Abstract
Rapid production of protein-based tumor-specific vaccines for the treatment of malignancies is possible with the plant-based transient expression system described here. We created a modified tobamoviral vector that encodes the idiotype-specific single-chain Fv fragment (scFv) of the immunoglobulin from the 38C13 mouse B cell lymphoma. Infected Nicotiana benthamiana plants contain high levels of secreted scFv protein in the extracellular compartment. This material reacts with an anti-idiotype antibody by Western blotting, ELISA, and affinity chromatography, suggesting that the plant-produced 38C13 scFv protein is properly folded in solution. Mice vaccinated with the affinity-purified 38C13 scFv generate >10 μg/ml anti-idiotype immunoglobulins. These mice were protected from challenge by a lethal dose of the syngeneic 38C13 tumor, similar to mice immunized with the native 38C13 IgM-keyhole limpet hemocyanin conjugate vaccine. This rapid production system for generating tumor-specific protein vaccines may provide a viable strategy for the treatment of non-Hodgkin’s lymphoma.
Most B cell malignancies are incurable and are increasing in frequency in the populations of industrial nations (1, 2). Although B cell tumors are characterized by extreme variability in treatment and prognosis (3), they share a common feature that makes them ideal for the development of patient-specific cancer vaccines. Each clone of malignant B cells expresses a unique cell surface immunoglobulin (Ig)–a tumor-specific marker. The tumor’s surface Ig, when made immunogenic by conjugation to keyhole limpet hemocyanin (KLH) and administered with an adjuvant, has been used to vaccinate patients in chemotherapy-induced remission (4, 5). When an immune response is triggered by such vaccination, patients have a superior clinical outcome (6, 7).
Unfortunately, Igs are difficult proteins to produce. Currently, Igs for patient therapies are created by fusion of tumor cells to a transformed human/mouse heteromyeloma cell (8, 9). Hybridomas are screened for secreted patient tumor-specific Ig and expanded for large-scale production of the protein. Besides the labor and expense involved, a drawback of hybridoma production systems is the unpredictable loss of chromosomes and of tumor-specific Ig protein expression over time. This phenomenon currently limits the application of the therapy, in terms of both the quantity of vaccine per patient and the total number of patients that can be treated. To expand the scope of individualized non-Hodgkin’s lymphoma (NHL) therapies, a source of abundant, safe, easily purified vaccine needs to be generated in a time frame of weeks rather than months or years.
An appealing alternative to multichain whole Ig vaccines is singe-chain variable region (scFv) vaccines. Consisting of just the hypervariable domains from the tumor-specific Ig, these proteins recreate the antigen-binding site of the native Ig (10–12), are a fraction of the size, and can be expressed in several expression systems (13–17), including transgenic plants (18–22). scFv vaccines, either as protein or DNA, are capable of eliciting anti-idiotype-specific responses in animals (23, 24) and are effective in blocking tumor progression in mouse models of lymphoma (23).
To extend the application of scFv vaccines for human NHL therapy, the expression systems used to produce individualized protein vaccines must accommodate a wide range of sequences and be capable of generating proteins that recapitulate the conformation of the variable region as it is presented by the malignant B cell. Although bacterial expression systems have the advantages of speed and abundant production, they are limited, in many instances, by their inability to produce properly folded soluble proteins without the need for denaturation and renaturation. Baculovirus expression systems offer the hope of better scFv solubility by driving expression through the secretory pathways of insect cells (14, 25); however, virus manipulation and growth of insect cells can be time consuming and costly, making the system less suitable for expression of tumor-specific scFv proteins.
We utilized a plant viral vector to transiently express an scFv protein derived from the 38C13 mouse B cell lymphoma. This viral system utilizes a hybrid tobacco mosaic virus (TMV) to introduce mammalian gene sequences into whole plants. TMV is a plus-strand RNA virus that replicates extrachromosomally, moves quickly from cell to cell from a site of local infection, and can redirect protein synthesis of host cells to express high levels of proteins of interest throughout the plant (26). By fusing a signal peptide sequence to the 38C13 scFv sequence, expressed protein can be directed to the plant secretory pathway, promoting proper protein folding and facilitating subsequent purification. The lack of nuclear integration, coupled to the kinetics of viral replication and protein expression, has allowed us to produce large quantities of 38C13 scFv protein within weeks of obtaining the gene. We show that plant-derived protein is soluble, generates an antibody response in vaccinated animals that is relevant to the tumor idiotype, and is an effective vaccine in a murine tumor challenge model of NHL.
MATERIALS AND METHODS
Cloning, Expression, and Purification 38C13 scFv in Plants.
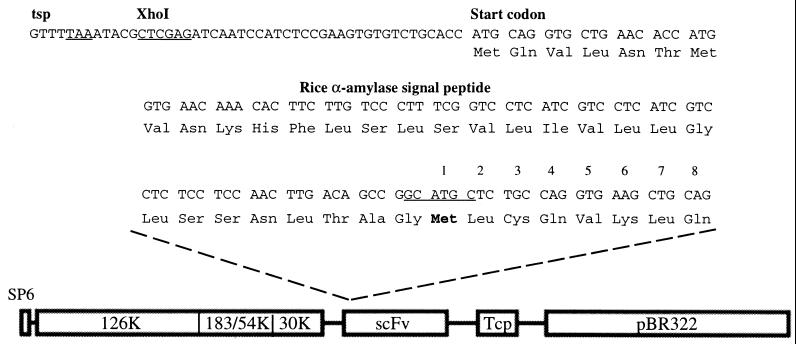
PCR primers specific for murine 38C13 sequences [GenBank accession nos. X14096–X14099 (27)] were used to amplify the 38C13 scFv coding sequence as described (23). 38C13 scFv insert was then cloned in-frame with the sequence encoding a rice α-amylase signal peptide into a modified TTO1A vector containing a hybrid fusion of TMV and tomato mosaic virus (28) (Fig. 1). Capped infectious RNA was made in vitro from 1 μg of PmeI-linearized plasmid by using an SP6 message kit from Ambion (Austin, TX). Synthesis of the approximately 7-kb message was quantified by gel electrophoresis. Approximately 2 μg of in vitro transcribed viral RNA was applied with an abrasive to the lower leaves (approximately 1–2 cm in size) of the tobacco-related species Nicotiana benthamiana as previously described (29). Subgenomic RNA encoding 38C13 scFv is initiated after infection at the indicated transcription start point (tsp; see Fig. 1).
Figure 1.
Schematic diagram of the viral expression vector used to produce murine 38C13 scFv NHL antigen in Nicotiana benthamiana host plants. Shown are the sequences encoding rice α-amylase signal peptide fused in-frame to the SphI site (underlined), introduced by PCR, 5′ of the 38C13 heavy chain. Also indicated in the linear map are the positions of the SP6 transcription start site, the 126-kDa, 183-kDa, and 30-kDa proteins from TMV, and the tomato mosaic virus coat protein, as well as pBR322 sequences for bacterial propagation.
Symptoms of infection were visible after 5–6 days as mild leaf deformation, with some variable leaf mottling and growth retardation. Eleven to 14 days after inoculation, leaf and stem material was harvested, weighed, and then subjected to a 700-mmHg (93-kPa) vacuum for 2 min in infiltration buffer (100 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/2 mM EDTA). Secreted proteins were then recovered from infiltrated leaves by mild centrifugation at 4,000 rpm (2,000 × g; Beckman JA-14) on supported nylon mesh discs (hereafter abbreviated interstitial fraction or IF), then filtered through a 0.2-μm membrane and stored at −80°C until purified. Ten microliters of secreted material from 38C13 scFv-infected leaves was separated by SDS/PAGE and stained by Coomassie brilliant blue, or 1 μl was separated by SDS/PAGE and transferred and analyzed by Western blotting using the 38C13-specific anti-idiotype monoclonal antibody S1C5. Enzyme-linked immunosorbent assays (ELISAs) were also performed on crude samples as described below. IF was also processed under mild reducing conditions in infiltration buffer containing 1 mM ascorbic acid and 0.04% sodium metabisulfite. The 38C13 scFv Cys− form was created through PCR by altering the 5′ primer to omit the three nucleotides encoding the third amino acid of 38C13 scFv.
Protein Analysis and Purification.
Protein levels were analyzed by ELISA as follows. The 38C13 idiotype-specific antibody S1C5 (30) was recovered from ascites fluid prepared in nude mice by standard procedures, and purified by staphylococcal protein A affinity chromatography. To detect plant-derived scFv proteins by ELISA, plates (Nunc, MaxiSorp) were coated overnight at 4°C with 2 μg/ml S1C5 in carbonate buffer (50 mM Na2CO3, pH 9), 50 μl per well. Plates were washed five times in wash buffer (150 mM NaCl/0.05% Triton X-100) and incubated for 20 min at room temperature in blocking buffer (100 mM Tris⋅HCl, pH 7.5/0.5% Tween 20/2% BSA). Plates were washed five times before incubation with 1:10 (vol/vol) starting dilutions of proteins in PBS/2% BSA. Bacterially produced 38C13-myc scFv at 300 ng/ml was used as a positive control and for quantitation (23). After a 1-hr room temperature incubation, plates were washed again five times, and incubated 1 hr at room temperature with 1 μg/ml protein A-horseradish peroxidase (HRP; Sigma), which recognizes a site in the heavy-chain variable (V) region of 38C13 scFv (31, 32). Plates were washed and then developed by a 10-min room temperature incubation with 0.15% 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS; Sigma) in 100 mM sodium citrate buffer, pH 8.5/0.001% hydrogen peroxide. Plates were read at 405 and 490 nm by an absorbance plate reader (Molecular Devices) and the data were analyzed by SoftMax.
SDS/PAGE was performed by a standard technique; 12% gels were purchased precast (Novex, San Diego). To visualize proteins, gels were stained for 1 hr in 0.2% Coomassie brilliant blue (Sigma) in 50% methanol, destained for 2 hr in 10% methanol/20% acetic acid/70% water, and air dried between cellophane sheets (Bio-Rad). Silver staining was performed as previously described (33). For Western analysis, proteins were transferred from an SDS gel by semidry transfer (Janssen Life Sciences) to nitrocellulose in standard Tris–glycine buffer with 20% methanol, 150 V for 1 hr. After transfer, blots were treated for 20 min at room temperature with blocking buffer (50 mM Tris⋅HCl, pH 8/150 mM NaCl/1 mM EDTA/2.5% nonfat dry milk/2.5% BSA/0.05% Tween 20) followed by 1-hr incubation in blocking buffer plus 1 μg/ml purified S1C5 antibody. After three 15-min washes (100 mM Tris⋅HCl, pH 8/150 mM NaCl/1 mM EDTA/0.1% Tween 20), blots were incubated for 1 hr in blocking buffer plus 1 μg/ml HRP-conjugated goat anti-mouse IgG (Southern Biotechnology). After three 15-min washes, Western blots were developed by enhanced chemiluminescence (ECL; Amersham). Exposure times were 1–5 sec.
For vaccination, crude secreted plant proteins were purified by affinity chromatography. S1C5 antibody (10 mg) was coupled to 1 g of CNBr Sepharose as described by the manufacturer (Pharmacia); all buffers for coupling, blocking, and washing were endotoxin-free as determined by a Limulus amebocyte assay (Associates of Cape Cod). Frozen plant extracts were thawed on ice, refiltered, and found to contain less than 0.06 endotoxin unit/ml. 38C13 scFv protein was then applied to an S1C5 column in infiltration buffer, washed with 50 ml of PBS, and eluted as 1-ml fractions in endotoxin-free 50 mM triethanolamine, pH 12.6, directly into 100 μl of 2 M Tris⋅HCl buffer, pH 8. Fractions containing 38C13 scFv protein were analyzed by S1C5 ELISA as described above, pooled, and dialyzed against PBS overnight, and S1C5 reactivity was verified by SDS/PAGE and Western analysis. This method yielded 38C13 scFv protein of approximately 90–95% purity as determined by ELISA and total protein determination.
Molecular Analysis of 38C13 scFv.
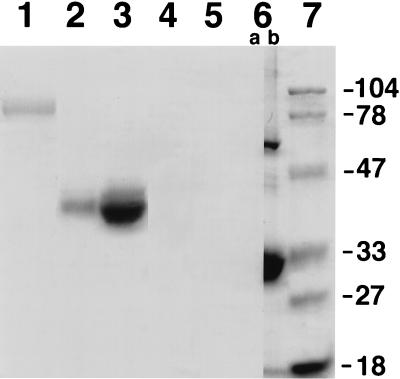
Total carbohydrate analyses of 38C13 scFv and controls were performed by the standard periodic acid–Schiff (PAS) method. Briefly, 10 μg of 38C13 IgM or 5 and 25 μg of HRP were separated on an SDS/12% acrylamide gel alongside one lane of 5 μg and two lanes of 25 μg of 38C13 scFv. The gel was divided at lane 6 (Fig. 5), which contained 25 μg of 38C13 scFv, and stained either by PAS (lanes 1–6a) or by Coomassie brilliant blue (lanes 6b and 7). After staining and destaining, the gel was reassembled and dried between two cellophane sheets.
Figure 5.
Plant-produced 38C13 scFv does not contain carbohydrates. PAS stain was used to reveal the total carbohydrate content of 10 μg of 38C13 IgM (lane 1) or 5 and 25 μg of HRP (lanes 2 and 3) but not in 5 or 25 μg of 38C13 scFv made in plants (lanes 4, 5, and 6a). Coomassie staining was done in parallel on lane 6b, which contains 25 μg of 38C13 scFv, as well as on size standards (lane 7, in kDa) as indicated.
N-terminal sequencing was performed on the 38C13 scFv 30-kDa dimer, which was further purified by Superose 12 HR 10/30 (Pharmacia) gel filtration after immunoaffinity purification. Enriched fractions were separated by SDS/PAGE and immobilized to a ProBlot (Applied Biosystems) membrane. Coomassie-stained bands were independently excised from the membrane and sent to the University of Michigan Protein Structure Laboratory for N-terminal sequencing.
Animal Vaccination, Analysis, and Tumor Challenge.
Purified 38C13 scFv protein, 15 μg alone, or with an additional 10 μg of QS-21 adjuvant (refs. 34 and 35; kindly provided by Aquilla, Framingham, MA) in a total volume of 200 μl of PBS, was used to vaccinate C3H mice (supplied by Harlan Sprague–Dawley) subcutaneously (s.c.) in the rear flank at 2-week intervals for a total of three vaccinations. Whole 38C13 IgM was conjugated to KLH by glutaraldehyde cross-linking (36), and 50 μg of 38C13 IgM-KLH was administered s.c. with QS-21 concurrent with the second and third scFv vaccinations. Vaccination with QS-21 alone was used as a negative control. Vaccinated animals were bled through the tail vein, and their responses to anti-38C13 serum were measured by anti-38C13 IgM ELISA 10 days after the second and third vaccinations as previously described (23). Ig isotype analysis was performed on pooled sera from each vaccine group after the third vaccination as described (23).
Two weeks after the last vaccination, animals were challenged with 200 38C13 tumor cells (37) that were grown and prepared for intraperitoneal (i.p.) administration as follows. Approximately 108 38C13 tumor cells were thawed, washed, resuspended in 10 ml of RPMI medium 1640 (Mediatech, Herndon, VA) supplemented with l-glutamine, 10% fetal calf serum (FCS), 1× penicillin/streptomycin, and 50 μM 2-mercaptoethanol, and grown in a 5% CO2/95% air atmosphere in a 37°C humidified incubator. One day later, cells were split 1:50 (vol/vol) into complete medium, and they were used the following day for tumor challenge. Cells were harvested, washed twice in RPMI medium to remove FCS, counted, and resuspended in RPMI medium at 4 × 102 cells per ml; 0.5 ml was administered i.p., yielding a total dose of 200 cells per animal. After 2 weeks, animals were checked for visible abdominal tumors, and they were checked daily thereafter for mortality.
RESULTS
Cloning, Expression, and Purification of 38C13 scFv in Plants.
DNA encoding the 38C13 scFv was inserted into a chimeric tobacco and tomato mosaic virus cDNA clone (28). In this vector, a TMV coat protein subgenomic promoter, upstream of the insertion site of the 38C13 scFv sequence, directs initiation of the 38C13 subgenomic RNA synthesis in plant cells at the transcription start point (tsp; Fig. 1). The rice α-amylase signal peptide (38) is fused in-frame to the 38C13 scFv sequence and encodes a 31-aa peptide that targets proteins to the secretory pathway (39) and is subsequently cleaved off between the last Gly of the signal peptide and the first Met of the expressed 38C13 scFv protein (in boldface, and annotated as amino acid 1 in Fig. 1). Also shown in Fig. 1 is the linear organization of the 11.2-kb plasmid in which the TMV cDNA is maintained; an SP6 promoter has been introduced upstream of the viral cDNA, allowing for transcription of infective genomic plus-strand RNA. The sequence encoding 38C13 scFv has been introduced between the 30-kDa movement protein and the tomato mosaic virus coat protein (Tcp) genes. High levels of subgenomic RNAs are synthesized in virus-infected plant cells (26), and they thus serve as templates for the translation and subsequent accumulation of 38C13 protein.
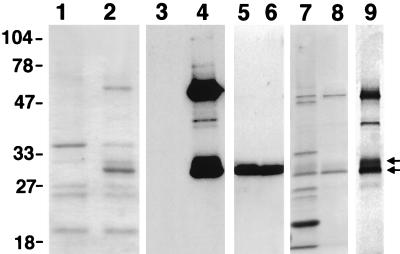
Secreted proteins were isolated by vacuum infiltration, centrifugation, and sterile filtration (IF extracts), and this material was analyzed for the presence of 38C13 scFv protein. SDS/PAGE of IF extracts showed strong Coomassie-stained bands at approximately 30 and 60 kDa (Fig. 2, lane 2) that are not present in an IF extract made from a control virus-infected plant (Fig. 2, lane 1). Several assays were employed to quantify expression and to determine whether the 38C13 scFv variable regions adopt a conformation in solution similar to that of the native IgM protein. S1C5, a monoclonal anti-idiotype antibody that recognizes native 38C13 IgM (30) and bacterially produced 38C13 scFv (23), binds to a determinant created by the association of heavy and light chains. This antibody reacts only with 38C13 IgM, or its derivatives, under nonreducing conditions, suggesting that the correct assembly of 38C13 variable regions must occur for recognition. We used this antibody to identify 38C13 scFv-specific bands in IF extracts by Western blotting (Fig. 2, lane 4). No cross-reactivity to plant proteins was seen in IF extracts prepared from control infected plants (Fig. 2, lane 3). Both the 30- and 60-kDa bands react strongly with S1C5 under nonreducing conditions, corresponding to the correct sizes for scFv monomer and a spontaneously assembling scFv dimer. A minor band at 40 kDa is also revealed by S1C5 Western analysis, most likely caused by proteolysis of the dimer. Mild disulfide reduction of crude extracts (Fig. 2, lane 5) or deletion of the cysteine at position 3 (lane 6) results in a single band of 38C13 scFv monomers in crude IF material.
Figure 2.
Protein analysis of N. benthamiana plants 2 weeks after inoculation. Lanes 1 and 2, Coomassie-stained SDS/PAGE of IF extract from viral constructs expressing rice α-amylase protein (lane 1) or of 38C13 scFv IF extract (lane 2) run under nonreducing conditions. Lanes 3 and 4, Western blot of a duplicate gel probed with anti-38C13-specific monoclonal antibody S1C5 containing the control extract (lane 3) or 38C13 scFv (lane 4). Lanes 5 and 6 show S1C5 Western blots of 38C13 scFv-containing IF prepared under reducing conditions (but separated by PAGE under nonreducing conditions) or IF containing an 38C13 scFv protein that has no cysteine at amino acid 3. Lanes 7 and 8, Coomassie stain of the starting IF extract (lane 7) and of single-pass, S1C5 affinity-purified 38C13 scFv protein (lane 8). Lane 9, silver stain of affinity-purified protein.
Approximately 90–95%-pure 38C13 scFv was recovered from plant IF extract by S1C5 affinity chromatography as determined by Coomassie and silver staining of SDS/PAGE. Lanes 7 and 8 of Fig. 2 show a Coomassie stain of crude IF before and after one-step purification of 38C13 scFv protein, respectively. A weaker band of slightly slower mobility is seen just above the 30-kDa band (double arrows in Fig. 2) by Coomassie and Western analysis in crude and S1C5-purified preparations, which is more clearly seen on a silver-stained gel of S1C5-purified material (Fig. 2, lane 9). Purification of 38C13 scFv was carried out under endotoxin-free conditions by sterile filtering plant extracts and using sterile-filtered and endotoxin-free buffers in all chromatography steps. Filtered IF protein extracts have very low endotoxin content initially (<0.06 endotoxin unit/ml; data not shown), and were considered endotoxin free after purification.
The 38C13 scFv protein continues to accumulate in the IF over the time period examined, 11–18 days, indicating that both the viral vector and protein are stable (data not shown). A summary of the purification results for two lots of 38C13 scFv is presented in Table 1. Plant-produced 38C13 scFv from two independent infections was quantified by anti-idiotype ELISA, and we found that IF extracts contain 20–60 μg/ml specific protein. By comparing quantitation by ELISA under conditions that favor anti-idiotype recognition in solution to quantitation by Coomassie and total protein determination (data not shown) we conclude that the major fraction of 38C13 scFv is soluble and properly folded in plant IF extracts. Protein yield is equivalent to or exceeds that of transgenic plants (18, 21, 22) and is similar to that of an scFv that has been optimized for expression in bacteria (40).
Table 1.
Purification of scFv proteins
| Harvest time | Leaf wt (wet), g | IF vol, ml | scFv recovered, μg/ml | Equivalent wt in plant, mg/kg |
|---|---|---|---|---|
| Day 11 | 205 | 110 | 22.95 | 12.3 |
| Day 14 | 206 | 100 | 62.20 | 30.2 |
Animal Vaccination, Analysis, and Tumor Challenge.
To determine if the plant-produced 38C13 scFv can elicit an anti-38C13 response and protect animals from 38C13 tumor challenge, we vaccinated C3H mice with 38C13 scFv protein, using a vaccination schedule that has been shown to be sufficient for tumor protection in a study with bacterially expressed 38C13 scFv (23). However, whereas the protein used by Hakim, et al. (23) included an immunoenhancing 9-aa peptide from interleukin 1β (41), we tested 38C13 scFv without the interleukin 1β peptide. The vaccine was administered in either the presence or the absence of QS-21 adjuvant, a purified derivative of saponin (34) that is now in use in some clinical vaccine treatments (42, 43). As a positive control, mice were vaccinated s.c. with 38C13 IgM coupled to KLH along with QS-21 concurrent with the second and third scFv vaccinations. Control animals received QS-21 in PBS.
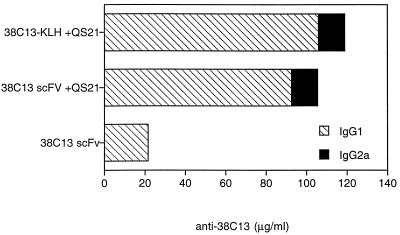
Specific anti-38C13 responses were detected by ELISA 10 days after the second (data not shown) and third vaccinations in sera of immunized mice (Fig. 3). After the third vaccination, the average IgG1 anti-38C13 titers increased from 3 to 21.6 μg/ml in animals receiving 38C13 scFv alone, and to 105 μg/ml with co-administration of QS-21 (Fig. 3, hatched bars). Anti-38C13 titers in mice vaccinated with 38C13 IgM-KLH + QS-21 were measured at 116 μg/ml. Administration of 38C13 scFv with QS-21 not only induced higher levels of antibody responses but also induced an IgG2a isotype response (13 μg/ml), which is similar to that seen for mice vaccinated with 38C13 IgM-KLH + QS-21 (Fig. 3, solid bars). Antibody titers of the IgG2a class have been correlated with augmented tumor protection; however, sufficient IgG1 responses can also be protective (44).
Figure 3.
Anti-38C13 titers in vaccinated animals. Ten days after the third vaccination, serum from each group of 10 C3H mice vaccinated with 38C13 scFv in the absence or presence of the adjuvant QS-21, or with 38C13-IgM coupled to KLH and injected with QS-21, was pooled and analyzed with the native 38C13 IgM as the target. Anti-38C13 IgG1 and IgG2a serum levels were quantitated by comparison with purified isotype-specific mouse anti-38C13 standards. Concentrations of IgG1 (hatched bars) and IgG2a (solid bars) are shown for all three 38C13 vaccine groups. No detectable anti-38C13 was measured in the control QS-21 vaccine group.
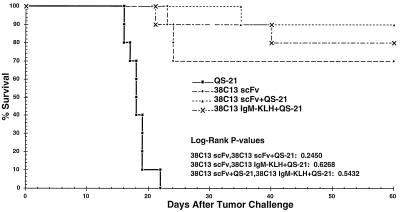
To assess the ability of the immune response to protect animals from tumor challenge, 38C13 tumor cells were injected into vaccinated mice or control animals 2 weeks after the third vaccination, and survival was monitored for 60 days thereafter (Fig. 4). Animals receiving QS-21 alone developed palpable abdominal tumors 15 days after implantation and died within 21 days. 38C13 scFv vaccine groups were statistically different from QS-21 alone (P < 0.00001). Animals receiving two vaccinations of 38C13 IgM-KLH + QS-21, the “gold standard” for idiotype vaccinations, gave 80% survival 60 days after tumor challenge. Groups of mice receiving plant-produced vaccine, either 38C13 scFv alone or 38C13 scFv + QS-21, showed a high degree of protection (70% and 90% survival 60 days after tumor challenge, respectively). The protective responses induced by plant-produced vaccines were statistically equivalent to the response of the gold standard (see tabulation within Fig. 4). Despite the lower levels of antibody responses in the mice vaccinated with 38C13 scFv, compared with mice vaccinated with either 38C13 scFv + QS-21 or 38C13 IgM-KLH, and despite the lack of IgG2a isotype, the mice receiving 38C13 scFv were protected equally well from tumor challenge. Immune serum from mice immunized with 38C13 scFv was used in a Western analysis on IF or purified 38C13 scFv; only monomer and dimer 38C13 scFv proteins were visualized, suggesting that the 38C13 scFv, not some plant contaminant, constituted the effective vaccine (data not shown).
Figure 4.
Mice immunized with plant-derived scFv protein are protected from tumor challenge. Tumor protection was measured from the time of tumor implantation (day 0) and is plotted as percent survival. These results are representative of two experiments. While all vaccinated groups statistically differed from the susceptible control (P < 0.00001), there was no statistical segregation among vaccinated groups (see tabulation in the graph).
Molecular Analysis of the scFv Protein.
Some plant glycans contain potentially immunogenic N-linked monosaccharides covalently attached to the chitobiose core (α1–3 fucose and β1–2 xylose) (45). Although there are no canonical N-glycosylation sites on 38C13 variable regions, alternative N-glycosylation site usage could not be ruled out. Therefore we performed total carbohydrate analysis of SDS-separated proteins by using PAS (Fig. 5). 38C13 IgM, which has five N-glycosylated sites (46), and HRP, which has one N-glycosylated site (47), were used as positive controls. PAS-positive material is detected in a lane containing 10 μg of 38C13 IgM (lane 1). Five and 25 μg of HRP (lanes 2 and 3) also show PAS-reactive bands at the expected molecular mass of 44 kDa, suggesting that a single N-linked carbohydrate molecule can clearly be detected at these protein concentrations. No PAS reactivity is seen with 5 or 25 μg of plant-produced scFv protein (lanes 4 and 5). Lane 6 has been split and shows both PAS stain (a) and Coomassie stain (b) of 25 μg of scFv; only Coomassie reactivity of 38C13 scFv appears in the expected position. Additionally, analysis kindly performed by Glyko (Novato, CA) found no O-linked carbohydrates in plant-produced scFv protein (data not shown).
To determine whether incomplete cleavage of the signal peptide might be the source of immunogenicity, the N-terminal region of both molecules in the 30-kDa region were sequenced (data not shown). Both species had the same Met start (amino acid 1) as shown in Fig. 1.
DISCUSSION
We present here evidence that a transient tobamoviral infection can successfully drive whole plant expression of a soluble scFv protein. Although scFv proteins have been produced by transgenic technology (18–22), we know of no other report of such immunogens being rapidly produced in plants by transient viral expression. In contrast to conventional plant transgenic approaches, which can take months or years, plant samples expressing the desired protein were positively identified by both ELISA and Western analysis approximately 4 weeks after molecular cloning. Because of the speed of expression and ease of isolation of proteins from enriched secretory fractions, our approach represents a dramatic improvement in the efficiency of producing complex, biologically active proteins in plants.
Recombinant production of properly folded scFv protein is a major advantage of the viral plant expression system. The literature cites many examples of proteins that can be refolded by denaturation/renaturation; however, in many cases only a small fraction of the protein adopts the correct conformation, a conformation that can be selected for by antigen affinity purification. scFv proteins that will be used for patient-specific vaccination will not be characterized, or purified, by antigen binding; therefore correct conformation in solution is extremely important to the successful application of scFv vaccines for human use. In our experience, scFv proteins that were not soluble or secreted from bacteria failed to react with anti-idiotype antibodies in solution, suggesting that they were not forming antigen-binding sites that were similar to those presented by the tumor cells. Two scFv proteins [one derived from BCL-1 (48) and one derived from human CJ sequences (17)] that were not soluble after expression in bacteria have been made by transient expression in plants, and both proteins are soluble, secreted, and react with anti-idiotype antibodies in solution (data not shown).
We have succeeded in rapid production of the murine 38C13 scFv, but more importantly, by both antibody responses in vaccinated mice and survival in a lethal tumor challenge, the conformation of the protein is relevant to the tumor idiotype. Molecular analysis has suggested that immunogenicity of the vaccine is not due to improper cleavage of the α-amylase signal peptide, based on N-terminal sequencing, nor is it likely due to the presence of immunogenic plant sugars, since 38C13 scFv protein is devoid of carbohydrate by several tests. The conformation of the 38C13 scFv protein in solution, as a dimer or multimer, may be an important determinant in generating immune responses in mice that are relevant to the tumor idiotype. Other studies comparing monomeric or oligomeric gp120 from HIV I have also shown that the oligomeric vaccine, which more closely resembles the viral presentation of gp120, is more effective in generating protective immune responses (49). Similarly, nonimmunogenic T cell epitopes could be made immunogenic by linear homopolymerization (50). In further support of this hypothesis, neither the monomeric 38C13 scFv made in bacteria [without the interleukin 1β tag (23)] nor the monomeric 38C13 scFv Cys− form produced in plants generates anti-idiotype reactivity in vaccinated mice (data not shown). These data suggest that presentation of the variable regions in a multimeric form, similar to that found on the surface of a B cell, may be a key to generating scFv vaccine efficacy. In addition, coadministration of immunostimulatory peptides and cytokines may enhance the vaccine efficacy above that of the current Ig vaccine.
The demonstrated ability of nontransgenic plants to produce functional scFv proteins quickly, abundantly, and in the correct conformation, may enable the treatment of human lymphomas through effective immunotherapy, and has broad-range implications for the production of other, medically important scFv proteins.
Acknowledgments
Excellent technical assistance was provided by Sherri Wykoff, Dave Clary, and Rhonda Bransteitter at Biosource. Thanks also go to the all the members of the Levy lab for advice and support for this project. This work was supported in part by Public Heath Service Grants CA33399 and AI37219, and R.L. is an American Cancer Society Clinical Research Professor.
ABBREVIATIONS
- NHL
non-Hodgkin’s lymphoma
- scFv
single-chain antibody
- KLH
keyhole limpet hemocyanin
- TMV
tobacco mosaic virus
- IF
interstitial fraction
- HRP
horseradish peroxidase
- PAS
periodic acid–Schiff
References
- 1.Ries L A G, Kosary C L, Hankey B F, Harras A, Miller B A, Edwards B K, editors. SEER Cancer Statistics Review, 1973–1993: Tables and Graphs. Bethesda: Natl. Cancer Inst.; 1996. [Google Scholar]
- 2.Parker S L, Tong T, Bolden S, Wingo P A. CA Cancer J Clin. 1997;47:5–27. doi: 10.3322/canjclin.47.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Armitage J O. CA Cancer J Clin. 1997;47:323–325. doi: 10.3322/canjclin.47.6.323. [DOI] [PubMed] [Google Scholar]
- 4.Kwak L W, Campbell M J, Czerwinski D K, Hart S, Miller R A, Levy R. N Engl J Med. 1992;327:1209–1215. doi: 10.1056/NEJM199210223271705. [DOI] [PubMed] [Google Scholar]
- 5.Hsu F J, Kwak L, Campbell M, Liles T, Czerwinski D, Hart S, Syrengelas A, Miller R, Levy R. Ann N Y Acad Sci. 1993;690:385–387. doi: 10.1111/j.1749-6632.1993.tb44039.x. [DOI] [PubMed] [Google Scholar]
- 6.Nelson E L, Li X, Hsu F J, Kwak L W, Levy R, Clayberger C, Krensky A M. Blood. 1996;88:580–589. [PubMed] [Google Scholar]
- 7.Hsu F J, Caspar C B, Czerwinski D, Kwak L W, Liles T M, Syrengelas A, Taidi-Laskowski B, Levy R. Blood. 1997;89:3129–3135. [PubMed] [Google Scholar]
- 8.Carroll W L, Thielemans K, Dilley J, Levy R. J Immunol Methods. 1986;89:61–72. doi: 10.1016/0022-1759(86)90032-3. [DOI] [PubMed] [Google Scholar]
- 9.Thielemans K, Maloney D G, Meeker T, Fujimoto J, Doss C, Warnke R A, Bindl J, Gralow J, Miller R A, Levy R. J Immunol. 1984;133:495–501. [PubMed] [Google Scholar]
- 10.Skerra A, Plückthun A. Science. 1988;240:1038–1041. doi: 10.1126/science.3285470. [DOI] [PubMed] [Google Scholar]
- 11.Plückthun A, Skerra A. Methods Enzymol. 1989;178:497–515. doi: 10.1016/0076-6879(89)78036-8. [DOI] [PubMed] [Google Scholar]
- 12.Winter G, Milstein C. Nature (London) 1991;349:293–299. doi: 10.1038/349293a0. [DOI] [PubMed] [Google Scholar]
- 13.Hayden M S, Linsley P S, Gayle M A, Bajorath J, Brady W A, Norris N A, Fell H P, Ledbetter J A, Gilliland L K. Ther Immunol. 1994;1:3–15. [PubMed] [Google Scholar]
- 14.Kretzschmar T, Aoustin L, Zingel O, Marangi M, Vonach B, Towbin H, Geiser M. J Immunol Methods. 1996;195:93–101. doi: 10.1016/0022-1759(96)00093-2. [DOI] [PubMed] [Google Scholar]
- 15.Bei R, Schlom J, Kashmiri S V. J Immunol Methods. 1995;186:245–255. doi: 10.1016/0022-1759(95)00149-5. [DOI] [PubMed] [Google Scholar]
- 16.Deshane J, Siegal G P, Alvarez R D, Wang M H, Feng M, Cabrera G, Liu T, Kay M, Curiel D T. J Clin Invest. 1995;96:2980–2989. doi: 10.1172/JCI118370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caspar C B, Levy S, Levy R. Blood. 1997;90:3699–3706. [PubMed] [Google Scholar]
- 18.Schouten A, Roosien J, van Engelen F A, de Jong G A, Borst-Vrenssen A W, Zilverentant J F, Bosch D, Stiekema W J, Gommers F J, Schots A, Bakker J. Plant Mol Biol. 1996;30:781–793. doi: 10.1007/BF00019011. [DOI] [PubMed] [Google Scholar]
- 19.Tavladoraki P, Benvenuto E, Trinca S, De Martinis D, Cattaneo A, Galeffi P. Nature (London) 1993;366:469–472. doi: 10.1038/366469a0. [DOI] [PubMed] [Google Scholar]
- 20.Artsaenko O, Peisker M, zur Nieden U, Fiedler U, Weiler E W, Muntz K, Conrad U. Plant J. 1995;8:745–750. doi: 10.1046/j.1365-313x.1995.08050745.x. [DOI] [PubMed] [Google Scholar]
- 21.Bruyns A M, Jaeger G, De Neve M, De Wilde C, Van Montagu M, Depicker A. FEBS Lett. 1996;386:5–10. doi: 10.1016/0014-5793(96)00372-9. [DOI] [PubMed] [Google Scholar]
- 22.Firek S, Draper J, Owen M R, Gandecha A, Cockburn B, Whitelam G C. Plant Mol Biol. 1993;23:861–870. doi: 10.1007/BF00021540. [DOI] [PubMed] [Google Scholar]
- 23.Hakim I, Levy S, Levy R. J Immunol. 1996;157:5503–5511. [PubMed] [Google Scholar]
- 24.Spellerberg M B, Zhu D, Thompsett A, King C A, Hamblin T J, Stevenson F K. J Immunol. 1997;159:1885–1892. [PubMed] [Google Scholar]
- 25.Brocks B, Rode H J, Klein M, Gerlach E, Dubel S, Little M, Pfizenmaier K, Moosmayer D. Immunotechnology. 1997;3:173–184. doi: 10.1016/s1380-2933(97)00011-0. [DOI] [PubMed] [Google Scholar]
- 26.Kumagai M H, Turpen T H, Weinzettl N, della-Cioppa G, Turpen A M, Donson J, Hilf M E, Grantham G L, Dawson W O, Chow T P, et al. Proc Natl Acad Sci USA. 1993;90:427–430. doi: 10.1073/pnas.90.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carroll W L, Starnes C O, Levy R, Levy S. J Exp Med. 1988;168:1607–1620. doi: 10.1084/jem.168.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumagai M H, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill L K. Proc Natl Acad Sci USA. 1995;92:1679–1683. doi: 10.1073/pnas.92.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawson W O, Beck D L, Knorr D A, Grantham G A. Proc Natl Acad Sci USA. 1986;83:1832–1836. doi: 10.1073/pnas.83.6.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maloney D G, Kaminski M S, Burowski D, Haimovich J, Levy R. Hybridoma. 1985;4:191–209. doi: 10.1089/hyb.1985.4.191. [DOI] [PubMed] [Google Scholar]
- 31.Potter K N, Li Y, Pascual V, Capra J D. Int Rev Immunol. 1997;14:291–308. doi: 10.3109/08830189709116521. [DOI] [PubMed] [Google Scholar]
- 32.Roben P W, Salem A N, Silverman G J. J Immunol. 1995;154:6437–6445. [PubMed] [Google Scholar]
- 33.Merril C R. Methods Enzymol. 1990;182:477–488. doi: 10.1016/0076-6879(90)82038-4. [DOI] [PubMed] [Google Scholar]
- 34.White A C, Cloutier P, Coughlin R T. Adv Exp Med Biol. 1991;303:207–210. doi: 10.1007/978-1-4684-6000-1_22. [DOI] [PubMed] [Google Scholar]
- 35.Kensil C R, Soltysik S, Wheeler D A, Wu J Y. Adv Exp Med Biol. 1996;404:165–172. doi: 10.1007/978-1-4899-1367-8_15. [DOI] [PubMed] [Google Scholar]
- 36.Kaminski M S, Kitamura K, Maloney D G, Levy R. J Immunol. 1987;138:1289–1296. [PubMed] [Google Scholar]
- 37.Bergman Y, Haimovich J. Eur J Immunol. 1977;7:413–417. doi: 10.1002/eji.1830070702. [DOI] [PubMed] [Google Scholar]
- 38.O’Neill S D, Kumagai M H, Majumdar A, Huang N, Sutliff T D, Rodriguez R L. Mol Gen Genet. 1990;221:235–244. doi: 10.1007/BF00261726. [DOI] [PubMed] [Google Scholar]
- 39.Firek S, Whitelam G C, Draper J. Transgenic Res. 1994;3:326–331. doi: 10.1007/BF01973593. [DOI] [PubMed] [Google Scholar]
- 40.Kipriyanov S M, Moldenhauer G, Martin A C, Kupriyanova O A, Little M. Protein Eng. 1997;10:445–453. doi: 10.1093/protein/10.4.445. [DOI] [PubMed] [Google Scholar]
- 41.Beckers W, Villa L, Gonfloni S, Castagnoli L, Newton S M, Cesareni G, Ghiara P. J Immunol. 1993;151:1757–1764. [PubMed] [Google Scholar]
- 42.Helling F, Zhang S, Shang A, Adluri S, Calves M, Koganty R, Longenecker B M, Yao T J, Oettgen H F, Livingston P O. Cancer Res. 1995;55:2783–2788. [PubMed] [Google Scholar]
- 43.Davis T A, Hsu F J, Caspar C B, Liles T M, Czerwinski D, Taidi B, Levy R. Blood. 1997;90:509A. (abstr.). [PubMed] [Google Scholar]
- 44.Kaminski M S, Kitamura K, Maloney D G, Campbell M J, Levy R. J Immunol. 1986;136:1123–1130. [PubMed] [Google Scholar]
- 45.Garcia-Casado G, Sanchez-Monge R, Chrispeels M J, Armentia A, Salcedo G, Gomez L. Glycobiology. 1996;6:471–477. doi: 10.1093/glycob/6.4.471. [DOI] [PubMed] [Google Scholar]
- 46.Anderson D R, Atkinson P H, Grimes W J. Arch Biochem Biophys. 1985;243:605–618. doi: 10.1016/0003-9861(85)90538-7. [DOI] [PubMed] [Google Scholar]
- 47.Wilson I B, Altmann F. Glycoconjugate J. 1998;15:203–206. doi: 10.1023/a:1006932725821. [DOI] [PubMed] [Google Scholar]
- 48.Slavin S, Strober S. Nature (London) 1978;272:624–626. doi: 10.1038/272624a0. [DOI] [PubMed] [Google Scholar]
- 49.Parren P W, Fisicaro P, Labrijn A F, Binley J M, Yang W P, Ditzel H J, Barbas C F, 3rd, Burton D R. J Virol. 1996;70:9046–9050. doi: 10.1128/jvi.70.12.9046-9050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borras-Cuesta F, Fedon Y, Petit-Camurdan A. Eur J Immunol. 1988;18:199–202. doi: 10.1002/eji.1830180203. [DOI] [PubMed] [Google Scholar]