concentration from 0.2 mM to 20 mM in the medium (Fig. 4B), suggesting a role for nitrogen availability in regulation of expression of these Opp proteins. Promoter activities for the other oppA genes showed minor increases in activity between 0.2 mM and 2 mM NH4Cl that may be due to cell viability at the extremes of nitrogen limitation. Only oppA-I promoter activity continued to increase with increases in NH4Cl concentration to 20 mM.
Regulation of Opp expression in other bacteria has been reported to be affected by phosphate concentration and leucine concentration (2, 19). However, we did not observe any changes in promoter activity in our constructs with changes in phosphate concentration (70 μM to 2 mM) or in leucine concentration (data not shown). Another environmental factor which has been shown to affect expression of other B. burgdorferi proteins is change in pH. Changes in pH are also physiologically relevant, as the bacteria must adapt to changes in pH as it moves from its tick to mammalian hosts. We did not observe any change in promoter activity for any of the oppA or oppB promoters with changes in pH over a range of 3.0 to 8.5 (data not shown).
Expression of opp genes in tick and mouse hosts.
We next chose to examine expression of oppA mRNA transcripts by B. burgdorferi in its tick host. B. burgdorferi residing within a tick must adapt to vast changes in nutrient ability during periods between blood meals. We examined gene expression from B. burgdorferi in nymphal Ixodes ticks in the unfed state and after allowing them to feed on an uninfected mouse for 60 h (Fig. 3).
Expression of oppA-II and oppA-IV mRNA was below the limit of reliable detection in both fed and unfed ticks (less than 150 copies determined, using known quantities of spiked DNA). oppA-I expression showed a trend to increased expression in the fed tick, but this was not significant. Overall, expression of oppA-I in either fed or unfed ticks did not differ significantly from oppA-I expression in in vitro-grown organisms relative to expression of recA. In contrast, expression of both oppA-III and oppA-V was significantly increased in the unfed ticks compared to the fed ticks (9.5- and 7-fold, respectively). oppA-V expression in both the fed and unfed ticks was significantly higher than that in in vitro-grown organisms at 25°C (18- and 32-fold, respectively). Expression of oppA-V in the fed tick approximates expression of oppA-V from in vitro-grown organisms at 37°C (a less than twofold difference) but is fourfold less than the expression seen in unfed ticks.
We also examined expression of the oppA genes from organisms in infected mice. Once again, expression of oppA-I remained relatively unchanged between organisms isolated from in vitro cultures, unfed and fed ticks, and mouse tissue. Expression of oppA-II was moderately increased in mouse tissue (approximately fivefold) compared with in vitro-grown organisms and significantly higher than in either fed or unfed ticks, where expression was not detected. Both oppA-III and oppA-V expression levels were significantly increased in mouse tissue compared with that seen for either fed or unfed ticks. Expression of oppA-V relative to recA was almost 500-fold higher in mouse tissue than that seen for cultured organisms grown at 25°C and 25- and 3.5-fold higher than that seen for fed and unfed ticks, respectively.
However, the most significant change in expression was seen with oppA-IV. Expression of oppA-IV, which was undetectable in organisms isolated from ticks, was increased more than 500-fold compared to expression in cultured organisms.
DISCUSSION
Sequence analysis of the upstream regions of B. burgdorferi oppA-I, oppA-II, oppA-III, oppA-IV, and oppA-V as well as oppB revealed little similarity, suggesting that expression of each gene is regulated by an independent mechanism(s). The data presented in this paper confirm that there are marked differences in the levels of expression of the various opp genes and that each may be subject to a different regulatory mechanism(s).
Our β-galactosidase reporter assays demonstrate that the upstream regions of oppA-I, oppA-II, and oppA-III all possess promoter activity. This represents further evidence to support the hypothesis that transcription can initiate at each oppA gene. Bono et al. have previously shown that the chromosomally encoded oppA genes, oppA-I, oppA-II, and oppA-III, may be transcribed as mono-, di,- or tricistronic messages (3). Based on our data, it appears that transcription of the chromosomally encoded oppA genes is initiated more frequently by the individual promoters rather than proceeding from the oppA-I promoter. If the majority of transcription of oppA-I, oppA-II, and oppA-III occurred through the activity of the oppA-I promoter, it would be expected that transcripts of the three genes would be equal or, if early termination occurred, that the amount of transcription would be largest for oppA-I, followed by those of oppA-II and, finally, oppA-III. On the other hand, if bicistronic messages originated from the oppA-II promoter, higher levels of oppA-II and oppA-III would be expected. However, in our studies of oppA transcription from in vitro-grown organisms, we have found that the amount of oppA-II transcription is almost 20-fold less than of that of oppA-III and over 200-fold less than that of oppA-I, as determined by RT PCR. This strongly suggests that transcription of oppA-I gene can be terminated before processing of oppA-II and that the majority of oppA-III transcription occurs through the activity of its own promoter.
After confirming that promoter activity measured in our E. coli reporter systems was consistent with the levels of transcription seen in B. burgdorferi under comparable conditions, we used this system to test the effects of various environmental changes. While to examine promoter activity in a heterologous system using a different host organism is certainly not optimal, the inability of B. burgdorferi to grow in defined media severely limits the ability to test the effects of these conditions on B. burgdorferi directly. Although we believe that the E. coli reporter system is reflecting activity of the B. burgdorferi opp promoters and have found this to be the case in some of the direct comparisons that are possible, it would be impossible to establish this for every condition. However, almost as important as the individual changes in promoter activity we described is the understanding that each of the promoter regions can respond differentially to the same environmental condition. For example, oppA-V expression is most affected by changes in temperature while oppA-I expression is the most affected by nitrogen concentration.
The ability to differentially regulate expression of its oppA genes could potentially provide B. burgdorferi with a mechanism for environmental adaptation. The organism is exposed to vast environmental changes as it moves from its tick and mammalian hosts. B. burgdorferi bacteria are acquired by larval ticks taking a blood meal from infected animals (4). The ticks do not take another meal until they molt into the nymphal stage months later. At the time of the next blood meal, when nutrients again become plentiful, the bacteria in the midgut of the organism begin to multiply rapidly and subsequently can infect the animal on which the tick is feeding. After the meal is complete, within days the number of organisms drops precipitously and remains low until the next blood meal. Narasimhan et al. have previously found that genes in the opp operon were differentially expressed in fed and unfed ticks but did not identify specific genes within the operon that accounted for the differences (13). We have found that expression of oppA-II and oppA-IV remains very low in the tick hosts and that oppA-III and oppA-V are up-regulated during periods of stable nutrient availability (unfed tick and mouse hosts). Expression of oppA-II appears to be specifically down-regulated in organisms growing in ticks compared with the activity of its promoter and expression in in vitro-grown organisms. In contrast, despite very weak promoter activity in vitro, oppA-IV undergoes tremendous up-regulation in organisms in the mouse host. The signals regulating these changes have not been identified and are likely to be complex. For example, it appears that factors other than temperature play a major role in determining oppA-V expression in fed and unfed ticks, since increases in temperature up-regulate oppA-V expression but expression in unfed ticks is higher than that in fed ticks. Increased nutrient availability appeared to play a role only for oppA-I, which was the only gene to show substantially increased promoter activity with increases in nitrogen availability.
Transport of specific peptides has been shown to mediate important bacterial adaptations such as chemotaxis, quorum sensing, and the feast or famine response (1, 8, 9, 11). It is tempting to speculate that the different OppA proteins are involved in separate adaptive functions. For example, could it be the case that OppA-I, which has the highest level of basal expression and which showed the greatest responsiveness to changes in nutrient availability, is involved in peptide transport for basic nutritional needs while OppA-4, which is expressed in high levels only in the mouse host, is involved in a more specific function such as peptide chemotaxis? Another possibility is that differences in expression and substrate specificity of the OppA proteins have evolved to allow the organism to adapt to different environmental niches, where the supplies of specific peptides can vary. Further studies of the conditions affecting expression of the individual OppA proteins as well as the substrate specificities of each protein will be necessary to achieve a better understanding of the role of each of the OppA proteins of this sole peptide transporter of B. burgdorferi.
Acknowledgments
We wish to thank Jenifer Coburn, Carla Cugini, Melissa Medrano, and Scott Samuels for their many helpful discussions with this project.
Work on this project was supported by grants from the National Institute of Allergy and Infectious Diseases (R01 AI44240 and R01 AI50043) to L.T.H.
REFERENCES
- 1.Abouhamad, W. N., M. Manson, M. M. Gibson, and C. F. Higgins. 1991. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol. Microbiol. 5:1035-1047. [DOI] [PubMed] [Google Scholar]
- 2.Austin, E. A., J. C. Andrews, and S. A. Short. 1989. Molecular genetics of bacteria and phages, p. 153. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 3.Bono, J. L., K. Tilly, B. Stevenson, et al. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144:1033-1044. [DOI] [PubMed] [Google Scholar]
- 4.Burgdorfer, W., S. F. Hayes, and D. Corwin. 1989. Pathophysiology of the Lyme disease spirochete, Borrelia burgdorferi, in ixodid ticks. Rev. Infect. Dis. 11:S1442-S1450. [DOI] [PubMed] [Google Scholar]
- 5.Fraser, C. M., S. Casjens, W. M. Huang, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 6.Hirshfield, I. N., and M. B. Price. 1975. Utilization of selected leucine peptide amides by Escherichia coli. J. Bacteriol. 122:966-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu, L. T., G. Perides, R. Noring, and M. S. Klempner. 1995. Binding of human plasminogen to Borrelia burgdorferi. Infect. Immun. 63:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazazzera, B. A., J. M. Solomon, and A. D. Grossman. 1997. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 89:917-925. [DOI] [PubMed] [Google Scholar]
- 9.Leonard, B. A. B., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgA, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, B., S. A. Short, M. Eskildsen, M. S. Klempner, and L. T. Hu. 2001. Functional testing of putative oligopeptide permease (Opp) proteins of Borrelia burgdorferi: a complementation model in opp(−) Escherichia coli. Biochim. Biophys. Acta 1499:222-231. [DOI] [PubMed] [Google Scholar]
- 11.Manson, M. D., V. Blank, G. Brade, and C. F. Higgins. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321:253-256. [DOI] [PubMed] [Google Scholar]
- 12.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Narasimhan, S., F. Santiago, K. R. Koski, B. Brei, J. F. Anderson, D. Fish, and E. Fikrig. 2002. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J. Bacteriol. 184:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payne, J. W. 1968. Oligopeptide transport in Escherichia coli. Specificity with respect to side chain and distinction from dipeptide transport. J. Biol. Chem. 243:3395-3403. [PubMed] [Google Scholar]
- 15.Payne, J. W. 1974. Peptide transport in Escherichia coli: permease specificity towards terminal amino group substituents. J. Gen. Microbiol. 80:269-276. [DOI] [PubMed] [Google Scholar]
- 16.Payne, J. W., B. M. Grail, S. Gupta, et al. 2000. Structural basis for recognition of dipeptides by peptide transporters. Arch. Biochem. Biophys. 384:9-23. [DOI] [PubMed] [Google Scholar]
- 17.Payne, J. W., and M. W. Smith. 1994. Peptide transport by micro-organisms. Adv. Microb. Physiol. 36:1-80. [DOI] [PubMed] [Google Scholar]
- 18.Schneider, K., and C. F. Beck. 1986. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene 42:37-48. [DOI] [PubMed] [Google Scholar]
- 19.Smith, M. W., and J. W. Payne. 1992. Expression of periplasmic binding proteins for peptide transport is subject to negative regulation by phosphate limitation in Escherichia coli. FEMS Microbiol. Lett. 79:183-190. [DOI] [PubMed] [Google Scholar]
- 20.Smith, M. W., D. R. Tyreman, G. M. Payne, et al. 1999. Substrate specificity of the periplasmic dipeptide-binding protein from Escherichia coli: experimental basis for the design of peptide prodrugs. Microbiology 145:2891-2901. [DOI] [PubMed] [Google Scholar]
- 21.Tame, J. R., G. N. Murshudov, E. J. Dodson, et al. 1994. The structural basis of sequence-independent peptide binding by OppA protein. Science 264:1578-1581. [DOI] [PubMed] [Google Scholar]