Abstract
Plasminogen activator inhibitor-2 (PAI-2), a member of the serpin gene family, is thought to serve as a primary regulator of plasminogen activation in the extravascular compartment. High levels of PAI-2 are found in keratinocytes, monocytes, and the human trophoblast, the latter suggesting a role in placental maintenance or embryo development. The primarily intracellular distribution of PAI-2 also may indicate a unique regulatory role in a protease-dependent cellular process such as apoptosis. To examine the potential functions of PAI-2 in vivo, we generated PAI-2-deficient mice by gene targeting in embryonic stem cells. Homozygous PAI-2-deficient mice exhibited normal development, survival, and fertility and were also indistinguishable from normal controls in response to a bacterial infectious challenge or endotoxin infusion. No differences in monocyte recruitment into the peritoneum were observed after thioglycollate injection. Epidermal wound healing was equivalent among PAI-2 −/− null and control mice. Finally, crossing PAI-2 −/− with PAI-1 −/− mice to generate animals deficient in both plasminogen activator inhibitors failed to uncover an overlap in function between these two related proteins.
Plasminogen activator inhibitor-2 (PAI-2), a member of the serine protease inhibitor (serpin) supergene family, exhibits inhibitory activity toward the plasminogen activators urokinase and tissue plasminogen activator (tPA) (1, 2). Although PAI-2 is presumed to function as a regulator of plasminogen activation based on this in vitro inhibitory activity, its protease target(s) and biologic function in vivo are still uncertain.
Based on gene structure and amino acid homology, PAI-2 belongs to a subfamily of serpins characterized by ovalbumin, which also includes maspin (a serpin with tumor suppressing activity in human mammary cells) (3), crmA (a viral encoded inhibitor of the interleukin-1β-converting enzyme) (4), and several potential inhibitors of granzyme B (5). Similar to ovalbumin, PAI-2 lacks a typical N-terminal secretory signal peptide and contains a relatively inefficient internal secretory signal sequence (6). Though a variable fraction is secreted as an ≈60- to 70-kDa glycosylated protein, the majority of PAI-2 appears to be retained in the cytoplasm in a nonglycosylated (47-kDa) form that is capable of spontaneous polymerization, particularly at high expression levels (7). The functional significance of this large intracellular pool of PAI-2 is unknown, though several studies suggest that it may play a role in protecting cells from apoptosis (8–11).
The regulation of PAI-2 gene expression is modulated by a variety of inflammatory mediators and other factors (1, 2) and involves both transcriptional and post-transcriptional mechanisms (12). The highest levels of PAI-2 gene expression are found in the macrophage and keratinocyte (13, 14), suggesting a role in the regulation of the inflammatory response, keratinocyte differentiation, and/or wound healing. The high levels of PAI-2 found within foam cells suggest a specific contribution to the pathogenesis of atherosclerosis (15). Alterations in PAI-2 also have been proposed to play a role in tumor progression and metastasis (1, 2, 16).
Deficiency of the related serpin PAI-1 in mice (17, 18) and humans (19) is associated with a moderate bleeding tendency as a result of increased fibrinolysis. In addition, PAI-1-deficient mice are resistant to pathologic pulmonary fibrosis after lung injury, presumably because of accelerated clearance of the provisional fibrin matrix (20). Though PAI-2 inhibits urokinase and two-chain tPA with rate constants of 9 × 105 M−1⋅s−1 and 2 × 105 M−1⋅s−1, respectively (21), these values are 20- to 100-fold less efficient than those observed for PAI-1. PAI-2 is also a very poor inhibitor of single chain tPA (21). PAI-2 is not generally detectable in human plasma, except during pregnancy, when levels rise as high as 250 ng/ml (22). The presumed source of PAI-2 in pregnancy is the trophoblastic epithelium (23).
Deficiency of PAI-2 has not been observed in humans or other mammals, suggesting a possible requirement for PAI-2 function for early embryonic development or placental function. To begin to address the potential contribution of PAI-2 to these and other processes in vivo, we generated PAI-2 deficient mice by gene targeting in embryonic stem (ES) cells. Our results demonstrate that PAI-2 is not required for normal murine development, survival, or fertility.
MATERIALS AND METHODS
Characterization of the Murine PAI-2 Gene.
Four independent λ phage-carrying segments of the murine PAI-2 gene (data not shown) were isolated by screening ≈106 clones of a murine strain 129SV genomic library in λFixII (Stratagene) by using an 889-bp probe predicted to span exon 8 of murine PAI-2, based on the human gene structure (24) (GenBank accession no. J04606). This probe was amplified by PCR from murine genomic DNA by using primers 5′-GCCAACTTCAACAAGTGGAT-3′ and 5′-GATGAGTTGCAGATACGTAC-3′, derived from the murine PAI-2 cDNA sequence (25) (GenBank accession no. X16490). The phage inserts were subcloned into pBluescript KS− (Stratagene), and the intron/exon boundaries of the murine PAI-2 gene (Fig. 1) were determined by direct sequence analysis, with additional primers designed by using the murine PAI-2 cDNA sequence and the human gene structure. Intron sizes were determined by PCR (introns 4–7), restriction mapping (intron 3), or Southern blotting (intron 2).
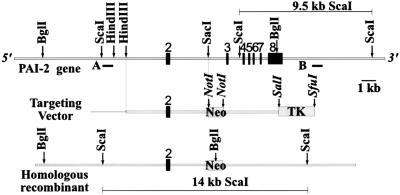
Figure 1.
Targeted disruption of the PAI-2 gene. The murine PAI-2 gene is depicted at the top, with the relative position of exons 2–8 indicated. Probe A (ScaI/HindIII fragment) and Probe B (XbaI/EcoRI fragment) are shown as black bars beneath the PAI-2 gene. The targeting vector (middle) consists of two homologous murine PAI-2 genomic fragments (5′, 5.5 kb; 3′, 3.8 kb) and neomycin resistance (neo) and thymidine kinase (TK) cassettes. Restriction sites shown in italics are derived from the vector. The expected homologous recombination between the endogenous gene and the targeting vector should result in the modified PAI-2 locus depicted at the bottom, with the neo cassette replacing 6.5 kb of the PAI-2 gene.
Construction of the Targeting Vector.
The targeting construct (Fig. 1) was assembled by inserting a 5.5-kb HindIII/SacI fragment of 5′ PAI-2 genomic DNA into the polylinker of the cloning vector pSL301 (Invitrogen). A neomycin (neo) cassette driven by the PGK promoter (26) was cloned into the NotI site of the polylinker, followed by a 3.8-kb 3′ NotI/SalI fragment of PAI-2 and a 3.3-kb SalI/SfuI fragment carrying a thymidine kinase cassette also under control of the PGK promoter (26). The NotI/SalI sites of the 3′ PAI-2 genomic fragment are derived from the pBluescript and λFixII vectors. The neo gene is oriented in the same transcriptional direction as the PAI-2 gene, with TK expression in the opposite orientation. A successful targeting event should delete the entire PAI-2 coding sequence except exon 2, replacing 6.5 kb of genomic DNA with the neo cassette (Fig. 1).
Targeting the PAI-2 Gene in ES Cells.
Two ES cell lines, D3 (obtained from T. Doetschman, University of Cincinnati) and CJ7 [obtained from T. Gridley, The Jackson Laboratory (27)], were grown on gelatin-coated plates in DMEM with high glucose (BioWhittaker) supplemented with 15% fetal calf serum (HyClone), 2 mM glutamine, 0.1 mM β-mercaptoethanol, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1,000 units/ml leukemia inhibitory factor (GIBCO/BRL) on a mitomycin C-inactivated, G-418-resistant feeder layer of primary murine fibroblasts isolated from embryos transgenic for a neo expression cassette (28). ES cells (1 × 107) were electroporated with 25 μg SfuI-linearized targeting vector DNA in 0.6 ml of culture medium by using a Bio-Rad Gene Pulser (320 V, 250 μF) and were plated on 2- × 100-mm gelatin-coated plates containing feeder cells. After 48 h, G-418 was added to the culture at 400 μg/ml for 24 h, then was decreased to 300 μg/ml for the duration of the selection. G-418 resistant clones were picked into 96-well plates after 8–9 days of selection. After growth to confluency, colonies were split into 3- × 96-well plates and were stored frozen or grown to confluency for DNA isolation.
Screening ES Colonies for Homologous Recombination.
DNA was prepared from ES colonies in 96-well plates (29) and was analyzed by Southern blotting using a 900-bp HindIII/ScaI 5′ genomic fragment (Fig. 1, probe A) after restriction with BglI or an 800-bp XbaI/EcoRI 3′ genomic fragment (Fig. 1, probe B) after ScaI digestion of cellular DNA. Probe A is outside and probe B within the targeting construct. DNA probe fragments were isolated in low melting point agarose (GIBCO/BRL) and were radiolabeled by using the Redi-prime kit (Amersham), according to the manufacturer’s instructions.
Generation of Chimeric and PAI-2 Null Mice.
Five independent, correctly targeted ES clones (four from the D3 cell line and one from CJ7) were identified after screening 507 G-418-resistant colonies (targeting efficiency ≈1.0%). These targeted clones were expanded without antibiotics on mitomycin C-treated feeder cells and were injected into C57BL/6J blastocysts, which were transferred to pseudopregnant CD1 females by using standard methods, as described (30, 31). Chimeric male offspring were tested for germ-line transmission by breeding with B6D2F1/J females. The resulting agouti pups were tested for the PAI-2 targeted allele by PCR of tail DNA preparations. The endogenous PAI-2 allele was identified by an 800-bp product spanning intron 6 (primer 1–5′-CAAACCTGCTACCCGAAGGT-3′, primer 2–5′-CAGTATGCGGAAGTTCTAGG-3′), and the targeted allele was identified by a 500-bp product from the neo cassette (primer 3–5′-GGACTGGCTGCTATTGGG CGAAGTG-3′, primer 4- 5′ TCTTGAGCAGTTCTTCCGCTATCTTCC-3′). Mice heterozygous for the PAI-2 targeted allele were bred to produce homozygous PAI-2 null mice. Genotype analysis of offspring was performed by PCR as above. The genotype of PAI-2 −/− animals was confirmed by Southern blotting or by an additional multiplex PCR using primers 5 (PAI-2 exon 8 5′-TTTGATAGGCGGGTTGTTTCTCTGT-3′), 6 (neo-specific 5′-CAGCCGAACTGTTCGCCAGG-3′), and 7 (3′ sequence flanking PAI-2 5′-GTTTGTCCACCATGCTCCCTCTA-3′). Primers 5 and 7 amplify a 500-bp product from the endogenous PAI-2 allele, and primers 6 and 7 amplify a 650-bp product from the targeted allele.
PAI-2 −/− mice were crossed with PAI-1 −/− mice (generous gift of P. Carmeliet and D. Collen, University of Leuven, Leuven, Belgium) to generate PAI-1 ±, PAI-2 ± double heterozygotes. These mice were intercrossed to produce PAI-1/PAI-2 double knockout mice. Offspring were genotyped by PCR of tail DNA by using four primer sets that specifically amplify the wild-type or mutant PAI-1 or PAI-2 allele (wild-type PAI-1, 5′ GACCGATCCTTTCTCTTTGTGGTT-3′ and 5′ AGGTCTGGGATGCTGGTTGGAAAG-3′; PAI-1 null, 5′ CGGTGGAGACATAACAGATGC-3′ and 5′ CAGCCGAACTGTTCGCCAGG-3′; wild-type PAI-2 primers 5 and 7 (see above); PAI-2 null, 5′ AGTTTGATAGGCGGGTTGTTTCTCT-3′ and 5′ ACAGCCTGATTGGGTCTTTCTTTAT-3′).
Generation of Antimurine PAI-2 Antibody and Western Blot Analysis.
A murine bone marrow cDNA library (gift of J. Lowe, University of Michigan, Ann Arbor) was screened by using the 889-bp PAI-2 exon 8 PCR product (see above). The longest cDNA isolated in this screen was lacking the first 60 nucleotides of coding sequence, based on the reported murine PAI-2 cDNA (25). An oligonucleotide containing a BamHI cloning site, the 60 missing nucleotides, and the next 20 nucleotides of PAI-2 sequence and a 3′ PAI-2 primer were used to amplify a 5′ extended product from the PAI-2 cDNA. A full length cDNA was assembled from a BamHI/PstI fragment of this PCR product and the original cDNA clone. The sequence of the original cDNA and the PCR-derived segment were confirmed by DNA sequencing.
The full length murine PAI-2 cDNA, digested with BamHI and HincII, was cloned into the BamHI and SmaI sites of pGEX-3X (Invitrogen), downstream and in frame of sequence-encoding glutathione S-transferase. Escherichia coli containing this plasmid were treated with 500 μM isopropyl β-d-thiogalactoside for 3 h at 37°C to induce expression of the glutathione S-transferase–PAI-2 fusion protein. The fusion protein, contained in the pellet of sonicated cells, was solubilized in 8 M urea and 100 μM phenylmethylsulfonyl fluoride by sonicating for 20 sec and incubating at 20°C for 30 min. After pelleting insoluble debris, the urea extract was dialyzed against three changes of 50 mM Tris (pH 8.0), 2 mM DTT, 1 mM EDTA, and 50 μM phenylmethylsulfonyl fluoride. The fusion protein was purified on a glutathione-Sepharose column following the manufacturer’s protocols (Amersham Pharmacia). The purified fusion protein was used as antigen for production of polyclonal rabbit anti-mouse PAI-2 sera through a commercial supplier (Rockland, Gilbertsville, PA).
Phenotypic Analysis.
Representative homozygote and heterozygote PAI-2 null mice were subject to routine autopsy examinations. For blood count surveys, whole blood was collected from the inferior vena cava into 1-ml syringes containing 0.1 ml 3.8% sodium citrate. Individual blood samples were evaluated from a total of eight PAI-2 −/− and eight control animals. After red blood cell lysis in platelet unopettes (Baxter Scientific Products, McGraw Park, IL), samples were diluted and counted by using a hemacytometer. Differentials were determined by evaluating 200 white blood cells/sample on a blood smear. Platelets were enumerated by dark field microscopy in a hemacytometer.
Response to a standardized skin wound was measured as follows: Mice were anesthetized with methoxyflurane, and their backs were shaved and wetted with 70% ethanol. The skin on the back was raised with forceps and was penetrated with a 4-mm tissue biopsy punch to produce two uniform, full-thickness wounds. The wounds were monitored daily, with the lesion area index calculated by multiplying the length of the wound by its width. These studies were performed in six PAI-2 −/−, eight PAI-2/PAI-1 double null, and eight wild-type control mice.
Macrophage recruitment in vivo was assayed by thioglycollate injection. Four percent thioglycollate (Difco) was prepared in boiling water, was stirred for 10 min, and was autoclaved, and 1 ml was injected i.p. into PAI-2 +/+ and PAI-2 −/− animals. At days 1, 3, 4, 6, and 7 after injection, mice were killed, and the peritoneum was lavaged with 10 ml Hank’s balanced salt solution (GIBCO/BRL), and the total number of white cells was counted in a hemacytometer. At least four mice of each genotype were studied per time point. For differential counts, lavage samples were cytospun onto slides, were stained with Wright’s stain, and were counted (100 cells/sample). To study macrophages in vitro, cells were recovered from thioglycollate-treated animals by peritoneal lavage with 10 ml DMEM containing 10% FCS and were plated in two tissue culture plates. One plate was treated with 1 mg/ml lipopolysaccharide (LPS) (E. coli serotype 0111:B4; Sigma) for 16 h. Cells were harvested from each plate by treatment with trypsin, and total cell lysates were prepared by pelleting cells and then solubilizing in SDS protein-loading buffer (32). For Western blot analysis, cell lysate samples were subjected to electrophoresis on a 12.5% SDS/PAGE gel and were electroblotted by using the Phast System (Amersham Pharmacia). PAI-2 protein was detected with polyclonal rabbit anti-mouse PAI-2 antisera (see above) followed by a peroxidase-tagged anti-rabbit Ig and was developed by using an enhanced chemiluminescence kit (Amersham Pharmacia).
Response to endotoxin challenge was determined by monitoring survival over time in animals injected i.p. with 1 ml of LPS (5 mg/kg). A total of six PAI-2 −/− and six controls (five wild-type and one heterozygote PAI-2 ±) were studied. Survivals after this dose of endotoxin ranged from 25 to 40%. As a standardized bacterial infection challenge, cecal ligation and puncture was performed as described by Remick and colleagues (33). Six PAI-2 −/− and six wild-type control mice were evaluated. Finally, four PAI-2 −/− and four wild-type mice received footpad injections of 50 μg of LPS. Six days after injection, animals were killed, and histologic analysis of the footpad was performed.
RESULTS
Targeting the Murine PAI-2 Gene.
The murine PAI-2 intron/exon boundaries were found to be identical to those previously reported for the human gene (24), with typical consensus splice acceptor and donor sequences (GenBank accession nos. AF069683–AF069695). The murine gene spans at least 11 kb, with the initiation codon in exon 2 and stop codon in exon 8. Intron sizes range from 421 bp to 4 kb, though the size and position of exon 1 (containing only the 5′ untranslated sequence) were not determined.
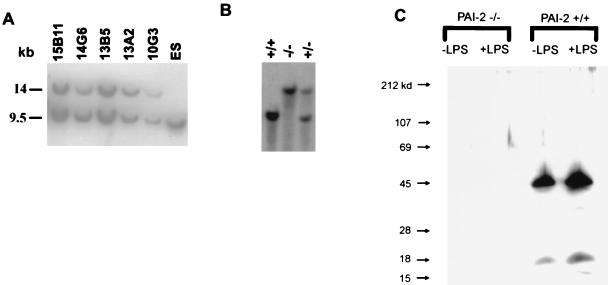
As shown in Fig. 1, the anticipated homologous recombination gene targeting event should remove exons 3–8, which includes the entire PAI-2 coding region except for the first 56 N-terminal amino acids encoded in exon 2. Neomycin-resistant ES colonies were screened by Southern blotting with probe A after restriction digestion with BglI (data not shown). Of 507 clones screened, 6 were identified as positive and were characterized further by additional restriction analyses using a probe at the 3′ end of the PAI-2 gene (Fig. 1, probe B). Five of six clones gave the expected pattern, as shown in Fig. 2A, with probe B detecting a 9.5-kb ScaI fragment from the endogenous PAI-2 gene and a 14-kb band from the targeted allele. Southern blot analysis using a neo gene probe confirmed the expected structure of the targeted allele and excluded the presence of any additional random integration events in these clones (data not shown).
Figure 2.
(A) Southern blot analysis of homologous recombinant ES clones. Genomic DNA was digested with ScaI and was hybridized with probe B (Fig. 1). The endogenous 9.5-kb band is seen in unmodified ES cells (last lane). The recombinant ES clones contain the novel 14-kb band predicted for the targeted allele, in addition to the 9.5-kb band corresponding to the unmodified normal allele. (B) Southern blot analysis of tail DNA from progeny of a PAI-2 ± × PAI-2 ± intercross. The DNA samples were analyzed as in A. The single wild-type 9.5-kb band in the first lane indicates a PAI-2 +/+ mouse, the 14-kb band in the middle lane indicates a homozygous null animal (PAI-2 −/−), and the two bands in the third lane indicate a heterozygote. (C). Western blot analysis of PAI-2 macrophages. Peritoneal macrophages elicited with thioglycollate from PAI-2 +/+ and PAI-2 −/− mice were grown in the presence (+) or absence (−) of LPS, and the cell lysate was analyzed by Western blotting with an antimurine PAI-2 antibody. A significant induction of PAI-2 expression by LPS is evident in wild-type macrophages, though no PAI-2 antigen is detectable in resting or stimulated macrophages obtained from the PAI-2 −/− mouse.
Generation of PAI-2-Deficient Mice.
The five ES clones analyzed in Fig. 2A were used for blastocyst injection with three lines producing germline transmission, two derived from the D3 ES cell line (13B5 and 15B11) and one from CJ7 (10G3). Intercrosses for all three cell lines produced viable PAI-2 −/− mice, with the expected structure at the targeted allele (deleting exons 3–8) confirmed by Southern blotting. Southern blot analysis of wild-type, heterozygous, and homozygous null mice derived from a 15B11 line intercross are shown in Fig. 2B. Subsequent routine genotype analysis generally was performed by PCR.
PAI-2 −/− mice obtained from the 15B11 line generally exhibited a runted phenotype that never was seen in PAI-2 −/− mice from the 10G3 and 13B5 lines and appears to be caused by an unknown, incidental genetic alteration outside of the PAI-2 locus. All of the additional analyses reported here were restricted to offspring of the latter two lines, which exhibited identical phenotypes. Genotype analysis of 309 F2 progeny from a PAI-2 ± intercross is shown in Table 1. The expected Mendelian ratios of offspring genotypes were observed, demonstrating that there is no preferential loss of PAI-2 −/− embryos in utero. Routine autopsy examination of PAI-2 null mice at 10 weeks and 13 months of age revealed no significant gross or microscopic abnormalities. Longevity of PAI-2 −/− mice appears to be normal, with survival observed up to an age of 24 months.
Table 1.
Genotype analysis of F2 progeny from 10G3 and 13B5 cell lines
| PAI-2 +/+ | PAI-2 +/− | PAI-2 −/− | Total | |
|---|---|---|---|---|
| Male | 40 (25.0%) | 72 (45.0%) | 48 (30.0%) | 160 |
| Female | 48 (32.2%) | 66 (44.3%) | 35 (23.4%) | 149 |
| Total | 88 (28.5%) | 138 (44.7%) | 83 (26.9%) | 309 |
An intercross of homozygous PAI-2 −/− mice demonstrated normal male and female fertility. Average litter size was 7.8 ± 2.0 mice (indistinguishable from the heterozygote intercross), with normal gestation duration for up to four successive litters. Routine histological examination of placenta from a PAI-2 −/− pregnancy in a PAI-2 −/− mother also revealed no obvious abnormality.
Absence of PAI-2 Antigen in PAI-2 −/− Monocytes.
Monocytes were collected after i.p. injection of thioglycollate from PAI-2 −/− and wild-type control mice and were examined for PAI-2 protein content by Western blot analysis after growth in culture in the presence or absence of LPS. A strong signal at the expected 42-kDa size was seen in control monocytes, with significant induction evident after exposure to LPS. No immunoreactive band was detectable in PAI-2 −/− monocytes in the presence or absence of LPS (Fig. 1C). Taken together with the Southern blot analysis (Fig. 1 A and B), these data document the complete absence of PAI-2 protein expression in these PAI-2 −/− mice.
Phenotypic Analysis of PAI-2 −/− Mice.
Routine peripheral blood counts in PAI-2 −/− mice demonstrated a trend toward a mild decrease in blood platelet count and total leukocyte count of borderline statistical significance (platelet count in PAI-2 +/+ mice was 8.6 ± 2.5 × 105 per microliter versus 6.5 ± 0.8 × 105 in PAI-2 −/− mice [P = 0.046], and white blood cell count was 4,600 ± 2,200 per microliter versus 2,900 ± 1,200 [P = .074]). Differential counts showed comparable slight decreases in lymphocytes and neutrophils, as well as monocytes. No gross abnormalities of blood cell morphology were evident, and the bone marrow also appeared normal. Monocyte recruitment into the peritoneum after thioglycollate also was examined, with no significant differences observed between PAI-2 −/− mice and wild-type controls. These recruited monocytes, as well as primary murine embryo fibroblasts and keratinocytes obtained from skin biopsies, all were grown in tissue culture. No obvious differences in morphology or growth in culture between wild-type and PAI-2 −/− cells were observed.
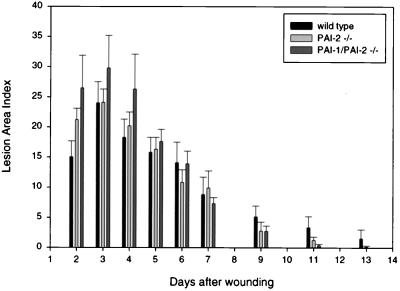
Mice also were examined for differences in survival after challenge with endotoxin or infectious challenge by using an established model of cecal ligation and puncture (33). In both cases, no significant differences between PAI-2 −/− mice and wild-type controls were observed. Injection of endotoxin into the footpad resulted in similar degrees of arterial and venous thrombosis, with no significant differences noted between PAI-2 null and wild-type mice. These results are in contrast to the response reported for PAI-1 −/− mice (18) in which a marked reduction in vascular thrombosis was observed. Finally, PAI-2 −/− mice were compared with wild-type littermates in a wound healing model after a uniform, bilateral, 4-mm punch biopsy injury to the dorsal skin. As shown in Fig. 3, no significant differences were seen in lesion size or rate of healing between wild-type and PAI-2 −/− mice.
Figure 3.
Time course of wound healing. Wild type, PAI-2 −/−, or PAI-1 −/−, PAI-2 −/− mice were wounded with a 4-mm biopsy punch and were followed over time. Lesion area index is the maximal length of wound multiplied by the maximal width of wound. No significant differences in the rate of wound healing were observed.
No Evidence for Overlap in Function Between PAI-1 and PAI-2.
To test the possibility that the related serpin, PAI-1, might potentially compensate for the absence of PAI-2 in PAI-2 −/− mice, PAI-2 null mice were crossed with PAI-1 null animals previously developed by Carmeliet et al. (17) to generate doubly deficient PAI-1 −/−, PAI-2 −/− animals. The doubly deficient mice again demonstrated normal survival and fertility without obvious gross developmental or other abnormalities. The skin wounding challenge to doubly deficient animals also revealed no difference in healing when compared with the singly PAI-2-deficient or wild-type controls (Fig. 3). No consistent histologic differences were observed on microscopic analysis of skin wounds from PAI-2 null, PAI-1/PAI-2 double null, or control mice.
DISCUSSION
The biologic function of PAI-2 has remained an unsettled question since its initial discovery in 1968 (1, 2, 34). Its inhibitory activity against urokinase suggests a primary role in the regulation of plasminogen activation, with the observation of elevated levels in pregnancy plasma further suggesting that this function may be particularly important in the maintenance of the placenta or, perhaps, for embryonic development.
The markedly elevated levels of PAI-2 observed in pregnancy plasma (rising from undetectable to as high as 250 ng/ml) appear to be of fetal origin, synthesized by the villous syncytiotrophoblastic epithelium. PAI-2 levels appear to be a marker for placental function, with decreased levels associated with intrauterine growth retardation (1, 2, 35). The observation of normal fertility in PAI-2 −/− mice and the ability of PAI-2 −/− female mice to support an apparently normal pregnancy, including an entirely PAI-2 −/− litter, exclude an essential function for PAI-2 in murine fetal development or the establishment or maintenance of the placenta in the mouse. However, we cannot exclude the possibility that PAI-2 plays an important role in human placental function. Though present in human trophoblasts, PAI-2 mRNA is not detected in the murine placenta at significant levels until very late in gestation (17–18 days postcoitum) (36).
Previous reports of human and mice genetically deficient in PAI-1 (17, 19) as well as the plasminogen activators themselves (37) or their target protease, plasminogen (38, 39), suggest that the primary function of the plasminogen activation system is in the regulation of hemostasis and the clearance of the fibrin blood clot. PAI-1 deficiency leads to enhanced fibrinolysis and a moderate bleeding tendency (18, 19). Although deficiency of either urokinase or tPA alone results in only a subtle phenotype, deficiency of both activators is associated with widespread fibrin deposition, similar to that observed in plasminogen deficient animals (37, 38). tPA appears to be the primary plasminogen activator in the vascular space, and, thus, it is perhaps not surprising that PAI-2 −/− mice do not exhibit a bleeding phenotype because PAI-2 is a poor inhibitor of tPA (21). Given the potential overlap in function between PAI-1 and PAI-2, we generated doubly deficient mice lacking both of these inhibitors. However, even in these doubly deficient animals, no obvious bleeding phenotype resulted from this potentially unopposed plasminogen activator activity. Though we cannot exclude a subtle abnormality caused by enhanced fibrinolysis in these animals, no unusual bleeding was observed after routine tail transection for DNA preparation or after the punch biopsy skin-wounding experiments. These results are consistent with the clinical observations of only moderate hemorrhage in a patient with a deficiency of α2 antiplasmin, the downstream direct inhibitor of plasmin (40).
PAI-2 appears to exist predominantly in a nonglycosylated intracellular form, and it has been suggested that it may participate in the regulation of an apoptosis pathway (8–11). Though no formal measurements of cell cycle parameters or doubling times were performed, we observed no obvious differences in organ histology or cell morphology and growth for cells grown in culture, including cell types known to exhibit high levels of PAI-2 expression. Nonetheless, alterations in the function of a cell death pathway in other cell types or in response to other specific stimuli cannot be excluded. It is also possible that PAI-2 could act in both survival and proliferation pathways in compensatory ways, resulting in no evident imbalance in unchallenged mice. Similarly, a number of previous studies (1, 2, 16) have suggested an important regulatory role for PAI-2 in tumor cell progression and metastasis, functions that have not yet been evaluated in this PAI-2 deficient animal model.
The localization of the highest levels of PAI-2 expression to the keratinocyte and macrophage suggest important roles in the maintenance or development of the integrity of the epidermis and possibly a role in host defense. Recent studies of PAI-2 gene expression have suggested that this protein may be required for specific steps in hair and skin development in the mouse (41). It is also a major component of the heavily cross-linked cornified envelope formed during the final stages of epidermal keratinocyte differentiation (42). The observation of normal development, survival, and fertility in PAI-2 deficient mice was thus unexpected. Although it is difficult to draw firm conclusions about specific gene function from the absence of an unidentifiable phenotype in knockout mice, (43) these results clearly demonstrate that PAI-2 function is not essential for cell survival or differentiation. Specifically, despite high levels of expression in keratinocytes and macrophages, skin development and integrity appeared unaltered in these animals, and no defect in host defense attributable to monocyte dysfunction was evident. In addition, normal numbers of monocytes were found in the circulation, and recruitment of macrophages in response to an inflammatory stimulus appeared to be intact. Although impaired wound healing has been observed in plasminogen-deficient mice (44), our studies failed to detect any differences in the repair process in PAI-2-deficient or doubly PAI-1- and PAI-2-deficient mice when compared with the wild type. However, a critical role in response to another specific challenge to epidermal integrity cannot be excluded.
The localization of PAI-2 to the outer layers of the epidermis and within activated macrophages, taken together with normal development of these tissues in the absence of PAI-2, might suggest a primary role in host defense, perhaps as an inhibitor of a specific microbial-associated protease. Indeed, a number of human pathogens are known to exhibit interactions with the plasminogen activator system, often with a critical role in disease pathogenesis (45, 46). Further analysis of PAI-2 null mice, as well as crosses of these mice with animals carrying targeted alterations in related genes, should provide further insights into the biological function of this unique class of intracellular serpins.
Acknowledgments
We thank T. L. Saunders, L. C. Samuelson, and S. A. Camper for valuable technical assistance with the generation of the PAI-2 null mice by gene targeting and S. E. Labun for help with manuscript preparation. Core support was provided by the University of Michigan Cancer Center, National Institutes of Health Grant CA 46592 and the University of Michigan Multipurpose Arthritis Center, National Institutes of Health Grant AR20557. Support also was provided by National Institutes of Health Grants HL49184-01A2 (to D.G.) and 1F32-HL09028-01 (to K.M.D.). D.G. is a Howard Hughes Medical Institute Investigator.
ABBREVIATIONS
- PAI-2
plasminogen activator inhibitor 2
- LPS
lipopolysaccharide
- tPA
tissue plasminogen activator
- ES
embryonic stem
- neo
neomycin
- kb
kilobase
Footnotes
References
- 1.Kruithof E K O, Baker M S, Bunn C L. Blood. 1995;86:4007–4024. [PubMed] [Google Scholar]
- 2.Dear A E, Medcalf R L. Fibrinolysis. 1995;9:321–378. [Google Scholar]
- 3.Zou Z, Anisowicz A, Hendrix M J C, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 4.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 5.Sun J, Ooms L, Bird C H, Sutton V R, Trapani J A, Bird P I. J Biol Chem. 1997;272:15434–15441. doi: 10.1074/jbc.272.24.15434. [DOI] [PubMed] [Google Scholar]
- 6.von Heijne G, Liljestrom P, Mikus P, Andersson H, Ny T. J Biol Chem. 1991;266:15240–15243. [PubMed] [Google Scholar]
- 7.Mikus P, Ny T. J Biol Chem. 1996;271:10048–10053. doi: 10.1074/jbc.271.17.10048. [DOI] [PubMed] [Google Scholar]
- 8.Jensen P H, Cressey L I, Gjertsen B T, Madsen P, Mellgren G, Hokland P, Gliemann J, Doskeland S O, Lanotte M, Vintermyr O K. Br J Cancer. 1994;70:834–840. doi: 10.1038/bjc.1994.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S, Baglioni C. J Biol Chem. 1991;266:20960–20964. [PubMed] [Google Scholar]
- 10.Gan H, Newman G W, Remold H G. J Immunol. 1995;155:1304–1315. [PubMed] [Google Scholar]
- 11.Dickinson J L, Bates E J, Ferrante A, Antalis T M. J Biol Chem. 1995;270:27894–27904. doi: 10.1074/jbc.270.46.27894. [DOI] [PubMed] [Google Scholar]
- 12.Maurer F, Medcalf R L. J Biol Chem. 1996;271:26074–26080. doi: 10.1074/jbc.271.42.26074. [DOI] [PubMed] [Google Scholar]
- 13.Chapman H A, Jr, Vavrin Z, Hibbs J B., Jr Cell. 1982;28:653–662. doi: 10.1016/0092-8674(82)90220-3. [DOI] [PubMed] [Google Scholar]
- 14.Saksela O, Hovi T, Vaheri A. J Cell Physiol. 1985;122:125–132. doi: 10.1002/jcp.1041220119. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie H, Jamieson A, Booth N A. Thromb Haemostasis. 1997;77:1168–1173. [PubMed] [Google Scholar]
- 16.Mueller B M, Yu Y B, Laug W E. Proc Natl Acad Sci USA. 1995;92:205–209. doi: 10.1073/pnas.92.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmeliet P, Kieckens L, Schoonjans L, Ream B, Van Nuffelen A, Prendergast G C, Cole M D, Bronson R, Collen D, Mulligan R C. J Clin Invest. 1993;92:2746–2755. doi: 10.1172/JCI116892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmeliet P, Stassen J M, Schoonjans L, Ream B, van den Oord J J, De Mol M, Mulligan R C, Collen D. J Clin Invest. 1993;92:2756–2760. doi: 10.1172/JCI116893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fay W P, Shapiro A D, Shih J L, Schleef R R, Ginsburg D. N Engl J Med. 1992;327:1729–1733. doi: 10.1056/NEJM199212103272406. [DOI] [PubMed] [Google Scholar]
- 20.Eitzman D T, McCoy R D, Zheng X, Fay W P, Shen T, Ginsburg D, Simon R H. J Clin Invest. 1996;97:232–237. doi: 10.1172/JCI118396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruithof E K O, Vassalli J-D, Schleuning W-D, Mattaliano R J, Bachmann F. J Biol Chem. 1986;261:11207–11213. [PubMed] [Google Scholar]
- 22.Kruithof E K O, Tran-Thang C, Gudinchet A, Hauert J, Nicoloso G, Genton C, Welti H, Bachmann F. Blood. 1987;69:460–466. [PubMed] [Google Scholar]
- 23.Astedt B, Hagerstrand I, Lecander I. Thromb Haemostasis. 1986;56:63–65. [PubMed] [Google Scholar]
- 24.Ye R D, Ahern S M, Le Beau M M, Lebo R V, Sadler J E. J Biol Chem. 1989;264:5495–5502. [PubMed] [Google Scholar]
- 25.Belin D, Wohlwend A, Schleuning W-D, Kruithof E K O, Vassalli J-D. EMBO J. 1989;8:3287–3294. doi: 10.1002/j.1460-2075.1989.tb08489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 27.Swiatek P J, Gridley T. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- 28.Gossler A, Doetschman T C, Korn R, Serfling E, Kemler R. Proc Natl Acad Sci USA. 1986;83:9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez-Solis R, Rivera-Pérez J, Wallace J D, Wims M, Zheng H, Bradley A. Anal Biochem. 1992;201:331–335. doi: 10.1016/0003-2697(92)90347-a. [DOI] [PubMed] [Google Scholar]
- 30.Zheng X, Saunders T L, Camper S A, Samuelson L C, Ginsburg D. Proc Natl Acad Sci USA. 1995;92:12426–12430. doi: 10.1073/pnas.92.26.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui J, O’Shea K S, Purkayastha A, Saunders T L, Ginsburg D. Nature (London) 1996;384:66–68. doi: 10.1038/384066a0. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 33.Eskandari M K, Bolgos G, Miller C, Nguyen D T, DeForge L E, Remick D G. J Immunol. 1992;148:2724–2730. [PubMed] [Google Scholar]
- 34.Kawano T, Morimoto K, Uemura Y. Nature (London) 1968;217:253–254. doi: 10.1038/217253a0. [DOI] [PubMed] [Google Scholar]
- 35.Estellés A, Gilabert J, Aznar J, Loskutoff D J, Schleef R R. Blood. 1989;74:1332–1338. [PubMed] [Google Scholar]
- 36.Kwata Y, Mimuro J, Kaneko M, Shimada K, Sakata Y. Thromb Haemostasis. 1996;76:569–576. [PubMed] [Google Scholar]
- 37.Carmeliet P, Schoonjans L, Kieckens L, Ream B, Degen J L, Bronson R, De Vos R, van den Oord J J, Collen D, Mulligan R C. Nature (London) 1994;368:419–424. doi: 10.1038/368419a0. [DOI] [PubMed] [Google Scholar]
- 38.Bugge T H, Flick M J, Daugherty C C, Degen J L. Genes Dev. 1995;9:794–807. doi: 10.1101/gad.9.7.794. [DOI] [PubMed] [Google Scholar]
- 39.Bugge T H, Kombrinck K W, Flick M J, Daugherty C C, Danton M J S, Degen J L. Cell. 1996;87:709–719. doi: 10.1016/s0092-8674(00)81390-2. [DOI] [PubMed] [Google Scholar]
- 40.Aoki N, Saito H, Kamiya T, Koie K, Sakata Y, Kobakura M. J Clin Invest. 1979;63:877–884. doi: 10.1172/JCI109387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavker R M, Risse B, Brown H, Ginsburg D, Pearson J, Baker M S, Jensen P J. J Invest Dermatol. 1998;110:917–922. doi: 10.1046/j.1523-1747.1998.00223.x. [DOI] [PubMed] [Google Scholar]
- 42.Robinson N A, Lapic S, Welter J F, Eckert R L. J Biol Chem. 1997;272:12035–12046. doi: 10.1074/jbc.272.18.12035. [DOI] [PubMed] [Google Scholar]
- 43.Erickson H P. J Cell Biol. 1993;120:1079–1081. doi: 10.1083/jcb.120.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romer J, Bugge T H, Pyke C, Lund L R, Flick M J, Degen J L, Dano K. Nat Med. 1996;2:287–292. doi: 10.1038/nm0396-287. [DOI] [PubMed] [Google Scholar]
- 45.Coleman J L, Gebbia J A, Piesman J, Degen J L, Bugge T H, Benach J L. Cell. 1997;89:1111–1119. doi: 10.1016/s0092-8674(00)80298-6. [DOI] [PubMed] [Google Scholar]
- 46.Boyle M D, Lottenberg R. Thromb Haemostasis. 1997;77:1–10. [PubMed] [Google Scholar]