Abstract
Whole genome sequences of Neisseria meningitidis strains Z2491 and MC58 and Neisseria gonorrhoeae FA1090 were analyzed for Correia repeats (CR) and CR-enclosed elements (CREE). A total of 533, 516, and 256 copies of CR and 270, 261, and 102 copies of CREE were found in these three genomes, respectively. The lengths of CREE range from 28 to 348 bp, and the lengths of multicopy CREE appear mainly in the ranges of 154 to 156 bp and 105 to 107 bp. The distribution of CREE lengths is similar between the two N. meningitidis genomes, with a greater number of 154- to 156-bp CREE (163 and 152 copies in N. meningitidis strain Z2491 and N. meningitidis strain MC58, respectively) than 105- to 107-bp CREE (72 and 77 copies). In the N. gonorrhoeae strain FA1090 genome there are relatively more 105- to 107-bp CREE (51 copies) than 154- to 156-bp CREE (36 copies). The genomic distribution of 107-bp CREE also shows similarity between the two N. meningitidis strains (15 copies share the same loci) and differences between N. meningitidis strains and N. gonorrhoeae FA1090 (only one copy is located in the same locus). Detailed sequence analysis showed that both the terminal inverted repeats and the core regions of CREE are composed of distinct basic sequence blocks. Direct TA dinucleotide repeats exist at the termini of all CREE. A survey of DNA sequence upstream of the sialyltransferase gene, lst, in several Neisseria isolates showed that 5 N. meningitidis strains contain a 107-bp CREE in this region but 25 N. gonorrhoeae strains show an exact absence of a 105-bp sequence block (i.e., the 107-bp CREE without a 5′ TA dinucleotide) in the same region. Whole-genome sequence analysis confirmed that this 105-bp indel exists in many homologous 107-bp CREE loci. Thus, we postulate that all CREE are made of target TA with indels of various lengths. Analysis of 107-bp CREE revealed that they exist predominantly in intergenic regions and are often near virulence, metabolic, and transporter genes. The abundance of CREE in Neisseria genomes suggests that they may have played a role in genome organization, function, and evolution. Their differential distribution in different pathogenic Neisseria strains may contribute to the distinct behaviors of each Neisseria species.
Since the report of repetitive extragenic palindromic (REP) sequences in Escherichia coli and Salmonella enterica serovar Typhimurium (25), short dispersed repetitive elements have been increasingly identified in prokaryotes (35). Genome analyses have confirmed the extensive repetition of REP sequences and REP elements in E. coli (4). Short dispersed repetitive elements have also been identified in other prokaryotes for which the complete genome sequences have been analyzed (for a review, see reference 44).
Correia et al. identified a 26-bp sequence as a repetitive element in the pathogenic Neisseria spp. (8). Subsequent studies showed that Correia repeats (CR) often constitute parts of longer repetitive sequence elements (9, 29). By using two-dimensional S1 nuclease heteroduplex mapping, Correia et al. estimated that in Neisseria gonorrhoeae there are ca. 20 copies of 152-bp elements whose ends are composed of inverted repeats of the 26-bp CR sequence (9). Gotschlich et al. identified a 105-bp sequence element that also contains the CR sequences as terminal inverted repeats, and these authors estimated that the 105-bp element is present at least 20 times in the genome of N. gonorrhoeae R10 (23). The 154-bp Correia element(s) (CE) was considered to be a transcriptional terminator in the division cell wall (dcw) cluster of N. gonorrhoeae CH811 and a search of the sequence databases revealed another 19 copies of similar elements adjacent to different neisserial genes (14). In genome sequences of Neisseria meningitidis, 163 copies of the 26-bp inverted repeat were found in N. meningitidis MC58 (49) and a total of 286 CE (sequences bounded by 26-bp inverted repeats) were found in N. meningitidis Z2491 (41). More recently, Mazzone et al. reported a total of 270, 259, and 110 copies of nemis (Neisseria miniature insertion sequences) in whole genomes of N. meningitidis Z2491 and MC58 and N. gonorrhoeae FA1090, respectively (37).
In the present study, we first analyzed the sequence conservation in all CR found in three completed neisserial genomes: N. meningitidis Z2491 (41), N. meningitidis MC58 (49), and N. gonorrhoeae FA1090 (http://www.genome.ou.edu/gono.html). We then used the most conserved region of the CR to identify those sequence elements that were enclosed by an inverted pair of the CR in these three complete genome sequences. We analyzed the detailed sequence features of CR-enclosed elements (CREE) and determined DNA sequences upstream of the sialyltransferase gene, lst, in several Neisseria strains to identify the size of indels in the loci of 107-bp CREE. The common sequence features identified among all CREE and the potential mechanisms for the formation of CREE may assist in an understanding of their origin, propagation, and function within the genome. The distinction between N. meningitidis and N. gonorrhoeae may supplement our understanding of the differential pathogenesis of these two clearly related but distinct pathogens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
N. gonorrhoeae F62 was obtained from P. Frederick Sparling of the University of North Carolina at Chapel Hill; N. meningitidis MC58 from E. Richard Moxon, University of Oxford, United Kingdom; N. meningitidis DNM51 from David Dyer, University of Oklahoma Health Sciences Center; and N. meningitidis NMB13, DNM3, and FAM18 from David S. Stephens (Emory University, Atlanta, Ga.). Clinical isolates of N. gonorrhoeae were obtained from MCP Hahnemann University Hospital in Philadelphia (labeled as the “A” series of strains) and from patients attending a sexually transmittted disease clinic in Baltimore (labeled as the “B” series of strains and provided to us by John Zenilman of Johns Hopkins University). To minimize genetic change, stocks of all strains and clinical isolates were stored at −70°C and experiments were started from frozen stocks with minimal passage (one to three times). Growth of Neisseria isoaltes was performed at 37°C in a 5%, humidified CO2 incubator (Forma Scientific, Marietta, Ohio) on GC agar (Difco, Detroit, Mich.) with supplement (30). Bacteria were suspended in sterile phosphate-buffered saline with 0.1% (wt/vol) gelatin and 0.01% (wt/vol) each of CaCl2 and MgCl2 (38).
PCR amplification of lst upstream regions (5′-lst).
To obtain template DNA for amplification of 5′-lst, several colonies of Neisseria were suspended in 20 μl of sterile H2O and boiled for 10 min, chilled on ice for 2 min, and then incubated with RNase A (final concentration of 16 μg/ml) at 37°C for 30 min. These suspensions were centrifuged at 8,000 × g for 5 min, and supernatants were collected and placed on ice for immediate use or stored at −20°C for later use.
To amplify the 5′-lst, we used primers synthesized according to sequence information derived from N. meningitidis MC58 (accession no. U60660) (18). The forward primer, FuseBam-For1 (5′-CGCTGGATCCGACATCAATATCGG), starts 124 bp from the initiation codon of putative isocitrate dehydrogenase gene icd on its minus strand and contains a BamHI restriction site at its 5′ end (underlined). The reverse primer, FuseBam-Rev1 (5′-CAAAGGATCCTTTTTCAAGCCC), starts 24 bp from the initiation codon of lst on its minus strand and also contains a BamHI site (underlined). PCR was performed in thin-wall glass capillary tubes by using an Air Thermo-Cycler (Idaho Technology Model 1605; Idaho Falls, Idaho) with a two-step program. Step one consisted of five cycles of 94°C for 0 s, 40°C for 0 s, and 72°C for 15 s. Step two consisted of 30 cycles of 94°C for 0 s, 55°C for 0 s, and 72°C for 15 s.
DNA sequencing.
DNA fragments amplified by PCR were cut from agarose gels after electrophoresis and purified by using a Wizard PCR Preps DNA purification system (Promega, Madison, Wis.). Nucleotide sequences were determined with the terminal primers described above by cycle sequencing with fluorescently labeled dideoxynucleotides (DyeDeoxy terminators; Perkin-Elmer) by using automated DNA sequencing facilities at the MCP Hahnemann School of Medicine.
Genome sequences.
The genome sequence of N. meningitidis MC58 (49) was obtained from The Institute of Genomic Research (ftp://ftp.tigr.org/pub/data/n_meningitidis/). The genome sequence of N. meningitidis Z2491 (41) was obtained from the Sanger Center (http://www.sanger.ac.uk/Projects/N_meningitidis/). The genome sequence of N. gonorrhoeae FA1090 was from the University of Oklahoma (http://www.genome.ou.edu/gono.html). The last search for CR and CREE in N. gonorrhoeae FA1090 was based on sequence released on 15 September 2000. It contains 2,154,110 sequence characters.
BLAST analysis of DNA sequences.
BLAST searches were performed by using BLASTN (version 1.4.7) in the version 9.1 Wisconsin package (Genetics Computer Group). The ungapped BLASTN program was used for searching homologues of the prototypic 26-bp CR (5′-GTACCGGTTTTTGTTAATTCACTATA) and a 105-bp sequence element was identified in the DNA sequence upstream first in N. meningitidis MC58. The GCG output information was parsed into a Microsoft Access database by using a custom written program. The copy number, length, and genomic location of the archived CR and CREE sequence information was analyzed by using the filtering, sorting, and querying functions of the Microsoft Access program. For sequence comparison, we converted all sequence elements to the same stranded orientation. Consensus sequences for CRs were found by using GCG's PILEUP and LINEUP programs. Consensus sequences for 105-bp sequence elements and other CREE were obtained by using the CLUSTALW multiple sequence alignment tool (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html) and the BoxShade program (http://www.ch.embnet.org/software/BOX_form.html). For analysis of the flanking sequences of the 105-bp elements, a custom written computer program was used to retrieve each 105-bp sequence and its 1-kb flanking region on each side. These retrieved 2,105-bp sequences were used as queries for BLASTN searches against the GenBank “nr” (nonredundant) database by using NCBI's BLAST 2.0 website (http://www.ncbi.nlm.nih.gov/BLAST/) with the default parameters.
FINDPATTERNS analysis of DNA sequences.
Single completed genomic sequences were divided into smaller segments of ca. 300 kb to comply with the length limitation of the GCG programs. These segments were then converted into a GCG-utilizable format by using the FROMFASTA and the data set programs in the GCG package. To avoid potential failure in detecting sequences at the ends of the divided segments, the complete genome sequences were divided into both eight and seven segments, and the number of findings were examined for agreements. The presentation of repeats in the genomes are based on original sequence coordinates indicated in the complete linear sequences. Searching for CREE was performed by using the FINDPATTERNS program of the GCG package. The search pattern used was TATAGTGGATT(N)[0, 328]AATCCACTATA, where “(N)[0, 328]” means any sequence with a length between 0 and 328 bp. This maximum limit of 328 bp in the core region is due to the total length limit of 350 bp in the FINDPATTERNS program. During the execution of the program, we used mismatch values from 0 to 6. The GCG output information was parsed into a Microsoft Access database by using a custom-written program. Then the copy number, length, and genomic location of the archived sequence information were analyzed by using the filtering, sorting, and querying functions of Microsoft Access.
Gene and ORF identification.
The matched segments were manually inspected for genes or open reading frames (ORFs) as identified in the genome reports for N. meningitidis Z2491 (41) and N. meningitidis MC58 (49). The distance of 105-bp elements relative to those identified genes or ORFs was determined by manual inspection of the genomic coordinates. Fine contextual analysis was then performed for N. meningitidis MC58 by using ACEDB (R. Durbin and J. T. Thierry-Mieg. 1991. A C. elegans database. Documentation, code, and data available at http://www.acedb.org), as used in the annotation of the genome sequence of this strain (49).
RESULTS
CR and CREE.
Using the 26-bp prototype CR (8) as a query, we found in ungapped BLAST analyses 533, 510, and 256 homologous sequence segments in the genomes of N. meningitidis Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090, respectively (Table 1). Among these matched segments, >75% are longer than 23 bp. Examination of alignments showed that all of these CR-homologous sequences have a conserved 11-bp terminal region: TATAGTGGTTT. Thus, in the present study, we refer to any sequence that contains this 11-bp sequence but is ≤26 bp as a CR. We refer to any sequence that contains this 11-bp sequence but is >26 bp as CEs. Since many CEs contain CR as terminal inverted repeats (TIR) (web table 1 [web tables and figures may be found at www.pages.drexel.edu/∼rr37/]), we used CREE to describe CR that are confined by two inverted CR.
TABLE 1.
Numbers of CR and CREE in three neisserial genomes
| Strain | No. of CR found by BLAST with matched length of the query (bp):
|
CREE found by FINDPATTERNS with mismatch alloweda:
|
% CR in CREEb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26 | 25 | 24 | 23-14 | 13 | 12-11 | All (11-26) | 0 | 1 | 2 | All (0-2) | ||
| N. meningitidis Z2491 | 381 | 14 | 5 | 18 | 103 | 12 | 533 | 183 (2) | +59 (1) | +28 (7) | 270 (10) | 99.4 |
| N. meningitidis MC58 | 377 | 7 | 2 | 25 | 94 | 11 | 516 | 176 (0) | +51 (2) | +34 (4) | 261 (6) | 100.3 |
| N. gonorrhoeae FA1090 | 153 | 13 | 26 | 21 | 37 | 6 | 256 | 59 (0) | +17 (2) | +26 (2) | 102 (4) | 78.1 |
Values indicate the number of additional CREE found upon increasing the number of allowed mismatches. The value in parentheses indicates the number of simple CREE contained inside complex CREE.
Calculation based on two and three CR in simple and complex CREE, respectively.
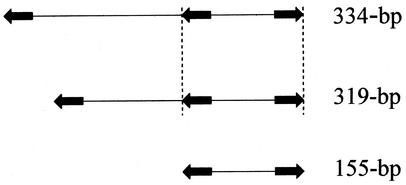
FINDPATTERNS analysis of the sequence pattern TATAGTGGTTT(core)AAACCACTATA yielded 270, 261, and 102 copies of CREE for N. meningitidis Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090, respectively (Table 1). Upon manual inspection of the CREE sequences, we noticed that some longer CREE contain an unpaired internal CR. This unpaired internal CR, when paired with a terminal CR, constitutes a shorter CREE that was also retrieved by the FINDPATTERNS program and listed in a shorter length category. For clarity, we refer to CREE enclosed by one pair of terminal CR as “simple” CREE and CREE containing an additional unpaired internal CR as “complex” CREE (Fig. 1). Since the number of nucleotides counted for a complex CREE contain the number of nucleotides of its enclosed simple CREE, the total nonredundant copies of CREE were adjusted by subtracting the number simple CREE from their parent complex CREE. The adjusted nonredundant copies of CREE are 260, 255, and 98 copies for N. meningitidis Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090, respectively (Table 1). In comparing the numbers of nonredundant CREE with the numbers of CR, we observed that almost all of the CR are contained in CREE in the two N. meningitidis genomes. In contrast, 22% of the CR are not contained in CREE in the N. gonorrhoeae FA1090 genome (Table 1).
FIG. 1.
Simple and complex CREE.
Length profiles of CREE.
CREE found in the above FINDPATTERNS analysis were sorted according to their length and copy number within each length (Table 2). In the three genomes, there are 50 categories of CREE based on length. N. meningitidis Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090 have 35, 26, and 19 of the 50 different CREE lengths, respectively. The total nucleotides in all CREE in N. meningitidis Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090 account for 1.67, 1.55, and 0.59% of their total genomic nucleotides, respectively.
TABLE 2.
Length profile of CREE in three neisserial genomes
| Length (bp) and other parameters |
N. meningitidis Z2491
|
N. meningitidis MC58
|
N. gonorrhoeae FA1090
|
|||
|---|---|---|---|---|---|---|
| Copy (n) | Internal CREE (n) | Copy (n) | Internal CREE (n) | Copy (n) | Internal CREE (n) | |
| Length | ||||||
| 348 | 1 | 105 | ||||
| 341 | 1 | 1 | 1 | |||
| 334 | 1 | 155 | ||||
| 329 | 1 | |||||
| 325 | 1 | 196 | ||||
| 319 | 1 | 155 | ||||
| 266 | 1 | 107 | ||||
| 245 | 1 | |||||
| 220 | 1 | 156 | ||||
| 219 | 1 | 155 | ||||
| 213 | 1 | 155 | 1 | 155 | ||
| 204 | 1 | 105 | ||||
| 199 | 1 | 155 | ||||
| 197 | 1 | 155 | ||||
| 185 | 1 | 156 | 1 | 156 | ||
| 182 | 1 | |||||
| 171 | 2 | 155 | ||||
| 169 | 1 | |||||
| 165 | 1 | 106 | ||||
| 159 | 2 | 105 (1) | ||||
| 157 | 1 | 3 | ||||
| 156 | 28 | 20 | 4 | |||
| 155 | 127 | 120 | 19 | |||
| 154 | 8 | 12 | 13 | |||
| 153 | 2 | 2 | ||||
| 150 | 1 | |||||
| 144 | 1 | |||||
| 142 | 1 | 2 | ||||
| 125 | 2 | |||||
| 116 | 1 | 71 | ||||
| 115 | 1 | 105 | ||||
| 111 | 1 | |||||
| 110 | 1 | 1 | ||||
| 108 | 1 | |||||
| 107 | 26 | 24 | 19 | |||
| 106 | 30 | 30 | 17 | |||
| 105 | 16 | 23 | 15 | |||
| 104 | 1 | 1 | ||||
| 101 | 1 | |||||
| 98 | 1 | |||||
| 97 | 1 | 2 | ||||
| 96 | 2 | 2 | ||||
| 92 | 1 | |||||
| 87 | 1 | |||||
| 73 | 2 | 2 | 1 | |||
| 72 | 1 | |||||
| 71 | 3 | 3 | 2 | |||
| 70 | 1 | 2 | ||||
| 69 | 1 | 1 | ||||
| 28 | 1 | 2 | ||||
| Length no. | 35 | 26 | 19 | |||
| Total copies | 270 | 10 | 261 | 6 | 102 | 4 |
| No. of nonredundant copiesa | 260 | 255 | 98 | |||
| % Nucleotide in genomeb | 1.67 | 1.55 | 0.59 | |||
Nonredundant CREE are calculated by subtracting the number of internal CREE from the total number of CREE directly counted by the FINDPATTERNS analysis.
The percentage of nucleotide in the nonredundant CREE in the total genome sequence.
Of the 50 CREE lengths, 9 are shared by all three neisserial genomes (Table 2) (341, 156, 155, 154, 107, 106, 105, 73, and 71 bp). Some CREE lengths are present in both meningococcal sequences but absent in the gonococcus (213, 185, 142, 110, 97, 96, 70, and 28 bp). CREE of 104 and 69 bp are present only in N. gonorrhoeae FA1090 and N. meningitidis Z2491. The 153-bp CREE is present only in N. gonorrhoeae FA1090 and N. meningitidis MC58. Fifteen CREE lengths are unique to N. meningitidis Z2491 (348, 325, 266, 219, 199, 182, 171, 159, 150, 144, 111, 108, 101, 98, and 72 bp). Seven CREE lengths are unique to N. meningitidis MC58 (334, 329, 220, 197, 125, 115, and 92 bp). N. gonorrhoeae FA1090 contains seven unique CREE lengths (319, 245, 204, 169, 165, 116, and 87 bp).
Most of the CREE lengths occur one to three times in each neisserial genome. Only the 154- to 156-bp and the 105- to 107-bp CREE have four or more copies per genome (Table 2). Interestingly, CREE of these lengths are also often found as simple CREE contained within complex CREE (Table 2). The longest simple CREE within the limitation of our FINDPATTERNS analysis is 341 bp, which was found only once in each of the three neisserial genomes. The 341-bp CREE of the two N. meningitidis strains are 100% homologous, whereas the 341-bp CREE of N. gonorrhoeae FA1090 shows little homology with the 341-bp CREE of N. meningitidis, having homologies of only 18 bp at the 5′ end and 11 bp at the 3′ end.
Sequence patterns of simple CREE.
To facilitate sequence analysis, we oriented all simple CREE into the same sequence direction. We then inspected all sequences to determine whether CREE of the same length belonged to one or more subtypes. Finally, we aligned each CREE length and subtype that had two or more copies in the genome. We collected all of the consensus CREE sequences and the original sequences of the single-occurrence CREE into a new data set for further analyses.
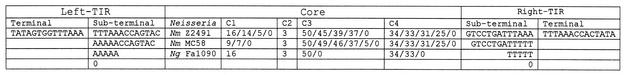
For the convenience of sequence comparison and presentation, we separated the symmetrical terminal inverted repeat regions from the asymmetrical core regions of each CREE and ordered them in separate tables. The terminal inverted repeats of simple CREE are listed in web tables 2 to 4 for N. meningitidis Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090, respectively, and the core regions of CREE from N. meningitidis strain Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090 are listed in web tables 5 to 7, respectively. Inspection of these consensus sequences further revealed the modular nature of the sequence composition in the TIR, as well as the core region of CREE (Fig. 2).
FIG. 2.
Common sequence blocks in CREE. The consensus sequences and common length of sequence fragments are presented in each sequence block. Slashes are used to separate alternative lengths of sequence fragments in each sequence block in the core region.
Short direct repeats flanking CREE.
We compared the 5-bp terminal sequences of every CREE with their flanking 5-bp sequences. We found that TA duplication is the most common direct repeat in the flanking sequences bordering CREE, followed by 3- to 5-bp direct repeats such as TAT or ATA, TATA, TATAG, or CTATA (Table 3). Because all CREE were identified by using a pair of 11-bp inverted repeats that contain terminal TATA, the terminal regions of all CREE comprise TA direct repeats.
TABLE 3.
Short direct repeats flanking CREE in the three neisserial genomes
| Neisserial strain | No. of short direct repeats upstream
|
CREE copya (TATAG***CTATA) | No. of short direct repeats downstream
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| TA | TAT | TATA | TATAG | TA | ATA | TATA | CTATA | ||
| N. meningitidis Z2491 | 12 | 2 | 1 | 0 | 270 | 10 | 6 | 3 | 1 |
| N. meningitidis MC58 | 16 | 2 | 1 | 1 | 261 | 17 | 4 | 2 | 0 |
| N. gonorrhoeae FA1090 | 8 | 3 | 1 | 0 | 102 | 3 | 2 | 2 | 0 |
The copy number of CREE includes simple CREE contained inside complex CREE in order to show the flanking sequence features of all CREE.
Genomic distribution of CREE.
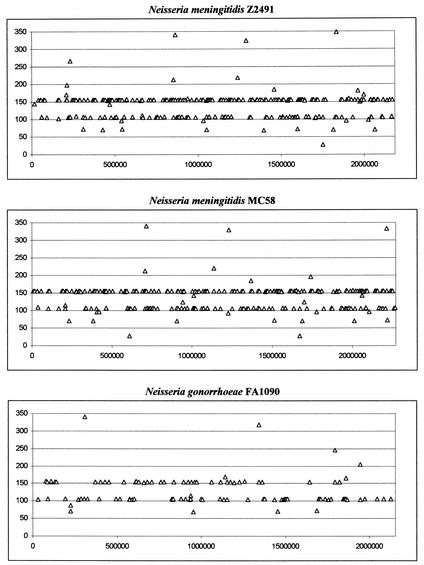
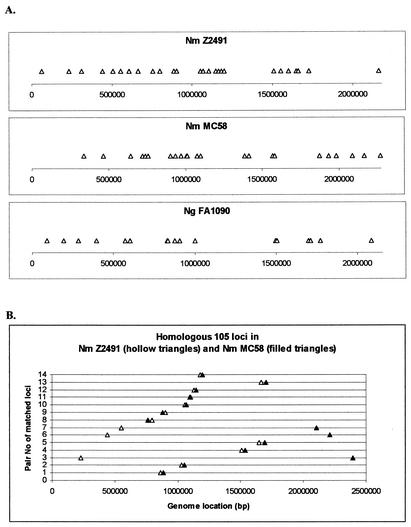
The distribution of all CREE in each genome of the three pathogenic Neisseria strains is presented in Fig. 3. From this presentation it is clear that CREE are distributed throughout the entire genome of each strain, although some local genomic regions have more of certain lengths of CREE than other regions. Whereas the overall distribution profiles are similar between the two N. meningitidis genomes, the CREE distribution in N. gonorrhoeae FA1090 is quite different from both N. meningitidis strains.
FIG. 3.
Genomic distribution of CREE.
CREE (107 bp) as 105-bp indels upstream of the sialyltransferase gene, lst.
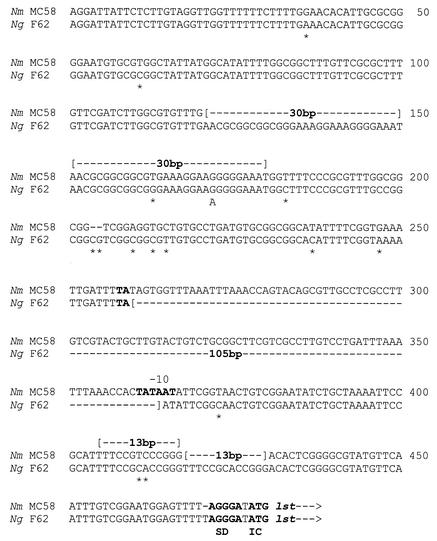
During our study of the contribution of variations in sequence upstream of the lst (5′-lst) to the differences in the amount of sialyltransferase activity expressed by N. meningitidis MC58 and N. gonorrhoeae F62, we noticed that a 105-bp sequence element is absent in the 5′-lst of N. gonorrhoeae F62 but present in the 5′-lst of N. meningitidis MC58 (Fig. 4). This 105-bp sequence element is a 107-bp CREE when combined with the TA dinucleotide at its 5′ flanking sequence. This result indicates that this 107-bp CREE reflects a 105-bp indel, a region of DNA that is present on the chromosome of an organism (insertion) but absent from closely related organisms (deletion), hence the word “indel.” Further investigation of more N. gonorrhoeae and N. meningitidis strains showed that this 105-bp indel (107-bp CREE) is present in the 5′-lst of all of the five N. meningitidis strains studied but in none of the 25 N. gonorrhoeae strains studied (web figure 1), suggesting the differential distribution of this 105-bp indel is a common difference between N. meningitidis and N. gonorrhoeae.
FIG. 4.
A 105-bp indel (107-bp CREE) upstream of the sialyltransferase gene, lst. DNA fragments absent or duplicated are indicated by “[--number of base pairs--]” within or above the sequence, respectively. The single base (A) in the imperfect repeat is written below the aligned sequences. Single nucleotide differences are indicated by an asterisk. The Shine-Dalgarno (SD) sequence, the −10 box, and the initiation codon (IC) for lst are in boldface. Nm, N. meningitidis; Ng, N. gonorrhoeae.
Identification of unique loci for 105-bp indels in three neisserial genomes.
The 105-bp indel upstream of lst in N. meningitidis MC58 was used as a query for ungapped BLASTN analysis against the databases of the three neisserial genomes. We identified 26, 23, and 17 copies of 105-bp indels in the genomes of N. meningitidis Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090, respectively (web tables 8 to 10). These numbers are in close agreement with those found in the FINDPATTERNS analysis (Table 2), except that the latter analysis identified one more 107-bp CREE in N. meningitidis MC58 and two more 107-bp CREE in N. gonorrhoeae FA1090. To reveal the genetic locations of the 105-bp indels in the three neisserial genomes, we used the 2,105-bp sequence segments representing each of the 105-bp indel plus 1 kb of flanking sequence on either side as queries and performed self- and reciprocal-BLASTN analyses (i.e., 2,105-bp sequences from one strain were used for BLASTN analysis against the genome sequence from which it was derived, as well as against the genome sequences of the other two strains). From these analyses, we identified 51 unique loci for a total of 66 copies of 105-bp indels in the three Neisseria genomes (Table 4). Among these 51 unique loci, only one is shared by all three pathogenic neisseriae. There are 14 more loci shared between the two N. meningitidis strains. The other loci are unique to each one of the three neisserial strains. Four 105-bp indel-associated loci of N. meningitidis Z2491 are unique, whereas three 105-bp indel-associated loci of N. meningitidis MC58 are unique. Two 105-bp indel-associated loci each from N. meningitidis Z2491 and N. meningitidis MC58 genomes do not show a significant match to the genomic of N. gonorrhoeae FA1090. Four 105-bp indel-associated loci of N. gonorrhoeae FA1090 are unique.
TABLE 4.
BLAST analysis of 105 bp loci among the three neisserial genomesa
| Locus |
N. meningitidis Z2491
|
N. meningitidis MC58
|
N. gonorrhoeae FA1090
|
|||
|---|---|---|---|---|---|---|
| 105No | Indel | 105No | Indel | 105No | Indel | |
| 1 | 1 | 105i | Not 105 | 103i | Not 105 | 104i |
| 2 | 2 | 105i | 23 | 105i | Not 105 | 105d |
| 3 | 3 | 105i | No match | NA | Not 105 | 105d |
| 4 | 4 | 105i | 21 | 105i | 9 | 105i |
| 5 | 5 | 105i | No match | NA | Not 105 | 105d |
| 6 | 6 | 105i | 20 | NA | Not 105 | 105d |
| 7 | 7 | 105i | No match | NA | No match | NA |
| 8 | 8 | 105i | No match | NA | Not 105 | 105d |
| 9 | 9 | 105i | Not 105 | Not 105 | 160i | |
| 10 | 10 | 105i | 3 | 105i | Not 105 | 92d |
| 11 | 11 | 105i | 5 | 105i | Not 105 | 105d |
| 12 | 12 | 105i | 6 | 105i | Not 105 | 105d |
| 13 | 13 | 105i | 7 | 105i | Not 105 | 105d |
| 14 | 14 | 105i | 8 | 105i | Not 105 | 31d |
| 15 | 15 | 105i | 9 | 105i | Not 105 | 105d |
| 16 | 16 | 105i | 11 | 105i | Not 105 | 105d |
| 17 | 17 | 105i | Not 105 | 174i | Not 105 | 176i |
| 18 | 18 | 105i | Q? | Not 105 | 105d | |
| 19 | 19 | 105i | Not 105 | 105d | Not 105 | 104i |
| 20 | 20 | 105i | 15 | Not 105 | 105d | |
| 21 | 21 | 105i | 12 | NA | Not match | NA |
| 22 | 22 | 105i | Not 105 | ND | Not 105 | 105d |
| 23 | 23 | 105i | 16 | Not 105 | 157i | |
| 24 | 24 | 105i | 17 | 105d | Not 105 | Q? |
| 25 | 25 | 105i | Not 105 | 155i | Not 105 | 105d |
| 26 | 26 | 105i | Not 105 | 239d | Not 105 | 103i |
| 27 | Not 105 | 106i | 1 | 105i | Not 105 | 105d |
| 28 | No match | 2 | 105i | Not 105 | ND | |
| 29 | Not 105 | 105d | 4 | 105i | Not 105 | 114d |
| 30 | No match | NA | 10 | 105i | Not 105 | Q? |
| 31 | Not 105 | 105d | 13 | 105i | No match | NA |
| 32 | Not 105 | 110d | 14 | 105i | Not 105 | 114i |
| 33 | Not 105 | Q? | 18 | 105i | Not 105 | ND |
| 34 | No match | NA | 19 | 105i | Not 105 | ND |
| 35 | Not 105 | Q? | 22 | 105i | Not 105 | ND |
| 36 | Not 105 | 105d | Not 105 | ND | 1 | 105i |
| 37 | Not 105 | ND | Not 105 | ND | 2 | 105i |
| 38 | Not 105 | 105d | Not 105 | 105d | 3 | 105i |
| 39 | Not 105 | Q? | Not 105 | Q? | 4 | 105i |
| 40 | Not 105 | 105d | Not 105 | 154i | 5 | 105i |
| 41 | No match | NA | No match | NA | 6 | 105i |
| 42 | No match | NA | No match | NA | 7 | 105i |
| 43 | Not 105 | 105d | Not 105 | 105d | 8 | 105i |
| 44 | Not 105 | 105d | Not 105 | 189i | 10 | 105i |
| 45 | Not 105 | 103i | Not 105 | 105d | 11 | 105i |
| 46 | Not 105 | 153i | Not 105 | 153i | 12 | 105i |
| 47 | Not 105 | ND | Not 105 | ND | 13 | 105i |
| 48 | Not 105 | 132i | Not 105 | 132i | 14 | 105i |
| 49 | No match | NA | No match | NA | 15 | 105i |
| 50 | No match | NA | No match | NA | 16 | 105i |
| 51 | Not 105 | 222i | Not 105 | 222i | 17 | 105i |
“Locus” refers to each unique 105-bp locus as represented by unique flanking sequence. “105No” reflects each 105 copy in each genome as originally assigned in web Tables 8 to 10. “Not 105” means the sequence match between two or three genomes does not include the 105 bp element. “No match” means there is no significant match (>80% nucleotide identity in over 351 bp). In the “Indel” columns, the “d” and “i” following a number indicate a “deletion” or an “insertion,” respectively. Here “deletion” and “insertion” merely means the presence or absence of a specific length of sequence element. “Q?” means the indel identification was not clear from the sequence alignment. “ND” means the indel identification was not done. NA, not applicable.
In most homologous loci, there is often an exact 105-bp indel that is either present or absent. However, some of the 105-bp indel-empty sites are filled with other lengths of DNA sequences (Table 4). These other indels range in length from 103 to 222 bp.
In general, 105-bp indels are scattered in each genome (Fig. 5A). However, the two N. meningitidis strains showed some similarity in the spacing pattern of some 105-bp elements. This similarity became clearer when the 14 matched 105-bp loci (Table 4) were plotted in pairs (Fig. 5B). Among these 14 pairs of matched 105-bp loci, 11 pairs are located in similar chromosomal locations. The other three pairs are in different chromosomal locations but the relative positions among these three loci are similar in the two N. meningitidis strains. When the DNA sequence segment containing these three 105-bp element loci was inverted, these three loci matched well between the two N. meningitidis genomes (data not shown). This result confirms that there is a major sequence inversion between the two N. meningitidis genomes (49).
FIG. 5.
Genomic locations of 105-bp indel and comparison between two N. meningitidis genomes. (A) 105-bp indels in each of the three neisserial genomes. (B) Fourteen homologous 105-bp indel loci in two N. meningitidis genomes when the two genomic sequences were aligned at the location of the 11th 105-bp indel.
TA direct repeats as a common feature in all loci of 105-bp indels.
Comparison of the 5′-lst regions from the various N. meningitidis and N. gonorrhoeae strains showed that a TA dinucleotide exists in target sequences where the 105-bp indel inserts (Fig. 4 and web figure 1). Further examination of all 105-bp indels in the genomes of the three Neisseria strains confirmed that a TA dinucleotide always exists in places where a 105-bp indel may be inserted (web table 11).
Genes and ORFs associated with or adjacent to 105-bp indels.
The homologous sequences matched by the BLAST analysis with the above 2,105-bp sequence queries were examined for genes and ORFs according to the published annotations for the two N. meningitidis genomes. Most 105-bp indels are intergenic (web tables 12 to 14), whereas some are located within ORFs; however, these ORFs are almost exclusively identified as transposases or insertion sequences except for two of the 17 copies of 105-bp indels in N. gonorrhoeae FA1090, which are inserted into ORFs other than those coding for transposases.
Importantly, some 105-bp indels are located close (<200 bp) to Neisseria virulence factors. For example, among the identifiable genes, 105-bp indels are associated with immunoglobulin A-specific serine endopeptidase (iga2), sialyltransferase (lst), outer membrane protein opcA, and class I outer membrane protein porA in the two N. meningitidis strains (web tables 12 and 13). The 105-bp indels are also associated with some metabolic genes such as glyceraldehyde-3-phosphate dehydrogenase (gapA and gapC) and acetate kinase (ackA) and also transporter genes for an ABC transporter ATP-binding protein. The 105-bp indels are adjacent to different sets of genes and ORFs in N. gonorrhoeae FA1090 (web table 14).
Detailed analysis of the N. meningitidis MC58 sequence using the ACEDB graphical interface revealed additional contextual information. In four instances (numbers 13, 16, 17 and 23) the 105-bp indels are located between convergent 3′ ends of genes, and a further six elements (numbers 3, 4, 5, 6, 11 and 19) are located remotely from likely promoter locations so that they are unlikely to affect expression. One (number 15) is inserted into a dead gene. Four elements (numbers 10, 12, 21, and 22) are present in the same location within copies of the IS1106 transposase. In seven instances (numbers 1, 2, 7, 8, 14, 18, and 20), the 105-bp indels are located such that they would be expected to form part of the promoter region of the associated gene, typically with the terminal TA forming part of a putative −10 element. The genes in which the 105-bp indels are likely to influence expression are (in the corresponding order): pilF (NMB0329), hypothetical proteins (NMB01387 and NMB0882), lst (NMB0922), a flavin reductase homologue (NMB1359), a hypothetical protein (NMB1782), and a peptidase homologue (NMB1877). The 105-bp indel associated with hypothetical protein NMB0882 is actually located between two divergent promoters but it is not located such that it would be expected to influence the adjacent cysT gene (NMB0881). Thus, 7 of 23 (ca. 30%) 105-bp indels in N. meningitidis MC58 may affect expression of associated genes.
DISCUSSION
We noticed discrepancies in the literature regarding CR and CE. For example, the 26-bp repetitive sequence originally reported by Correia et al. (8) was also named “N. gonorrhoeae inverted repeats” (49). CE (9) are sometimes termed “Correia full” (150 to 159 bp) or “Correia internal deletion” (∼104 bp) (41) and sometimes “unit-length (154 to 158 bp) nemis” (for Neisseria miniature insertion sequences) and “internally rearranged (104 to 108 bp) nemis” (37). Estimates of the abundance of CR and CE also vary depending on the methodologies and query sequences used (37, 41, 49).
In the present study, we first identified the most conserved region of CR as an 11-bp terminal sequence. Then we used this 11-bp sequence to formulate a sequence pattern that allows retrieval of any sequence bracketed by an inverted pair of this 11-bp sequence (within a specified overall length range and mismatch in the 11-bp sequence region). The FINDPATTERNS analysis performed in this way is much easier than BLASTN analysis for identifying CREE. This FINDPATTERNS analysis also gives more consistent and comparable data than the equivalent BLASTN analysis since it uses the same sequence pattern for identifying related sequences of varied lengths compared to using different queries in the BLASTN analysis. In fact, this FINDPATTERNS analysis is more powerful than the equivalent BLASTN analysis because it allows detection of greater diversity in the core regions and includes a wider range of lengths, i.e., more overall hits of more complex CREE. The accuracy of the FINDPATTERNS-based analysis was tested in a direct comparison with the BLASTN-based analysis of 107-bp CREE (105-bp indel). FINDPATTERNS analysis detected 26, 24, and 19 copies of 107-bp CREE, whereas BLASTN analysis retrieved only 26, 23, and 17 copies of 105-bp indels in N. meningitidis Z2491, N. meningitidis MC58, and N. gonorrhoeae FA1090, respectively.
In comparing the number of CR and the number of CREE, it is apparent that CREE are indeed the most common form of CE. This is especially true for the two N. meningitidis genomes in which almost all identified CR are found in CREE, usually as pairs of TIR. We made several interesting new observations in the present study. First, some longer CREE contain CREE of shorter lengths. Second, although the length profile of CREE shows several discrete clusters, there is often a continuous spreading of lengths within each cluster. Third, both the terminal regions of CREE and the core regions of CREE consist of distinctive sequence blocks. Fourth, the shortest CREE can be as short as 28-bp and comprise just the inverted CR.
Our analysis revealed that variations of CREE length reflect various combinations of several common sequence blocks (Fig. 2; see also web tables 2 to 7) and that CREE of the same length do not necessarily share the same sequence. In fact, many CREE lengths contain subtypes of sequences. These lengths include 159-, 156-, 155-, 106-, 96-, and 73-bp CREE in N. meningitidis Z2491 (web tables 2 and 5); 156-, 154-, 106-, and 105-bp CREE in N. meningitidis MC58 (web tables 3 and 6); and 155- and 105-bp CREE in N. gonorrhoeae FA1090 (web tables 4 and 7). The differences in the subtype sequences within these CREE are often due to the different combinations of the terminal inverted repeat sequence blocks and the core sequence blocks. However, the differences are sometimes also due to the presence of completely different core sequences. For example, the 341-bp CREE of N. gonorrhoeae FA1090 is different from the 341-bp CREE of the two N. meningitidis strains.
The CREE described in the present study do not contain any ORFs and cannot have any transposase activity. Thus, they are not insertion sequences by conventional definition (36). In this sense, we feel that it may be misleading to call CE (most of them are shown in the present study as CREE) as “nemis (Neisseria miniature insertion sequences)” (37). Instead, CREE may be better treated as indels, i.e., regions of DNA that are present on the chromosome of an organism but absent from closely related organisms (7). The experimental determination of DNA sequences upstream of sialyltransferase gene lst in multiple strains of N. meningitidis and N. gonorrhoeae revealed that a 107-bp CREE is actually a combination of a 105-bp indel with a TA dinucleotide in the flanking target sequence upstream of the 105-bp indel. Since all CREE have TA dinucleotides at their termini, it is reasonable to assume that all CREE are also made of target TA dinucleotides and indels of different lengths. This is supported by the observations of Abadi et al. that a 157-bp indel is located between the divergently transcribed mtrR and mtrC genes in N. meningitidis but not in the same region in N. gonorrhoeae (1). Upstream of this 157-bp indel in the N. meningitidis sequence is a TA dinucleotide. Thus, a 159-bp CREE is formed when this 157-bp indel combined with this flanking TA.
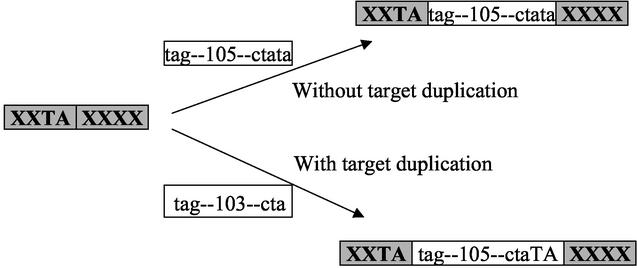
Currently, little is known about the origin or propagation of CREE. The discrete clustering of CREE around several lengths and the modular nature of CREE sequences suggest that the sequence diversity of CREE did not arise through sequential single base additions or deletions. Rather, different CREE might form via insertion or deletion of smaller sequence blocks plus some single-base-pair mutations. The question remains whether all CREE are mobile or whether some CREE are simply “genetic fossils” of past DNA mobilization events. It also remains to be determined how frequent and by what mechanism this mobilization occurs. For example, the 105-bp indel found upstream of the lst gene of N. meningitidis strains might be formed through an insertion of a 103-bp or a 105-bp sequence element (Fig. 6). If the inserted sequence originally exists as a 103-bp sequence element, then a duplication event of the target TA sequence must occur to add another TA to the 3′ end of the 103-bp element and form a 105-bp indel. The fact that all CREE contain a TA direct repeat at both ends supports the possibility of TA duplication. The observation of TA dinucleotides as the most common direct repeat found in the target sequence flanking CREE further suggests that repeated TA duplication may occur as multiple insertion events in the same target sites and that repeated insertion and target duplication events account for the spreading or diffusion of CREE length within each length cluster. Although target duplication during the insertion of noncoding short sequence elements has not been directly demonstrated in prokaryotes, such events have been described for short interspersed elements in eukaryotes (33). A recent study showed that a 107-bp interspersed repeat in Streptococcus pneumoniae could be mobilized via trans-mobilization by using the transposase of IS630-Spn1 and, interestingly, a TA dinucleotide exists upstream of this 107-bp repeat (39). Thus, it is possible for the noncoding CREE to be passively mobilized and short sequences such as a TA dinucleotide in the insertion target to be duplicated in the insertion process.
FIG. 6.
Hypothetical mechanism of the formation of a 105-bp indel. Shaded boxes represent “target” sequences with uppercase sequence letters. Hollow boxes represent “insertion” sequences with lowercase sequence letters. X, unspecified target sequence.
The phenomenon of shorter repetitive sequences being enclosed within longer repetitive sequences is widely recognized. The REP/palindromic units (19, 25, 47) enclose some core sequences to form REP elements such as the IRU or ERIC (28, 45) and bacterial interspersed mosaic elements (BIME) (22). In E. coli K-12 there are 581 REP sequences and 314 REP elements (4). Therefore, most REP sequences are components of longer repeats. REP sequences may share similar functions such as binding chromoid-associated protein (20), DNA gyrase (DNA topoisomerase) (51) and DNA polymerase I (21) and affecting mRNA stability (26). One subclass of BIME, called RIB (reiterative ihf BIME) (5) or RIP (repetitive IHF-binding palindromic elements) (40) is capable of binding integration host factor (IHF).
The relationship between CR and CREE resembles that between REP sequences and REP elements. The conservation of terminal regions in CR, and thus all CREE may reflect a highly conserved function for this DNA sequence fragment such as serving as a binding site for transposase. The 28-bp shortest CREE may represent the simplest indel structure on which a transposase can work. The diversification of core region may form the structural basis for the diverse function for various CREE. In this regard, there have been increasing reports of associating CREE with distinctive functions. For example, an earlier report demonstrated that a 106-bp CREE has promoter activity for the uvrB gene in N. gonorrhoeae (3). A study found that a 154-bp CREE appears to act as a transcriptional terminator (14). A recent study demonstrated that nemis (corresponding to some CREE described here) are cotranscribed with nearby cellular genes and subsequently processed at either one or both TIR (37). Since many CREE are located upstream of the coding regions of genes (web tables 1 and 12 to 14), the possibility for their involvement in a much wider range of gene regulation exists. For example, a 107-bp CREE is located near the regF-regG gene cluster (10), a 154-bp CREE is located downstream of the mtrE gene in N. gonorrhoeae (11), a 156-bp CREE exists in N. gonorrhoeae between divergently transcribed frpB and groES (48), a 152-bp CREE is located between carA and carB in both pathogenic and commensal Neisseria strains (34), and a 159-bp CREE is located between divergently transcribed mtrR and mtrC genes in N. meningitidis but not in the same region in N. gonorrhoeae (1). A total of 20 copies of CE were found in intergenic regions adjacent to different neisserial genes, including some virulence genes (14). However, the functional significance of these CREE remains to be experimentally determined.
Pathogenic Neisseria strains contain very “plastic” genomes (2, 6, 17) and are naturally competent for transformation (46), and they show evidence of extensive horizontal gene transfer (15, 32). The presence of a large number of interspersed DNA repeats in pathogenic genomes could affect the functional and evolutionary behaviors of these pathogens. The abundance of CREE (260, 255, and 98 nonredundant copies) and the percentage of nucleotides contained in these CREE (1.67, 1.55, and 0.59%) in the genomes of N. meningitidis Z1491, N. meningitidis MC58, and N. gonorrhoeae FA1090, respectively, are higher than that described for comparable intergenic repeats in other prokaryotic species. For example, the best-studied REP sequences and REP elements account for ca. 0.54% of the E. coli K-12 genome (4). The impact of these dispersed CREE in pathogenic neisserial genomes may be greater than currently realized because of their abundance and proximity to several virulence genes.
Pathogenic Neisseria strains present an interesting example of morphologically and biochemically similar organisms that cause very distinctive diseases, including life-threatening disseminated septicemia and meningitis caused by N. meningitidis and localized urogenital tract disease caused by N. gonorrhoeae. Analyses of the physical chromosomal maps of N. meningitidis (13, 16) and N. gonorrhoeae (12) show a high degree of conservation in overall gene organization between N. meningitidis Z2491 and N. gonorrhoeae FA1090 (13) and between N. meningitidis B1940 and N. gonorrhoeae FA1090 and MS11 (16). Previous DNA hybridization studies estimated that N. meningitidis and N. gonorrhoeae are 90% homologous in genes that are common to both species (24, 27). Whole genome sequence comparison showed that 91.2% of the 2,158 ORFs of N. meningitidis MC58 are similar to the ORFs of N. meningitidis Z2491 (49). There is no doubt that differences in the genes or ORFs (31, 42, 50) are important in determining the pathogenic differences between different Neisseria strains. However, the question is whether the distinctive pathogenic behaviors of different pathogens can be completely explained by these differences in the genes or ORFs. In this regard, the characterization of significant extragenic differences in one major family of neisserial repeat, CREE, among three strains of pathogenic Neisseria strains may offer some valuable additional insights.
A better understanding of CR and CREE may help the study of the evolutionary history and the phylogenetic classification of Neisseria. It is known that DNA loss via indel mutation is a determining factor for genome size reduction in eukaryotes (43). The relative genome sizes of three pathogenic Neisseria strains are 100% (N. meningitidis MC58), 96.1% (N. meningitidis Z2491), and 94.8% (N. gonorrhoeae FA1090). The contribution of CREE to the size variations of these three genomes is 0.08% between the two N. meningitidis genomes and ca. 1% between N. gonorrhoeae and N. meningitidis genomes. Thus, deletion or insertion of CREE alone can be a significant factor in altering the genome size of pathogenic Neisseria strains. CREE may serve as hot spots for genomic recombination and rearrangements, which may involve even larger segments of DNA and affect many different sets of genes, making CREE an important extragenic DNA component in the study of genome function.
Acknowledgments
N. meningitidis Z2491 sequence data were determined by the N. meningitidis strain Z2491 Sequencing Group at the Sanger Centre (http://www.sanger.ac.uk/Projects/N_meningitidis/). N. meningitidis MC58 sequence data were determined by the N. meningitidis MC58 Sequencing Group at the Institute of Genome Research (ftp://ftp.tigr.org/pub/data/n_meningitidis/). We acknowledge the Gonococcal Genome Sequencing Project (supported by USPHS/NIH grant AI38399) and B. A. Roe, L. Song, S. P. Lin, X. Yuan, S. Clifton, Tom Ducey, Lisa Lewis, and D. W. Dyer at the University of Oklahoma. N. gonorrhoeae FA1090 sequence data can also be obtained online (ftp://ftp.genome.ou.edu/pub/gono). The GenBank accession number for the completed N. gonorrhoeae FA1090 genome is AE004969, and this link gives the GenBank sequence file containing the sequence as deposited in GenBank. We thank B.-J. Tang for writing computer codes for the custom programs.
This work was supported in part by grants AI33505 and AI20897 to R.F.R. from the National Institutes of Health. N.J.S. is supported by a Wellcome Trust Research Fellowship in Medical Microbiology.
REFERENCES
- 1.Abadi, F. J. R., P. E. Carter, P. Cash, and T. H. Pennington. 1996. Rifampin resistance in Neisseria meningitidis due to alterations in membrane permeability. Antimicrob. Agents Chemother. 40:646-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bautsch, W. 1998. Comparison of the genome organization of pathogenic neisseriae. Electrophoresis 19:577-581. [DOI] [PubMed] [Google Scholar]
- 3.Black, C. G., J. A. M. Fyfe, and J. K. Davies. 1995. A promoter associated with the neisserial repeat can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J. Bacteriol. 177:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 5.Boccard, F., and P. Prentki. 1993. Specific integration of IHF with RIBs, a class of bacterial repetitive DNA elements located at the 3′ end of transcription units. EMBO J. 12:5019-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, J. G., and P. F. Sparling. 1984. Genetics of the gonococcus. Annu. Rev. Microbiol. 38:111-134. [DOI] [PubMed] [Google Scholar]
- 7.Comeron, J. M. 2001. What controls the length of noncoding DNA? Curr. Opin. Genet. Dev. 11:652-659. [DOI] [PubMed] [Google Scholar]
- 8.Correia, F. F., S. Innoye, and M. Inouye. 1986. A 26-base-pair repetitive sequence specific for Neisseria gonorrhoeae and Neisseria meningitidis genomic DNA. J. Bacteriol. 167:1009-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Correia, F. F., S. Innoye, and M. Inouye. 1988. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 263:12194-12198. [PubMed] [Google Scholar]
- 10.Delahay, R. M., B. D. Robertson, J. T. Balthazar, W. M. Shafer, and C. A. Ison. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143:2127-2133. [DOI] [PubMed] [Google Scholar]
- 11.Dempsey, J. A., and J. G. Cannon. 1994. Locations of genetic markers on the physical map of the chromosome of Neisseria gonorrhoeae FA1090. J. Bacteriol. 176:2055-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dempsey, J. A., A. B. Wallace, and J. G. Cannon. 1995. The physical map of the chromosome of a serogroup A strain of Neisseria meningitidis shows complex rearrangements relative to the chromosomes of the two mapped strains of the closely related species N. gonorrhoeae. J. Bacteriol. 177:6390-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Reuse, H., and M.-K. Taha. 1997. RegF, an SspA homologue, regulates the expression of the Neisseria gonorrhoeae pilE gene. Res. Microbiol. 148:289-303. [DOI] [PubMed] [Google Scholar]
- 14.Francis, F., S. Ramirez-Arcos, H. Salimnia, C. Victor, and J.-A. R. Dillon. 2000. Organization and transcription of the division cell wall (dcw) cluster in Neisseria gonorrhoeae. Gene 251:141-151. [DOI] [PubMed] [Google Scholar]
- 15.Frosch, M., and T. F. Meyer. 1992. Transformation-mediated exchange of virulence determinants by cocultivation of pathogenic neisseriae. FEMS Microbiol. Lett. 79:345-349. [DOI] [PubMed] [Google Scholar]
- 16.Gaher, M., et al. 1996. A physical and genetic map of Neisseria meningitidis B1940. Mol. Microbiol. 19:249-259. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs, C. P. 1996. Genome plasticity in Neisseria gonorrhoeae. FEMS Microbiol. Lett. 145:173-179. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, M., D. C. Watson, A. M. Cunningham, M. P. Jennings, N. M. Young, and W. W. Wakarchuk. 1996. Cloning of the lipooligosaccharide α-2,3-sialyltransferase from the bacterial pathogens Neisseria meningitidis and Neisseria gonorrhoeae. J. Biol. Chem. 271:28271-28276. [DOI] [PubMed] [Google Scholar]
- 19.Gilson, E., J.-M. Clement, S. Perrin, and M. Hifnung. 1987. Palindromic units a case of highly repetitive DNA sequences in bacteria. Trends Genet. 3:226-230. [Google Scholar]
- 20.Gilson, E., D. Perrin, J. M. Clement, S. Szmelcman, E. Dassa, and M. Hofnung. 1986. Palindromic units from E. coli as binding sites for a chromoid-associated protein. FEBS Lett. 206:323-328. [DOI] [PubMed] [Google Scholar]
- 21.Gilson, E., D. Perrin, and M. Hofnung. 1990. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 18:3941-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilson, E., W. Saurin, D. Perrin, S. Bachellier, and M. Hofnung. 1991. Palindromic units are part of a new bacterial interspersed mosaic element (BIME). Nucleic Acids Res. 19:1375-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gotschlich, E. C., M. Seiff, and M. S. Blake. 1987. The DNA sequence of the structural gene of gonococcal protein III and the flanking region containing a repetitive sequence. J. Exp. Med. 165:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guibourdenche, M., M. Y. Popoff, and J. Y. Riou. 1986. Deoxyribonucleic acid relatedness among Neisseria gonorrhoeae, N. meningitidis, N. lactamica, N. cinerea, and “Neisseria polysaccharea.” Ann. Inst. Pasteur Microbiol. 137B:177-185. [DOI] [PubMed] [Google Scholar]
- 25.Higgins, C. F., G. F.-L. Ames, W. M. Barnes, J. M. Clemen, and M. Hofnung. 1982. A novel interastronic regulatory element of prokaryotic operons. Nature 298:760-762. [DOI] [PubMed] [Google Scholar]
- 26.Higgins, C. F., R. S. McLaren, and S. F. Newbury. 1988. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene 72:3-14. [DOI] [PubMed] [Google Scholar]
- 27.Hoke, C., and N. A. Vedros. 1982. Taxonomy of the Neisseriae: deoxyribonucleic acid base composition, interspecific transformation, and deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 32:57-66. [Google Scholar]
- 28.Hulton, C. S., C. F. Higgins, and P. M. Sharp. 1991. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium, and other enterobacteria. Mol. Microbiol. 5:825-834. [DOI] [PubMed] [Google Scholar]
- 29.Kahler, C. M., and D. S. Stephens. 1998. Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin). Crit. Rev. Microbiol. 24:281-334. [DOI] [PubMed] [Google Scholar]
- 30.Kellogg, D. F., Jr., W. L. Peacock, Jr., L. Brown, and C. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klee, S. R., et al. 2000. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immun. 68:2082-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroll, J. S., K. E. Wilks, J. L. Farrant, and P. R. Langford. 1998. Natural genetic exchange between Haemophilus influenzae and Neisseria: intergenetic transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. USA 95:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labuda, D., E. Zietkiewicz, and G. A. Mitchell. 1995. Alu elements as a source of genomic variation: deleterious effects and evolutionary novelties, p. 1-24. In R. Maraia (ed.), The impact of short interspersed elements on the host genome. Springer Publishing, New York, N.Y.
- 34.Lawson, F. S., F. M. Billowes, and J.-A. R. Dillon. 1995. Organization of carbamoyl-phosphate synthase genes in Neisseria gonorrhoeae includes a large, variable intergenic sequence which is also present in other Neisseria species. Microbiology 141:1183-1191. [DOI] [PubMed] [Google Scholar]
- 35.Lupski, J. R., and G. M. Weinstock. 1992. Short, interspersed repetitive DNA sequences. J. Bacteriol. 174:4525-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzone, M., E. De Gregorio, A. Lavitola, C. Pagliarulo, P. Alifano, and P. P. Di Nocera. 2001. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic neisseriae. Gene 278:211-222. [DOI] [PubMed] [Google Scholar]
- 38.McGee, D. J., and R. F. Rest. 1996. Regulation of gonococcal sialyltransferase, lipooligosaccharide, and serum resistance by glucose, pyruvate, and lactate. Infect. Immun. 64:4630-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oggioni, M. R., and J. P. Claverys. 1999. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 145(Pt. 10):2647-2653. [DOI] [PubMed] [Google Scholar]
- 40.Oppenheim, A. B., K. Rudd, I. Mendelson, and D. Teff. 1993. Integration host factor binds to a unique class of complex repetitive extragenic DNA sequences in Escherichia coli. Mol. Microbiol. 10:113-122. [DOI] [PubMed] [Google Scholar]
- 41.Parkhill, J., et al. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 42.Perrin, A., X. Nassif, and C. Tinsley. 1999. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect. Immun. 67:6119-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrov, D. A., T. A. Sangster, J. S. Johnston, D. L. Hartl, and K. L. Shaw. 2000. Evidence for DNA loss as a determinant of genome size. Science 287:1060-1062. [DOI] [PubMed] [Google Scholar]
- 44.Rocha, E. P., A. Danchin, and A. Viari. 1999. Functional and evolutionary roles of long repeats in prokaryotes. Res. Microbiol. 150:725-733. [DOI] [PubMed] [Google Scholar]
- 45.Sharples, G. J., and R. G. Lloyd. 1990. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 18:6503-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stein, D. C. 1991. Transformation of Neisseria gonorrhoeae: physical requirements of the transforming DNA. Can. J. Microbiol. 37:345-349. [DOI] [PubMed] [Google Scholar]
- 47.Stern, M. J., et al. 1984. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37:1015-1026. [DOI] [PubMed] [Google Scholar]
- 48.Tauschek, M., C. W. Hamilton, L. A. Hall, C. Chomvarin, J. A. Fyfe, and J. K. Davies. 1997. Transcriptional analysis of the groESL operon of Neisseria gonorrhoeae. Gene 189:107-112. [DOI] [PubMed] [Google Scholar]
- 49.Tettelin, H., et al. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 50.Tinsley, C. R., and X. Nassif. 1996. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc. Natl. Acad. Sci. USA 93:11109-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, Y., and G. F. Ames. 1988. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc. Natl. Acad. Sci. USA 85:8850-8854. [DOI] [PMC free article] [PubMed] [Google Scholar]