Abstract
Together, the biosyntheses of histidine, purines, and thiamine pyrophosphate (TPP) contain examples of convergent, divergent, and regulatory pathway integration. Mutations in two purine biosynthetic genes (purI and purH) affect TPP biosynthesis due to flux through the purine and histidine pathways. The molecular genetic characterization of purI mutants and their respective pseudorevertants resulted in the conclusion that <1% of the wild-type activity of the PurI enzyme was sufficient for thiamine but not for purine synthesis. The respective pseudorevertants were found to be informational suppressors. In addition, it was shown that accumulation of the purine intermediate aminoimidazole carboxamide ribotide inhibits thiamine synthesis, specifically affecting the conversion of aminoimidazole ribotide to hydroxymethyl pyrimidine.
Cellular metabolism requires the control of flux through a complex network of pathways. This control is accomplished by regulation at several levels. Regulation of gene transcription or mRNA translation can control individual pathways. Allosteric control of the enzyme catalyzing the first committed step of a pathway is another mechanism used by cells to regulate biosynthetic pathways. Regulatory effects resulting from interactions between pathways are less well understood, despite the fact that many metabolites are present in multiple pathways in the cell.
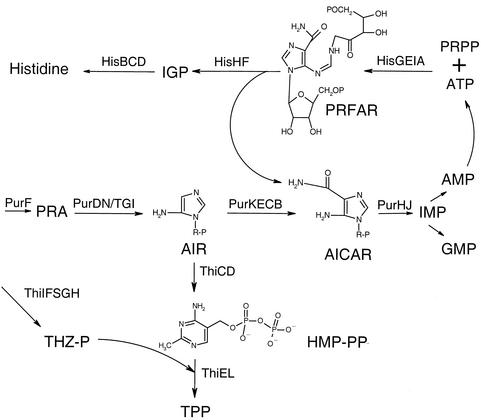
The expanding network of defined pathways that affect the biosynthesis of thiamine provides an attractive model system to investigate metabolic integration. As depicted in Fig. 1, the biosynthetic pathway for the hydroxymethylpyrimidine (HMP) moiety of thiamine pyrophosphate (TPP) branches off from that for purine mononucleotides at the metabolite aminoimidazole ribotide (AIR). Relevant to the work presented here is a subsequent purine intermediate, aminoimidazole carboxamide ribotide (AICAR), that is also a by-product of the biosynthesis of histidine.
FIG. 1.
Model pathways for metabolic integration studies. Schematically shown are the pathways for purine mononucleotide, histidine, and thiamine biosynthesis. The gene products involved are indicated in the relevant part of each pathway. Abbreviations: IGP, imidazoleglycerol phosphate; PRFAR, phosphoribulosyl-formimino-5-aminoimidazole carboxamide ribonucleotide; PRPP, 5′-phosphoribosyl-1-pyrophosphate; GTP, GMP; THZ-P, 4-methyl-5(β-hydroxyethyl)thiazole phosphate; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate.
The distribution of AIR at the purine-HMP branch point must account for the different level of purine mononucleotides and TPP required for growth (purines/TPP ratio, 1,000:1, based on auxotrophic requirements). The distribution of AIR between these pathways could be accomplished by kinetic parameters of the relevant enzymes; however, the first dedicated HMP biosynthetic enzyme is poorly understood, thus hampering direct tests of this possibility.
While thiamine and purine mononucleotides are synthesized by pathways diverging at the metabolite AIR, the cellular pool of AICAR arises from both purine and histidine biosynthesis and is used exclusively for purine mononucleotide synthesis (28, 29). Since AICAR enters purine mononucleotide synthesis after the purine-HMP branch point, no interaction between AICAR and HMP synthesis was anticipated. Hence, it was surprising to find that mutants blocked in the utilization of AICAR (purH mutants) required both purines and thiamine to grow (16, 58). The hypothesis proposed to explain this requirement posited that inactivation of purH would result in the accumulation of AICAR, which would be a strong negative allosteric effector of the PurF enzyme, which catalyzes the first step in purine mononucleotide biosynthesis. The demonstration that PurF was not always required for HMP synthesis prompted further analyses of these mutants (18, 40).
The studies reported in this paper were initiated to explain unanticipated phenotypes caused by mutations in the purine biosynthetic genes purI and purH. We show here that these phenotypes result from the integration between the purine, histidine, and TPP biosynthetic pathways. Our data predicted kinetic properties of the first committed step of HMP synthesis, identified two informational suppressors that had not been previously described, and explained previously observed phenotypes on the basis of pathway flux.
MATERIALS AND METHODS
Bacterial strains and culture media.
All strains used in this study are derivatives of Salmonella enterica LT2 and are listed with their genotypes in Table 1. Tn10d(Tc) refers to the transposition-defective mini-Tn10 described by Way et al. (55). MudJ refers to the defective transposon Mu dI1734 described elsewhere (12). Unless otherwise indicated, all strains were part of the lab collection or were constructed during the course of this work. The isolation of a mutant requiring less AIRs to satisfy a thiamine requirement has been reported (40). This phenotype has been shown to be caused by derepression of a kinase (STM4066) that is able to phosphorylate AIR riboside (AIRs). The causative mutation (stm4068-1) is a null allele of the Salmonella-specific open reading frame STM4068, a transcriptional repressor at ∼88 min on the S. enterica chromosome (M. Dougherty and D. M. Downs, submitted for publication). Multiply mutant strains were constructed and verified using classical and molecular genetic techniques. Transductions were performed as described previously (17). Nutritional requirements were determined by (i) liquid growth curves, (ii) replica plating on solid medium, and/or (iii) soft agar overlay analyses. Each of these techniques has been described previously (20, 41).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or description |
|---|---|
| Strains | |
| LT2 | Wild type |
| TT248 | hisC527(Am)cysA1348(Am) |
| DM2 | purH355 |
| DM42 | purI2152::MudJ |
| DM521 | zfb-8017::Tn10d(Tc)apurI2937 |
| DM531 | zfb-8017::Tn10d(Tc) purI2944 |
| DM539 | zfb-8017::Tn10d(Tc) purI2945 |
| DM929 | purF2085 purE884::Tn10d(Tc) |
| DM1216 | zfb-8017::Tn10d(Tc) purI2944 zje-8058::MudJbalaW221 |
| DM1472 | zfb-8017::Tn10d(Tc) purI3004 |
| DM1473 | zfb-8017::Tn10d(Tc) purI3005 |
| DM1474 | zfb-8017::Tn10d(Tc) purI3006 |
| DM1476 | zfb-8017::Tn10d(Tc) purI3008 |
| DM1479 | zfb-8017::Tn10d(Tc) purI3011 |
| DM1481 | zfb-8017::Tn10d(Tc) purI3013 |
| DM1482 | zfb-8017::Tn10d(Tc) purI3014 |
| DM1483 | zfb-8017::Tn10d(Tc) purI3015 |
| DM1936 | purF2085 |
| DM2093 | zfb-8017::Tn10d(Tc) purI3006 zxx-8087::MudJ sup-5851 |
| DM2095-DM2102 | zfb-8017::Tn10d(Tc) purI2944 alaW231-alaW238 |
| DM2625 | zfb-8017::Tn10d(Tc) purI3006 sup-5852 |
| DM2626 | zfb-8017::Tn10d(Tc) purI3006 sup-5853 |
| DM2627 | zfb-8017::Tn10d(Tc) purI3006 sup-5854 |
| DM2628 | zfb-8017::Tn10d(Tc) purI3006 sup-5855 |
| DM2629 | zfb-8017::Tn10d(Tc) purI3006 sup-5856 |
| DM2630 | zfb-8017::Tn10d(Tc) purI3006 sup-5857 |
| DM2631 | zfb-8017::Tn10d(Tc) purI3006 sup-5858 |
| DM2633 | zfb-8017::Tn10d(Tc) purI3006 sup-5859 |
| DM5976 | purH355 purE884::Tn10d(Tc) |
| DM6068 | purH355 hisG1102 |
| DM6069 | purH355 |
| DM6123 | purG2324::MudJ zxx9126::Tn10d(Tc) stm4068-1 purH355 |
| DM6124 | purG2324::MudJ zxx9126::Tn10d(Tc) stm4068-1 |
| Plasmids | |
| pJZ531 | Cmr 1.3-kb insert with alaW in HindIII site of pSU19 |
| pJZala1 | Cmr 0.6-kb insert containing alaW in HindIII/SmaI of pSU19 |
| pJZG252A | Cmr 1.2-kb insert of G252A purI gene in SmaI site of pSU19 |
| pLUX1c | Cmr wild-type luxAB genes in pCAM203 |
| pLUX2c | Cmr includes an amber codon at position 13 of luxB gene |
| pLUX3c | Cmr includes an ochre codon at position 13 of luxB gene |
| pLUX4c | Cmr includes an opal codon at position 13 of luxB gene |
No-carbon E medium supplemented with 1 mM MgSO4 (15, 53) and a carbon source (11 mM) was used as a minimal medium, and Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) was used as a rich medium. Unless indicated otherwise, glucose was used as sole carbon source. Difco BiTek agar was added (15 g/liter) for solid media. Adenine and thiamine were included in media as needed, to final concentrations of 0.4 mM and 0.5 μM, respectively. Chloramphenicol was added as needed to the following concentrations: 20 μg/ml (rich) or 4 μg/ml (minimal). Unless noted, chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.). AIRs was provided by M. Dougherty and synthesized by the protocol of Bhat and colleagues (7, 26, 34).
PCR amplification and DNA sequencing.
The wild-type purI from S. enterica was sequenced from a previously identified clone (p42) at least twice on both strands (GenBank accession number U68765) (J. L. Zilles and D. M. Downs, unpublished data). purI was amplified from the chromosome of strains containing purI mutations by using primers based on the wild-type sequence. Amplification was performed using Vent (exo−) polymerase (New England Biolabs) in a Thermolyne Temperature-Tronic thermocycler. The PCR products were purified using the Qiaquick gel extraction kit (Qiagen) and sequenced by the University of Wisconsin Biotechnology Center Nucleic Acid and Protein Facility (Madison, Wis.). Additional primers used in sequencing were also generated from known sequences and were synthesized either by Genosys (The Woodlands, Tex.) or by the University of Wisconsin Biotechnology Center Nucleic Acid and Protein Facility. Sequence data were examined using EditView (ABI Prism; Perkin-Elmer) and aligned using SeqEd (Applied Biosystems). For each strain, the complete coding region of purI was sequenced from both strands from at least two independent PCRs. The purI mutants sequenced are shown, grouped by phenotypic class, in Table 2 with the mutation(s) and the predicted amino acid changes.
TABLE 2.
purI mutations and predicted amino acid changes, by mutant class
| Class and allele | Mutation, at nucleotide no.a | Amino acid change |
|---|---|---|
| Pur− Thi−, class 1 | ||
| purI2944 | G-A at 755 | G252D |
| purI2945 | G-A at 755 | G252D |
| purI3005 | G-A at 386 | G129D |
| purI3008 | G-A at 386 | G129D |
| purI3013 | G-A at 386 | G129D |
| purI3015 | G-A at 755 | G252D |
| Pur− Thi−, class 2 | ||
| purI2937 | C-T at 292 | Q98UAG |
| purI3004 | C-T at 245 & C-T at 850 | A82V and Q284UAA |
| purI3006 | C-T at 625 | Q209UAA |
| purI30014 | C-T at 823 | Q275UAG |
| Pur− Thi+ | ||
| purI2938 | G-A at 828 | W276UGA |
| purI3011 | C-T at 677 | T226I |
Numbering began with the first nucleotide of the purI coding region or the first amino acid of the PurI protein.
Cloning an alaW mutation.
A MudJ insertion 98% linked to a suppressor mutation designated apbB76 (and referred to as alaW throughout this report) was described previously (60). This zje-8058::MudJ was replaced by a MudP and a MudQ, allowing isolation of chromosomal DNA enriched for the region around alaW as previously described (6, 57). The DNA was digested with HindIII and ligated into HindIII-digested pSU19 (Cmr). Restriction enzymes and DNA ligase were purchased from Promega (Madison, Wis.). The ligation mix was introduced into DM531 (purI2944) by electroporation using an Escherichia coli Pulser (Bio-Rad), selecting Cmr and scoring Thi+ (growth in the absence of thiamine). In both the initial selection and after reconstruction of the strain, pJZ531 allowed thiamine but not purine mononucleotide synthesis in DM531 (purI2944). pJZ531 was sequenced by the dideoxy method (44), using a Sequenase 2.0 kit (U.S. Biochemical Corp.). The sequence was compared to known sequences by using the BLAST program (1).
Sequences downstream of alaW included on pJZ531 were amplified from the chromosome from wild-type and mutant strains and sequenced as described above for purI. Primers were designed based on the sequence of the pJZ531 insert.
pJZala1 was constructed by PCR amplification of alaWX using pJZ531 as a template. A primer hybridizing to pSU19 sequence was used upstream of alaWX (−40 primer, 5′ GTTTTCCCAGTCACGAC) and the downstream primer was designed from pJZ531 sequences (5′ GCAGAAGGCAGTAAGAAT). The resulting PCR product, containing alaWX and 200 bp of downstream sequence, was blunt-end ligated into SmaI-digested pSU19, and the ligation mix was electroporated into DM531 (purI2944), selecting Cmr and scoring Thi+.
Site-directed mutagenesis.
The glycine codon at position 252 of the PurI protein was changed to an alanine codon by site-directed mutagenesis using the mega-primer method as described previously (3). (Primers were as follows: no. 1, 5′ AAGCTTTTCAATAACCACACGCTG; no. 2, 5′ GGCGGCTTTCTGCGTCGG; and no. 3, 5′ CCTGTCGTAAACCAAGTGCTG.) The final resulting PCR product was blunt-end ligated into SmaI-digested pSU19 and electroporated into a purI insertion mutant (DM42), selecting Cmr and replica printing to assess growth on minimal medium supplemented with adenine. A resulting clone (pJZG252A) was confirmed by sequencing. Additional phenotypic tests were performed by replica printing.
Mapping supUAA/UAG mutations.
A MudJ insertion linked to the supUAA/UAG locus (previously designated apbD) had been identified previously (60). A MudJ-specific primer and an arbitrary primer were used to amplify sequences flanking this MudJ as previously described (11, 38). The resulting PCR product was sequenced and compared to the E. coli (and subsequently the S. enterica) genome sequence by using the BLAST program (1). The position of the MudJ in the envD gene was confirmed by cotransduction of the MudJ and an insertion mutation in the fis gene.
Western blot analysis.
An amino-terminal histidine tag was added to the wild-type S. enterica PurI protein by using the pET14-b vector, and the protein was purified via a nickel affinity column as described by Novagen. The purified protein was used to generate polyclonal antibodies at the animal care center at the University of Wisconsin Medical School. Western blotting was performed and detection used the colorimetric alkaline phosphatase reaction as described elsewhere (2, 27). Comparison of strains containing purI in multicopy with purI null mutants confirmed the specificity of the antibodies.
RESULTS
Analysis of purI mutants determined the maximum flux requirement for thiamine synthesis.
Previous work identified 18 independent purI point mutants with a Pur− phenotype (60). These mutant strains fell into three phenotypic classes. Two of the purI mutants required purines but not thiamine for growth (phenotypically Pur− Thi+). The other sixteen mutants required both purines and thiamine for growth, but were further classified based on suppression analysis. Ten purI mutants became phenotypically Pur− Thi+ in the presence of one of two extragenic suppressor mutations, indicating a restored ability to synthesize thiamine. The remaining six Pur− Thi− purI mutants were not affected by either suppressor mutation and reverted only to Pur+ Thi+ by mutation(s) in purI. Molecular characterization of the purI mutants was initiated to determine the mechanism of thiamine synthesis in the suppressed strains.
Class 1 suppressible Pur− Thi− purI mutants contain missense alleles.
Six independently isolated purI mutants with a Pur− Thi− phenotype regained the ability to synthesize thiamine if a single additional mutation was present (60). Work presented here demonstrated that the lesions responsible for suppression are alleles of alaW and they are referred to as such throughout. The purI gene in each of these strains contained a single base pair change, from codon GGC to GAC, resulting in a glycine-to-aspartate substitution in the protein sequence. Three of the mutations affected amino acid 129 (purI3005, purI3008, and purI3013) and three affected amino acid 252 (purI2944, purI2945, and purI3015) (Table 2). The independent isolation of each lesion three times was surprising, since only 18 Pur− mutants were examined and their selection was based solely on the inability to grow in the absence of purines.
Missense suppressor mutations mapped in alaW.
A suppressor locus was defined by spontaneous mutations that eliminated the thiamine requirement in the purI2944 mutant (class I) (60). Nine independent suppressor mutations were isolated; all nine mapped in the same chromosomal region and suppressed the same purI mutations. Using a variation of MudP/Q technology (57), a clone (pJZ531) that could suppress the thiamine requirement of the relevant purI mutants was identified, confirmed by reconstruction, and sequenced. Sequence analysis determined that the insert in pJZ531 began 8 bp upstream of alaW, containing all of alaW and alaX in addition to downstream sequences for a total insert size of 1.3 kb. The chromosomal location of the cloned DNA (based on the E. coli and S. enterica genome sequence) was consistent with previous linkage data and suggested that pJZ531 contained the mutant suppressor locus, not a multicopy suppressor. These results also indicated that the suppressor mutation was dominant.
The only complete genes within the insert of pJZ531 were promoterless alaWX, a tandem repeat of an alanine tRNA gene (10). The orientation of alaWX was such that transcription could result from the lacZ promoter on the original vector. Compared to both the E. coli and S. enterica sequences, the alaW tRNA gene cloned from the suppressed strain had a single base change in the second position of the anticodon (GGC to GTC). This mutation would result in a tRNA that should insert alanine at GAC codons. This finding was consistent with the fact that the relevant purI mutants had GAC codons at the mutant position in purI. A plasmid was constructed that contained the mutant copy of alaW without downstream sequences (pJZAla1). This plasmid was able to confer the Thi+ phenotype in strains containing purI2944 (G252D), demonstrating that the mutation in alaW was sufficient to restore thiamine synthesis in this class I strain.
Missense suppression results in functional PurI.
A simple interpretation of the above data was that the G252D PurI protein was not functional, while the suppressor-generated G252A PurI protein was at least partially functional. A strain containing a plasmid-encoded G252A protein as the only source of PurI was prototrophic (Pur+ Thi+) (data not shown). This result supported a conclusion that the suppressed phenotype was the result of a low level of functional PurI protein. An efficient missense suppressor would be expected to cause significant growth defects not found with the alaW suppressor. From the above results we concluded that a low level of functional PurI protein was sufficient to meet the cellular thiamine but not purine requirement.
Class 2 suppressible Pur− Thi− purI mutants carry nonsense alleles.
The purI gene was sequenced from four mutants able to synthesize thiamine only in the presence of an extragenic suppressor mutation (supUAA/UAG) distinct from the alaW lesion. Three of the suppressed strains contained mutations changing glutamine codons to termination codons at different residues in the PurI protein (Q98UAG, Q209UAA, and Q275UAG in purI2937, purI3006, and purI3014, respectively). The fourth mutant contained two mutations in the purI gene, predicted to cause A82V and Q284UAA changes in the protein sequence (purI3004). Because the alanine at position 82 was not conserved across species and other mutants in this phenotypic class had premature termination codons, we assumed, but did not rigorously show, that the Q284UAA change was the causal mutation.
Identification of a low-efficiency nonsense suppressor.
Each of the four mutants suppressed by supUAA/UAG contained a UAG (amber) or a UAA (ochre) termination codon in the purI coding sequence (Table 2), mutant codons not found in purI mutants of any other phenotypic class. From this we presumed that supUAA/UAG was an ochre suppressor. However, the presence of the supUAA/UAG mutation did not eliminate the requirement for either histidine or cysteine in strain TT248, which contains both hisC527(Am) and cysA1348(Am) mutations. This result was significant, since previously described S. enterica ochre suppressors were isolated in this strain (56). More efficient suppression might be required to restore amino acid synthesis than for thiamine, so the effect of the well-characterized amber suppressor supD501 on the phenotype of two purI amber mutants was assessed. Both purI2937 supD501 and purI3014 supD501 double mutants were prototrophic. When supD501 was introduced into strains carrying the ochre mutations (purI3004, purI3006), neither the thiamine nor the purine requirement was eliminated.
If the supUAA/UAG mutations were nonsense suppressors with a lower efficiency than supD501, it would explain why (i) supD mutations were not identified in our screen for Pur− Thi+ revertants, and (ii) supUAA/UAG mutations were not identified in previous screens utilizing suppression of an amber mutation in an amino acid biosynthetic gene. Suppression by the supUAA/UAG mutation was quantified in a luciferase system, using constructs containing different termination codons at the same position in the luxB gene (45). Results from these analyses, shown in Table 3, demonstrated that the supUAA/UAG mutation allowed readthrough with an efficiency of below 1%. As expected, the supD allele, which restored both purine mononucleotide and thiamine synthesis to purI amber mutants, caused readthrough with a much higher efficiency (74%). These data demonstrated that a low level of functional PurI (perhaps ≤1%) was sufficient to result in prototrophy for thiamine but not purine. Nucleotide sequences flanking a linked MudJ insertion placed the supUAA/UAG locus at 73.4 min on the S. enterica chromosome, in close proximity to the rrnD rRNA operon. In S. enterica this operon encodes two tRNAs: thrV, also found in E. coli, and a glutamate tRNA (STM3397) which is not present in E. coli. The glutamate tRNA was assumed to be the site of the lesion, since a single base substitution in the TTC anticodon to a TTA anticodon would generate a tRNA able to read the UAA/UAG codons in the relevant mutant class.
TABLE 3.
Quantification of nonsense suppression by the supUAA/UAG mutation
| Codon | Efficiency of suppressiona (%) in:
|
||
|---|---|---|---|
| Wild type | supUAA/UAG | supD | |
| UAG (amber) | 0.13 | 0.91 | 74 |
| UAA (ochre) | 0.02 | 0.83 | 0.03 |
| UGA (opal) | 0.64 | 0.59 | 1.23 |
Efficiency of suppression was calculated as the luminescence of a luxB nonsense mutant/luminescence of a wild-type lux allele in the same background.
Mutations in purI alone can result in a Pur− Thi+ phenotype.
The sequence of the purI gene from mutants with a Pur− Thi+ phenotype (containing mutant alleles purI2938 and purI3011) was determined. In each case a single nucleotide change was found in the purI coding sequence, resulting in predicted changes in the PurI protein of W276UGA and T226I, respectively. S. enterica is known to contain weak UGA suppressor activity (42). As shown in Table 3, the background level of UGA suppression in a wild-type strain was roughly the level of UAG suppression by the supUAA/UAG mutations (0.64 versus 0.91%). Therefore, the W276UGA mutant was likely to contain enough functional PurI protein to allow thiamine synthesis; growth experiments supported this conclusion (data not shown).
Lesions in purH result in a requirement for the pyrimidine moiety of thiamine.
From the data above, we concluded that the Pur− Thi+ phenotypes reported here and elsewhere for point mutations in purI and purG (54, 60) reflected the ability to maintain low-level flux through the purine mononucleotide biosynthetic pathway and were not a consequence of additional metabolic redundancy. While this explanation was sufficient for lesions in the steps prior to the AIR (Fig. 1), it failed to explain a similar phenotype reported for mutations in purH, a gene whose product functions past the branch point to thiamine synthesis (16, 32, 54, 58). Phenotypes associated with multiple independent mutations in purH were assessed, and the allele used herein (purH355) was found to be representative. From our work and other reports in the literature, the following phenotypes have been identified for mutations (including null mutations) in purH: (i) requirement for a source of purines, (ii) requirement for the pyrimidine moiety of thiamine, and (iii) ability of histidine or pantothenate to satisfy the thiamine requirement. Since purH is the first gene in an operon with purD, encoding an enzyme active prior to the branch point, it is important to note that a plasmid containing purD did not affect the phenotypes of purH mutants reported here. Thus, polarity was not considered a feasible explanation for the thiamine requirement of purH mutants (data not shown).
Accumulation of AICAR in a purH mutant results in a thiamine requirement.
Mutants lacking purH accumulate the purine intermediate AICAR, a metabolite also generated as a by-product of histidine biosynthesis (46) (Fig. 1). Exogenous histidine has been reported to eliminate the thiamine requirement of a purH mutant (16, 30). It was formally possible that histidine could interact directly with the biosynthetic pathway to restore thiamine synthesis. Alternatively, the allosteric inhibition of HisG caused by histidine (31) could reduce the AICAR accumulation in a purH mutant. An allele of hisG that renders the enzyme insensitive to allosteric regulation by histidine (hisG1102) (47) was used to distinguish between these possibilities. Isogenic purH strains with or without the hisG1102 allele were generated. Growth experiments with both liquid and solid media showed that histidine did not restore growth when the hisG1102 allele was present (data not shown). This result was consistent with a model in which (i) accumulated AICAR caused the thiamine requirement of purH mutants, and (ii) histidine prevented this accumulation by reducing the flux through the histidine biosynthetic pathway. The presence of pantothenate (in addition to adenine) restored growth of both strains, suggesting that pantothenate antagonized AICAR activity, not its accumulation (data not shown).
The thiamine requirement of a purH mutant is not mediated through PurF.
The position of AICAR in the purine pathway indicated its effect on thiamine synthesis was indirect. Past workers suggested that the site of this effect was PurF, which was shown to be inhibited allosterically by a number of purine nucleotides (32, 35, 36). In vitro studies with purified PurF (33, 52, 59) have not directly addressed an allosteric role for AICAR (H. Zalkin, personal communication). The finding that mutants lacking PurF are conditional HMP auxotrophs (40), in addition to work here demonstrating that ≤1% of wild-type levels of flux provided sufficient thiamine synthesis for growth, prompted us to readdress the thiamine requirement caused by purH lesions.
If inhibition of PurF activity was responsible for the thiamine requirement, purH mutants should be able to synthesize thiamine under conditions where PurF is dispensable for this synthesis. The thiamine requirement of a purH mutant was assessed under three conditions that allow thiamine-independent growth of a purF mutant. In each case the purH mutant retained a requirement for thiamine, while the purF mutant did not.
(i) Nonglucose carbon sources.
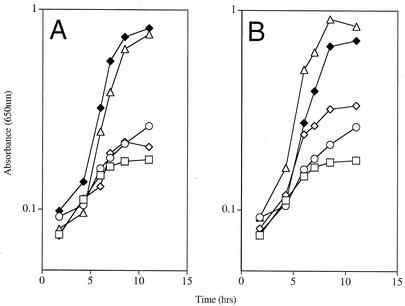
A purF mutant can grow in the absence of thiamine on a number of nonglucose carbon sources due to the contribution from the oxidative pentose phosphate pathway (20, 41). Under these conditions, PurF-independent phosphoribosylamine (PRA) synthesis is increased. Data shown in Fig. 2A demonstrated that purH mutants retained a requirement for thiamine when gluconate was used as sole carbon source. A number of additional carbon sources that support PurF-independent thiamine synthesis were tested, and purH mutants were also unable to grow in the absence of exogenous thiamine on these sources (data not shown).
FIG. 2.
purH mutants require thiamine under broad conditions. (A) The effect of carbon source on the thiamine requirement of DM2 (purH355) and DM1936 (purF2085) in minimal media and adenine is shown. Cultures of DM2 were grown with glucose (□) or gluconate (⋄) as sole carbon source. Also shown is growth when gluconate medium was supplemented with thiamine (♦). Cultures of DM1936 were grown with glucose (○) or gluconate (▵) as sole carbon source. (B) The effect a purE null mutation on the thiamine requirement of a purH355 and a purF2085 mutant in glucose medium is shown. Cultures of DM929 (purF purE mutant) (▵) and DM5976 (purH purE mutant) (⋄) were grown in the absence of thiamine. Growth of DM5976 in the presence of thiamine is shown (♦). Included for comparison are the purH (□) and purF (○) mutants in glucose medium, as in panel A.
(ii) Blocking diversion of AIR to purines.
Blocking PurE restores thiamine synthesis in a purF mutant on glucose medium due to an increased accumulation of AIR that is sufficient to satisfy the cellular requirement for thiamine (Fig. 1) (41). The same purE mutation was unable to fully restore thiamine-independent growth to a purH mutant (Fig. 2B). This result suggested that either PurF-independent PRA synthesis was decreased or the conversion of AIR to HMP was constrained in the purH mutants.
(iii) Extragenic suppressors.
In previous work, a mutation in yjgF was identified that restored thiamine synthesis in purF mutants on glucose medium and eliminated the requirement for the oxidative pentose phosphate pathway in this synthesis under other conditions (21). A mutation in yjgF had no effect on the thiamine-independent growth of a purH mutant (data not shown), suggesting that lack of PRA was not the problem in the purH mutant strains.
Elevated levels of AICAR reduce the conversion of AIR to HMP.
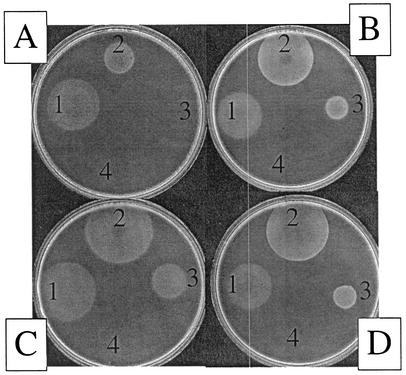
The above results showed that the thiamine requirement of a purH mutant could not be due to inhibition of PurF. The step(s) involved in the conversion of AIR to HMP was genetically isolated from the rest of the pathway, allowing us to test this step(s) as a possible target for the effect of AICAR. A purG mutation, introduced to eliminate de novo synthesis of AIR, made growth of the resulting strains dependent on both purines and thiamine. In these strains, the role of purine biosynthetic steps in synthesis of HMP can be bypassed by exogenously providing AIR riboside (AIRs) (Fig. 1) (37, 41). A recent result showing that overexpression of a cellular kinase, by mutating its repressor (stm4068-1), reduced the AIRs needed to satisfy an HMP requirement 94-fold and made these experiments feasible (39; Dougherty and Downs, submitted). Starting in a double mutant background (purG stm4068-1), isogenic strains differing by a purH mutation were generated. The resulting strains DM6124 (purG stm4068-1) and DM6123 (purG stm4068-1 purH) required both purines and thiamine. In each strain, exogenous AIRs could be provided to satisfy the HMP requirement. The amount of AIRs required to satisfy the thiamine requirement in these strains was titrated, and the results are shown in Fig. 3. Both strains responded similarly to thiamine on all growth media (Fig. 3 and data not shown). However, the purH-containing strain (DM6123) required ∼10-fold more AIRs to satisfy the thiamine requirement than did the isogenic strain DM6124 when both were plated on medium with adenine (compare Fig. 3A and D). Significantly, when plated on medium with adenine and histidine, the AIRs requirement of the purH mutant was reduced to the level required by the isogenic wild-type strain (compare Fig. 3B and D). Addition of pantothenate similarly reduced the level of AIRs required by DM1623 to that of the wild-type strain (Fig. 3C). This latter result narrowed the site of the previously described effect of coenzyme A on thiamine synthesis (17, 24, 43) to a role in the conversion of AIR to HMP, possibly due to its structural similarity to AIR.
FIG. 3.
Accumulation of AICAR impairs the conversion of AIR to HMP. The nutritional phenotype of strains DM6123 (purG2324:: MudJ stm4068-1 zxx9126::Tn10d purH355) and DM6124 (purG2324::MudJ stm4068-1 zxx9126::Tn10d) were determined. In each case soft agar containing cells of DM6123 (A to C) or DM6124 (D) was overlaid on a minimal glucose plate with adenine (0.4 mM). In addition to adenine, plate B contained histidine (50 μM) and plate C contained pantothenate (0.7 μM). On top of the overlay, additional supplementation was provided; the numbers 1 to 4 represent the position of thiamine (50 pmol in 0.5 μl) (1), 1 μl of a stock solution of AIRs (SS) (2), 1 μl of 0.1× SS (3), and 1 μl of 0.01× SS (4). A positive growth response is indicated by turbidity after overnight incubation at 37°C. The concentration of AIRs was estimated to be ∼300 mM using both bioassay (39) and the Bratton and Marshall method of quantification (9).
DISCUSSION
Connections among the biosynthetic pathways for purine mononucleotides, histidine, and thiamine have been recognized for decades. Subsequent work defined the biochemical basis for this integration (Fig. 1) and explained the nutritional phenotypes of most mutant strains. In addition, a number of recent studies aimed at identifying metabolic pathways or processes that affect the ability of the cell to synthesize thiamine have defined a larger metabolic network centered around these pathways (4, 5, 13, 14, 19, 23-25, 39, 40, 48). In reviewing past literature and our recent studies, the phenotypes of several mutant strains could not be explained simply on the basis of the biochemical integration that had been described. The work presented here began as an investigation of phenotypically anomalous purI mutants and the unexpected thiamine requirement of purH mutants. These studies have defined previously reported phenotypes in the context of the integration between the purine mononucleotide, histidine, and thiamine biosynthetic pathways. In addition, this work identified two new informational suppressor mutations.
AIR levels control the flux through the purine-thiamine branch point.
At the time these studies began it was formally possible that redundancy in the purine mononucleotide biosynthetic pathway went beyond that already described for PurF (18, 41). In fact, evidence of thiamine synthesis in purI mutants requiring purines for growth initially seemed to support this possibility (60). The molecular characterization of purI mutants described here clarified the mechanism of thiamine synthesis in these mutants, determining that cells could be proficient at thiamine, but not purine, biosynthesis with a low level of metabolic flux through the pathway. This work supported the conclusion that ≤1% of the wild-type levels of PurI are enough to allow sufficient thiamine, but not purine, synthesis for growth.
AICAR has a regulatory effect on synthesis of HMP.
Previous work had defined a biochemical connection between the purine mononucleotide and histidine biosynthetic pathways through the metabolite AICAR. Data presented here suggest that this metabolite also affects the synthesis of thiamine. Previous work indicated that strains blocked in purH accumulated AICAR and required the HMP moiety of thiamine (16, 54). Results presented here showed that AICAR does not mediate its effect on thiamine synthesis by regulating PurF activity but by negatively affecting the conversion of AIR to HMP. Since the transcription of the thi operon was not increased in a purH mutant (data not shown), we suggest that the effect of AICAR is posttranscriptional. Neither the biochemistry of this conversion nor the enzyme(s) involved has been rigorously defined, and thus it is difficult to suggest a mechanism or more precise site for the target or the AICAR effect. However, AICAR has been implicated previously in regulation, both as a negative effector of cytochrome terminal oxidase production in Rhizobium etli (50) and as a proposed alarmone for C-1-folate deficiency (8). The work presented here has expanded our model system for metabolic integration by defining additional parameters of the interaction between thiamine and purine mononucleotide synthesis, identifying a new connection between histidine and thiamine biosynthesis, and defining the part of the thiamine pathway that is affected by coenzyme A levels in the cell.
Selection results in the ability to differentiate between weak and strong informational suppressors.
Characterization of alaW alleles described here was the first demonstration of a missense suppressor derived from an alanine tRNA. The efficiency of suppression by the alaW alleles was not quantified, but it is likely to be low since the mutant strains showed no significant growth defect. Previously described missense suppressor alleles of glyV and glyW were not recovered in our screen, presumably because they suppress too efficiently and would thus cause a Pur+ Thi+ phenotype (22, 51). Despite repeated attempts, we were unable to assess the effects of existing glyV and glyW suppressors due to their mutator phenotype (49). An additional mutation that suppressed UAA and UAG but not UGA codons was identified by this work. This mutation mapped to a region that, in S. enterica, carries a glutamate tRNA, and thus a single base substitution in the anticodon would result in a suppressor with the noted specificity.
Though not the original intent of this work, pursuing the metabolic phenotypes ultimately uncovered a powerful genetic selection to identify weak informational suppressors, based on the role of the PurI enzyme in two biosynthetic pathways. This selection allows one to rapidly eliminate strong suppressors as well as true revertants by the resulting Pur+ phenotype. The work described here identified two such informational suppressors and further application of this selection, with the construction of the appropriate purI alleles, could aid in studies of codon context effects or translational accuracy.
Acknowledgments
We acknowledge John Roth, in whose laboratory some of the initial observations about purH mutants were made during the doctoral work of D.D. During this time, we also became aware of similar observations by C. Drabble that were communicated to us by B. Bochner. As far as we are aware, these observations have not been published elsewhere, but they contributed to the background for the work on purH described herein.
This work was supported by National Institutes of Health grant GM47296, a Shaw Scientist award from the Milwaukee Foundation, and a 21st Century Scientist Award from the J.S. McDonnell Foundation. J.L.Z. was supported by a National Science Foundation Graduate Fellowship and a Wisconsin Alumni Research Foundation Annual Fellowship.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Barik, S. 1995. Site-directed mutagenesis by double polymerase chain reaction. Mol. Biotechnol. 3:1-7. [DOI] [PubMed] [Google Scholar]
- 4.Beck, B., L. Connolly, A. De Las Peñas, and D. Downs. 1997. Evidence that rseC, a gene in the rpoE cluster, has a role in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 179:6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck, B. J., and D. M. Downs. 1998. The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 180:885-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, N. P., and B. S. Goldman. 1992. Rapid mapping in Salmonella typhimurium with Mud-P22 prophages. J. Bacteriol. 174:1673-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat, B., M. P. Groziak, and N. J. Leonard. 1990. Nonenzymatic synthesis and properties of 5-aminoimidazole ribonucleotide (AIR). Synthesis of specifically 15N-labeled 5-aminoimidazole ribonucleoside (AIRs) derivatives. J. Am. Chem. Soc. 112:4891-4897. [Google Scholar]
- 8.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257:9759-9769. [PubMed] [Google Scholar]
- 9.Bratton, A. C., and E. K. Marshall. 1939. A new coupling component for sulfanilamide determination. J. Biol. Chem. 128:537-550. [Google Scholar]
- 10.Brun, Y. V., R. Breton, P. Lanouette, and J. Lapointe. 1990. Precise mapping and comparison of two evolutionarily related regions of the Escherichia coli K-12 chromosome. J. Mol. Biol. 214:825-843. [DOI] [PubMed] [Google Scholar]
- 11.Caetano-Annoles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 12.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christian, T., and D. M. Downs. 1999. Defects in pyruvate kinase cause a conditional increase of thiamine synthesis in Salmonella typhimurium. Can J. Microbiol 45:565-572. [PubMed] [Google Scholar]
- 14.Claas, K., S. Weber, and D. M. Downs. 2000. Lesions in the nuo operon, encoding NADH dehydrogenase complex I, prevent PurF-independent thiamine synthesis and reduce flux through the oxidative pentose phosphate pathway in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:228-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Downs, D. M. 1987. Ph.D. thesis. University of Utah, Salt Lake City.
- 17.Downs, D. M., and L. Petersen. 1994. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 176:4858-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downs, D. M., and J. R. Roth. 1991. Synthesis of thiamine in Salmonella typhimurium independent of the purF function. J. Bacteriol. 173:6597-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enos-Berlage, J. L., and D. M. Downs. 1996. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 178:1476-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enos-Berlage, J. L., and D. M. Downs. 1997. Mutations in sdh (succinate dehydrogenase genes) alter the thiamine requirement of Salmonella typhimurium. J. Bacteriol. 179:3989-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enos-Berlage, J. L., M. J. Langendorf, and D. M. Downs. 1998. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J. Bacteriol. 180:6519-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleck, E. W., and J. Carbon. 1975. Multiple gene loci for a single species of glycine transfer ribonucleic acid. J. Bacteriol. 122:492-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frodyma, M., and D. M. Downs. 1998. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J. Bacteriol. 180:4757-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frodyma, M., A. Rubio, and D. M. Downs. 2000. Reduced flux through the purine biosynthetic pathway results in an increased requirement for coenzyme A in thiamine synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gralnick, J., E. Webb, B. Beck, and D. Downs. 2000. Lesions in gshA (encoding gamma-l-glutamyl-l-cysteine synthetase) prevent aerobic synthesis of thiamine in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:5180-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groziak, M. P., B. Bhat, and N. J. Leonard. 1988. Nonenzymatic synthesis of 5-aminoimidazole ribonucleoside and recognition of its facile rearrangement. Proc. Natl. Acad. Sci. USA 85:7174-7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 28.Henderson, J. F. 1972. Inhibition and simulation of purine biosynthesis by drugs, p. 218-251. In F. M. Beringer (ed.), Regulation of purine biosynthesis. American Chemical Society, Washington D.C.
- 29.Hoffmeyer, J., and J. Neuhard. 1971. Metabolism of exogenous purine bases and nucleosides by Salmonella typhimurium. J. Bacteriol. 106:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magasanik, B., and D. Karibian. 1960. Purine nucleotide cycles and their metabolic role. J. Biol. Chem. 235:2672-2681. [PubMed] [Google Scholar]
- 31.Martin, R. G. 1963. The first enzyme in histidine biosynthesis: the nature of feedback inhibition. J. Biol. Chem. 238:257-262. [Google Scholar]
- 32.Mehra, R. K., and W. T. Drabble. 1981. Dual control of the gua operon of Escherichia coli K-12 by adenine and guanine nucleotides. J. Gen. Microbiol. 123:27-37. [DOI] [PubMed] [Google Scholar]
- 33.Messenger, L. J., and H. Zalkin. 1979. Glutamine phosphoribosyl pyrophosphate amidotransferase from Escherichia coli. J. Biol. Chem. 254:3382-3392. [PubMed] [Google Scholar]
- 34.Meyer, E., N. J. Leonard, B. Bhat, J. Stubbe, and J. M. Smith. 1992. Purification and characterization of the purE, purK, and purC gene products: identification of a previously unrecognized energy requirement in the purine biosynthetic pathway. Biochemistry 31:5022-5032. [DOI] [PubMed] [Google Scholar]
- 35.Newell, P. C., and R. G. Tucker. 1966. The control mechanism of thiamine biosynthesis. A model for the study of control of converging pathways. Biochem. J. 100:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newell, P. C., and R. G. Tucker. 1966. The derepression of thiamine biosynthesis by adenosine. A tool for investigating this biosynthetic pathway. Biochem. J. 100:512-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newell, P. C., and R. G. Tucker. 1968. Precursors of the pyrimidine moiety of thiamine. Biochem. J. 106:271-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 39.Petersen, L., and D. M. Downs. 1996. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J. Bacteriol. 178:5676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen, L., J. Enos-Berlage, and D. M. Downs. 1996. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics 143:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen, L. A., and D. M. Downs. 1997. Identification and characterization of an operon in Salmonella typhimurium involved in thiamine biosynthesis. J. Bacteriol. 179:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth, J. R. 1970. UGA nonsense mutations in Salmonella typhimurium. J. Bacteriol. 102:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubio, A., and D. M. Downs. 2002. Elevated levels of ketopantoate hydroxymethyltransferase (PanB) lead to a physiologically significant coenzyme A elevation in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:2827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz, D. W., and M. Yarus. 1990. A simple and sensitive in vivo luciferase assay for tRNA-mediated nonsense suppression. J. Bacteriol. 172:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shedlovsky, A. E., and B. Magasanik. 1962. A defect in histidine biosynthesis causing an adenine deficiency. J. Biol. Chem. 237:3725.. [PubMed] [Google Scholar]
- 47.Sheppard, D. E. 1964. Mutants of Salmonella typhimurium resistant to feedback inhibition by l-histidine. Genetics 50:611-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skovran, E., and D. M. Downs. 2000. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J. Bacteriol. 182:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slupska, M. M., C. Baikalov, R. Lloyd, and J. H. Miller. 1996. Mutator tRNAs are encoded by the Escherichia coli mutator genes mutA and mutC: a novel pathway for mutagenesis. Proc. Natl. Acad. Sci. USA 93:4380-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soberon, M., O. Lopez, J. Miranda, M. L. Tabche, and C. Morera. 1997. Genetic evidence for 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) as a negative effector of cytochrome terminal oxidase cbb3 production in Rhizobium etli. Mol. Gen. Genet. 254:665-673. [DOI] [PubMed] [Google Scholar]
- 51.Squires, C., and J. Carbon. 1971. Normal and mutant glycine transfer RNAs. Nature (London) 233:272-277. [DOI] [PubMed] [Google Scholar]
- 52.Tso, J. Y., M. A. Hermodson, and H. Zalkin. 1982. Glutamine phosphoribosylpyrophosphate amidotransferase from cloned Escherichia coli purF. J. Biol. Chem. 257:3532-3536. [PubMed] [Google Scholar]
- 53.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 54.Watson, M. D., and W. T. Drabble. 1975. Relationship between purine nucleotide biosynthesis and requirement for thiamine in Escherichia coli K-12. Proc. Soc. Gen. Microbiol. 2:44-45. [Google Scholar]
- 55.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 56.Winston, F., D. Botstein, and J. H. Miller. 1979. Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 137:433-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Youderian, P., P. Sugiono, K. L. Brewer, N. P. Higgins, and T. Elliot. 1988. Packaging specific segments of the Salmonella chromosome with locked-in Mud-P22 prophages. Genetics 118:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yura, T. 1956. Evidence of nonidentical alleles in purine requiring mutants of Salmonella typhimurium. Publ. Carnegie Inst. 612:63-75. [Google Scholar]
- 59.Zalkin, H. 1983. Structure, function, and regulation of amidophosphoribosyltransferase from prokaryotes. Adv. Enzyme Regul. 21:225-237. [DOI] [PubMed] [Google Scholar]
- 60.Zilles, J. L., and D. M. Downs. 1996. A novel involvement of the PurG and PurI proteins in thiamine synthesis via the alternative pyrimidine biosynthetic (APB) pathway in Salmonella typhimurium. Genetics 144:883-893. [DOI] [PMC free article] [PubMed] [Google Scholar]