Abstract
We show that a single clinical isolate of the human opportunistic pathogen Pseudomonas aeruginosa (strain PA14), which previously was shown to be pathogenic in mice and plants, also kills Caenorhabditis elegans. The rate of PA14-mediated killing of C. elegans depends on the composition of the agar medium on which PA14 is grown. When PA14 is grown on minimal medium, killing occurs over the course of several days and is referred to as “slow” killing. When PA14 is grown on high-osmolarity medium, killing occurs over the course of several hours and is referred to as “fast” killing. Several lines of evidence, including the fact that heat-killed bacteria are still capable of fast but not slow killing of C. elegans, indicate that fast and slow killing occur by distinct mechanisms. Slow killing involves an infection-like process and correlates with the accumulation of PA14 within worm intestines. Among 10 PA14 virulence-related mutants that had been shown previously to affect pathogenicity in plants and mice, 6 were less effective in killing C. elegans under both fast- and slow-killing conditions, indicating a high degree of commonalty among the P. aeruginosa factors required for pathogenicity in disparate eukaryotic hosts. Thus, we show that a C. elegans pathogenicity model that is genetically tractable from the perspectives of both host and pathogen can be used to model mammalian bacterial pathogenesis.
Host–pathogen interactions are antagonistic relationships in which the success of each organism depends on its ability to overcome the other. The outcome of any particular interaction depends on the status of a coevolutionary “arms race” in which advances in pathogen virulence factors are overcome by counteradvances in host defense responses. Recently, a variety of observations suggest that some of the underlying mechanisms of defense against pathogens by plant, invertebrate, and mammalian hosts may be similar. For example, in response to microbial infection, both mammals and insects use a conserved signal-transduction pathway to activate defense-related genes. In the mammalian innate immune response, the activation of NF-κB in the presence of a pathogen-generated signal is mediated by the interleukin-1 or TOLL receptors (1, 2). In adult Drosophila melanogaster, the TOLL receptor signals via Dorsal, the NF-κB homologue, to activate an antifungal response (3). Interestingly, three plant proteins, N, L6, and RPP5, which are involved in the activation of the plant defense response, share homology with the cytoplasmic domains of the mammalian interleukin-1 receptor and the mammalian and Drosophila TOLL receptors (4). Effector molecules used by the hosts to kill pathogens directly are also similar. Invertebrates, vertebrates, and plants all produce a similar class of membrane-active small cationic peptides, the arginine-rich three disulfide-containing β-sheet defensins, in a pathogen-inducible manner (5–7).
Because of these universal host responses, a coevolutionary arms race between hosts and pathogens could select for a common strategy of microbial pathogenesis. Indeed, plant and animal bacterial pathogens seem to share a set of similar strategies in invading their hosts. First, several of the secretory apparatuses used by both plant and animal bacterial pathogens to export virulence factors are similar (8). Second, the signal-transduction pathways and regulatory genes by which pathogens respond to environmental or host stimuli and the subsequent transcriptional activation of virulence genes are similar in plant and animal bacterial pathogens (9). Third, as plant and animal hosts deploy similar antimicrobial peptides against invading pathogens, plant and animal pathogens use the same mechanism, the Sap system, to nullify the cytotoxic peptides produced by their respective host’s defense system (10).
Recently, by using Pseudomonas aeruginosa strain PA14, which is capable of causing disease in both mice and Arabidopsis thaliana, our laboratory showed that mutations in a variety of PA14 virulence-related genes resulted in significantly reduced virulence in both plants and mice, further suggesting that the molecular basis for bacterial pathogenesis may be conserved in evolutionarily divergent hosts (11, 12). P. aeruginosa is a ubiquitous Gram-negative bacterium commonly found in aquatic environments, in soil, and in association with many eukaryotic organisms. P. aeruginosa has been reported to cause disease in plants (13, 14), insects (15), and a variety of vertebrates (16). In humans, P. aeruginosa causes a broad spectrum of opportunistic infections and is one of the most important causes of Gram-negative bacteremia in major medical centers in the United States (17).
Much remains to be understood about animal–pathogen interactions at the molecular level. Moreover, the use of mammalian hosts makes it difficult to dissect genetically the host pathways that are targeted by bacterial virulence factors because of a lack of a genetically tractable metazoan model system. We reasoned that if some aspects of both offensive and defensive features of vertebrate, invertebrate, and plant pathogenesis are universal, then we should be able to model certain aspects of mammalian pathogenesis by using a simple and genetically tractable host such as Caenorhabditis elegans. In this paper, we describe the development of a pathogenesis model that involves the killing of the soil nematode C. elegans by the P. aeruginosa strain PA14. Hitherto, except for reports that Bacillus megaterium kills C. elegans (18) and that beta exotoxin from Bacillus thuringiensis exhibits contact nematicide activity against all stages of C. elegans (19), to the best of our knowledge, no pathogenic interaction between C. elegans and a bacterium has been described in the literature.
We show that one mechanism by which P. aeruginosa kills C. elegans involves an infection-like process that correlates with the accumulation of P. aeruginosa in worm intestines. In addition, we show that many of the P. aeruginosa factors (genes) required for this killing are also required for pathogenesis in mammalian or plant hosts. Because this metazoan system allows for genetic dissection of both pathogen virulence and host responses, in the future, the C. elegans–P. aeruginosa pathogenesis model can be used to dissect genetically the interactions between pathogen virulence factors and host defense mechanisms.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Media.
The P. aeruginosa strains PA14 (11), PAO1 (20), PAK (21), PA29 (11), and PO37 (22), the Pseudomonas syringae pv. maculicola strain ES4326 (23), the Pseudomonas fluorescens strains WCS365 (24) and 2-79 [Northern Utilization Research and Development (USDA) B-15132; ref. 25], and the Escherichia coli strains DH5α (Bethesda Research Laboratories) and OP50 (26) have been described. P. fluorescens strain 55 was obtained from E. Schott (Massachusetts General Hospital, Boston). The P. aeruginosa PA14 derivatives containing a cassette encoding gentamycin in the toxA, plcS, and gacA genes (11), as well as the nonpolar PA14 gacA mutation SW7-4 and the PA14 TnphoA derivatives 33C7, 25A12, 33A9, 16G12, 34H4, 25F1, pho15, ID7, and pho34B12 (12), have been described. Plasmids pRR54 (27) and pGFP-19-1 (28) have been described. Complete media for bacteria culture and maintenance were Luria–Bertani broth and King’s broth (29, 30), and minimal medium was M9 (30). PA14 was cultured on nematode growth (NG; modified from nematode growth medium agar described in ref. 31; 0.35% instead of 0.25% peptone was used) or peptone-glucose-sorbitol (PGS; 1% Bacto-Peptone/1% NaCl/1% glucose/0.15 M sorbitol/1.7% Bacto-Agar) media for nematode killing assays.
Nematode Strains.
All strains were cultured as described (31). Bristol N2 is the parental wild-type strain (26) of the following strains used in this study: CB1489, him-8(e1489); BA1, fer-1(hc1ts)I; and DA531, eat-1(ad427)IV. The Caenorhabditis Genetics Center provided some of these strains. J. Kaplan (University of California, Berkeley, CA) provided DA531.
Construction of PA14 and E. coli DH5α Expressing Green Fluorescent Protein (GFP).
The superglow GFP derivative containing the substitution Val163 to alanine and Ser65 to threonine was used in this study (28). A 1.0-kb fragment containing the entire GFP ORF, as well as the ribosome binding site and the T7 termination site, was cloned from pGFP-19-1 into the XbaI site of the broad host-range vector pRR54 downstream of the lacZ promoter of pRR54. This construct, pRR54GFP19-1, was subsequently transformed into E. coli DH5α and mated into PA14 and PA14 gacA SW7-4 by the method of triparental mating by using pRK2013 (32) to make strains DH5α/GFP, PA14/GFP, and gacA SW7-4/GFP, respectively.
C. elegans Killing Assays.
An overnight King’s broth culture (5–10 μl) of the test bacterial strain was spread on a 3.5-cm diameter NG (slow killing) or PGS (fast killing) agar plate and incubated at 37°C for 24 h. After 8–24 h (slow killing) or 8–12 h (fast killing) at room temperature (23–25°C), each plate was seeded with 40–50 (slow killing) or 30–40 (fast killing) L4 stage or adult hermaphrodite worms. Plates were incubated at 25°C and scored for live worms every 4–6 h. For statistical purposes, 3–4 replicates per trial were carried out. E. coli OP50 was used as a negative control. A worm was considered dead when it no longer responded to touch. Any worms that died as a result of getting stuck to the wall of the plate were excluded from the analysis. To determine time required to kill 50% of the nematodes (LT50) in the slow-killing assay a curve was fitted to the data by using the systat (version 5.2.1) computer program: proportion of worms killed = A + (1 − A)/{1 + exp[B − G × log (hours after exposure)]}, where A is the fraction of worms that died in a OP50 control experiment and B and G are parameters that are varied to fit the curve. Once B and G have been determined, LT50 is calculated by the formula LT50 = exp (B/G) × (1 − 2 × A)(1/G).
C. elegans Shifting Experiments.
About 60 L4 worms were seeded on PA14 lawns and allowed to feed. After 18, 24, or 30 h, one half the total number (between 20 and 30 per plate) was transferred to plates containing E. coli OP50; the other half was transferred to plates containing PA14. For each time point, the experiment was repeated three times. Before transfer, worms were washed briefly in M9 buffer containing 200 μg/ml neomycin to kill any PA14 cells that were not ingested. To ensure no further transfer of noningested PA14, 200 μg/ml streptomycin was added to OP50 plates. Worm mortality was scored every 6–8 h for the next 30–40 h.
RESULTS
P. aeruginosa Kills C. elegans.
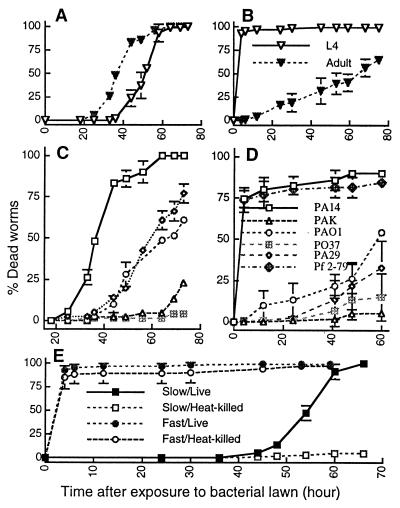
Because we had shown previously that P. aeruginosa strain PA14 was both a pathogen of plants and mice (11), we tested PA14 for its ability to kill C. elegans under different media conditions. As shown in Fig. 1A, both L4 larval stage and 1-day-old adult hermaphrodite C. elegans die over a period of 2.5–3 days when feeding on a lawn of PA14 grown on NG agar. Similar results were obtained on minimal M9 agar (not shown). The rate of killing was quantified by determining the LT50; adults are more susceptible than L4 stage worms (LT50 = 37.8 ± 2.4 h and 48.3 ± 1.8 h, respectively; Fig. 1A).
Figure 1.
Kinetics of the killing of C. elegans by P. aeruginosa under slow-killing and fast-killing conditions. (A) L4 stage (open inverted triangles) and 1-day-old adult hermaphrodite (closed inverted triangles) worms feeding on P. aeruginosa PA14 grown on NG (slow killing) medium. (B) L4 stage (open inverted triangles) and 1-day-old adult hermaphrodite (closed inverted triangles) worms exposed to P. aeruginosa PA14 grown on PGS (fast killing) medium. (C) Adult (1-day-old) hermaphrodite C. elegans feeding on P. aeruginosa PA14 (open squares), PAO1 (open circles), PA29 (open diamonds), PAK (open triangles), and PO37 (crossed squares) grown on NG (slow killing) medium. (D) L4 stage C. elegans feeding on P. aeruginosa PA14 (open squares), PAO1 (open circles), PA29 (open diamonds), PAK (open triangles), and PO37 (crossed squares), and P. fluorescens 2-79 (crossed diamonds) grown on PGS (fast killing) medium. (E) L4 stage worms feeding on heat-killed PA14 (30 min at 65°C) grown on NG (open squares) or PGS (open circles) media or L4 stage worms feeding on live PA14 grown on NG (closed squares) or PGS (closed circles) media.
In contrast to the killing of C. elegans observed on NG and M9 media, when PA14 is grown on PGS agar, a richer medium of higher osmolarity, the L4 larval stage worms die within 4–24 h (Fig. 1B). We define the killing of C. elegans described in the previous paragraph that uses NG and M9 media as slow killing and the more rapid killing on PGS agar as fast killing. In contrast to slow killing, L4 worms are dramatically more susceptible to fast killing than 1-day-old adults (Fig. 1B).
We tested several P. aeruginosa strains for their ability to kill C. elegans under both fast-killing (PGS media) and slow-killing (NG media) conditions. As shown in Fig. 1C, among the P. aeruginosa strains tested for slow killing, PA14 was the most lethal to adult C. elegans (LT50 = 37.8 h); PAO1 (LT50 = 68.2 h) and PA29 (LT50 = 61.5 h) were moderately lethal; and PO37 and PAK did not kill. No slow killing was also observed with E. coli OP50, with the phytopathogen P. syringae pv. maculicola strain ES4326, or with several P. fluorescens strains (WCS365, 55 and 2-79; not shown). In the case of the nonlethal strains, including P. aeruginosa strains PO37 and PAK, C. elegans was able to undergo normal development and reproduction; a healthy brood consisting of hundreds or thousands of progeny was produced by day 4 from an initial founding population of 10 worms, and the entire lawn of bacteria was consumed. In contrast, the few progeny that were seen in populations of worms fed on P. aeruginosa PA14 eventually died, and the bacterial lawn remained intact.
As shown in Fig. 1D, PA14 was also the most effective P. aeruginosa strain tested at fast killing L4 stage C. elegans. Similarly to slow killing, E. coli OP50, P. syringae pv. maculicola E4326, and P. fluorescens 55 also did not kill under fast-killing conditions (not shown). On the other hand, P. fluorescens strains 2-79 (Fig. 1D) and WCS365 (not shown) killed C. elegans at the same rate as PA14 on PGS medium. We chose P. aeruginosa strain PA14 for further studies, because it was the most lethal in both the fast- and slow-killing assays.
No killing of C. elegans was observed under slow-killing conditions when they were fed heat-killed (Fig. 1E) or antibiotic-killed (not shown) PA14; the worms produced normal broods, which completely consumed the bacterial lawn, showing that PA14 is nutritionally adequate to support the growth and development of C. elegans. In contrast, heat-killed bacteria still killed as efficiently as the nonheated controls under fast-killing conditions (Fig. 1E), suggesting that fast killing may be mediated at least in part by an excreted heat-stable PA14 toxin(s).
Some Virulence Determinants Required for P. aeruginosa Pathogenesis in Plants and Mice Are Also Essential for Killing C. elegans.
To determine whether the molecular mechanisms underlying slow and fast killing of C. elegans by P. aeruginosa PA14 are similar to the mechanisms involved in mammalian and plant pathogenesis, we tested two categories of PA14 mutants in the nematode-killing models. The first category of mutants contained mutations in three known P. aeruginosa virulence factors, gacA, toxA, and plcS, that we had constructed previously by a marker-exchange process (11). plcS encodes a hemolytic phospholipase C (33), and toxA encodes exotoxin A, an ADP-ribosyltransferase that inactivates eukaryotic elongation factor 2 (34). GacA is a two-component response regulator and in P. aeruginosa serves as a global activator of the synthesis of cyanide, lipase, pyocyanin, and the autoinducer N-butyrylhomoserine lactone (35). We showed previously that gacA, toxA, and plcS are required for full pathogenicity of PA14 in both a mouse-burn model and in an Arabidopsis-leaf infiltration model (11).
The second category of PA14 mutants that we tested in the C. elegans killing assays consisted of a set of nine TnphoA-generated PA14 mutants that had been identified on the basis of reduced pathogenicity in a plant-leaf infection assay. Two of these nine mutants, ID7 and pho15, correspond to TnphoA insertions in the gacA and dsbA genes, respectively. dsbA encodes a periplasmic disulfide bond-forming enzyme (36). The other seven mutants contained TnphoA insertions in genes that either had not been described previously or that were not known to be involved in virulence. All nine TnphoA mutants, with the possible exception of 16G12, were reduced significantly in pathogenicity in the mouse-burn model (12).
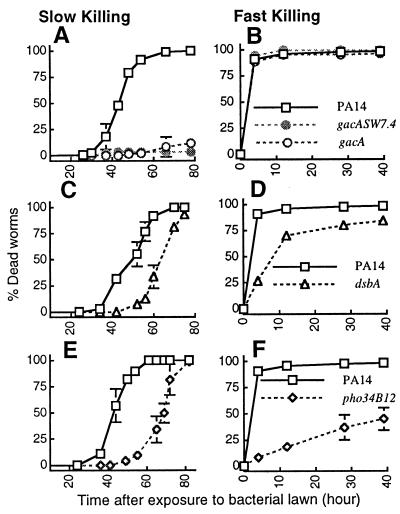
As summarized in Table 1 and shown in part in Fig. 2, mutations in six different PA14 virulence-related genes [toxA, 34H4, 25F1, dsbA(pho15), gacA (ID7), and pho34B12] affected slow killing of C. elegans, and two mutants [dsbA(pho15) and pho34B12] affected fast killing. Fig. 2 A and B shows that mutations in the PA14 gacA gene, including a construct (SW7-4) that is nonpolar on the downstream uvrC gene (12), were severely defective in their ability to kill C. elegans under slow-killing conditions (LT50 > 90 h) but had no detectable effect on the rate of fast killing. Fig. 2 C and D shows that the dsbA mutant is delayed moderately in both slow and fast killing, and Fig. 2 E and F shows that mutant pho34B12 is affected severely in fast killing and affected moderately in slow killing. The toxA, 34H4, and 25F1 mutants exhibited highly reproducible but moderate delays in slow killing and no differences from wild type in fast killing (not shown).
Table 1.
Killing of C. elegans by P. aeruginosa PA14 mutants
| Strain | Slow-killing conditions* | Fast-killing conditions† |
|---|---|---|
| PA14 | ++ | ++ |
| 33A9 | ++ | ++ |
| 33C7 | ++ | ++ |
| 25A12 | ++ | ++ |
| 16G12 | ++ | ++ |
| plcS | ++ | ++ |
| toxA | + | ++ |
| 25F1 | + | ++ |
| 34H4 | + | ++ |
| gacA (polar insertion‡) | − | ++ |
| gacA SW7.4 (nonpolar insertion§) | − | ++ |
| ID7 (gacA∷TnphoA) | − | ++ |
| pho15 (dsbA∷TnphoA) | + | + |
| pho34B12 | + | − |
Rate of slow killing relative to wild-type PA14: ++, indistinguishable (LT50 = 48–53 h); +, moderately reduced (LT50 = 55–70 h); and −, severely reduced (LT50 > 90 h).
Rate of fast killing relative to PA14: ++, indistinguishable (percentage of mortality at 12 h, %12 > 80); +, moderately reduced (%12 = 25–80); and −, severely reduced (%12 < 25).
The gacA gene is interrupted by a cassette conferring gentamycin resistance and is polar to the downstream uvrC gene (11).
A kanamycin cassette is inserted in-frame into the gacA gene, such that the translation of uvrC is reinitiated by the 3′ end of the cassette (12).
Figure 2.
The kinetics of killing by P. aeruginosa mutants, gacA (A and B), gacASW7-4 (A and B), dsbA (C and D), and pho34B12 (E and F) compared with the parental wild-type PA14 (open squares) under slow-killing (Left) and fast-killing (Right) conditions.
The observations that mutations in gacA affected only slow killing (Fig. 3 A and B) and that the TnphoA insertion in pho34B12 primarily affected fast killing (Fig. 3 E and F) suggest that fast killing is not simply an acceleration of the process seen in slow killing. The data indicating that heat-killed bacteria are still capable of fast but not slow killing are consistent with the conclusions that fast and slow killing may be mechanistically distinct and that PA14 employs different mechanisms in killing C. elegans depending on the medium in which the bacteria are grown. To explore the mechanism of PA14-mediated slow killing of C. elegans further, we designed experiments described below to determine whether slow killing involves an active infectious process. We have shown that fast killing is mediated at least in part by low molecular-weight diffusible toxins (37).
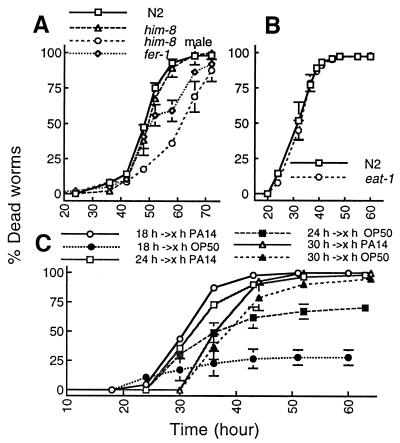
Figure 3.
Mechanisms of slow killing. (A) Death rates of nongravid adult worms. him-8 males (open circles) and fer-1 hermaphrodites (open diamonds) were compared with gravid adult worms, wild-type N2 hermaphrodites (open squares), and him-8 hermaphrodites (open triangles) feeding on PA14 grown on NG (slow killing) medium. (B) Mortality rates of 1-day-old adult wild-type N2 (open squares) and 1-day-old adult eat-1 (ad427) feeding on PA14 grown on NG medium. (C) C. elegans shifting experiments (see Materials and Methods) showing the average percentages of dead worms after transfer from PA14 to either OP50 or PA14: worms transferred to PA14 (open circles) or OP50 (closed circles) after feeding for 18 h on PA14; worms transferred to PA14 (open squares) or OP50 (closed squares) after feeding for 24 h on PA14; and worms transferred to PA14 (open triangles) or OP50 (closed triangles) after feeding for 30 h on PA14. Similar results were obtained when PA14 or PA14/GFP were shifted to DH5α or DH5α/GFP (not shown).
Slow Killing Correlates with Proliferation of PA14 in the Gut.
The events leading to L4 stage worm mortality under slow-killing conditions are as follows. Within the first 18–24 h of feeding on PA14, the rate of pharyngeal pumping and defecation intervals are not significantly different compared with worms feeding on E. coli OP50. Between 24 and 48 h, however, the motility of the worms and the rate of pharyngeal pumping gradually decline, until the nematodes become immobile and die. In most cases, animals become laden with eggs and embryos hatched internally, suggesting an egg-laying defect.
To determine whether slow killing is simply a result of matricide caused by the hatching of embryos within adult worms, we tested males and hermaphrodites of two C. elegans mutants, him-8(e1489) and fer-1(hc1ts). In him-8 mutant populations, 37% of the adults are males, whereas in wild-type N2, males occur at a frequency of only 0.2% (38). The temperature-sensitive mutation of the fer-1 gene results in sterile adults at 25°C (39), the temperature used for the slow-killing assay. As shown in Fig. 3A, him-8 males and fer-1 hermaphrodites were also killed by PA14, albeit at a slower rate than N2 or him-8 hermaphrodites. Two conclusions can be drawn. First, because males and sterile hermaphrodites were also killed, the hatching of eggs within adult hermaphrodites is not the sole cause of death. Second, embryo hatching from gravid adults increases their susceptibility to PA14-mediated killing.
To investigate the time course of PA14 pathogenesis, L4 stage worms were allowed to feed on PA14 for 18, 24, or 30 h. At each time point, half of the initial number of worms were transferred to plates containing E. coli or to fresh lawns of PA14. As shown in Fig. 3C, the proportion of worms that survived after the transfer to E. coli was inversely proportional to the time spent feeding on the pathogenic PA14. At the 60 h time point, only 25% of the worms that had been transferred to the E. coli plates after feeding on PA14 for 18 h died. The proportion of dead worms increased to 70% when worms were fed on PA14 for 24 h before transfer and to 95% when worms were transferred after 30 h. In all of these cases, 100% of the worms died when transferred from PA14 to fresh lawns of PA14. All worms transferred to E. coli after feeding on PA14 for 12 h or less survived (not shown). These data show that C. elegans can overcome a relatively brief exposure to PA14, but that once a worm has been exposed to PA14 for a sufficient time or ingested a sufficient number of PA14, they are unable to overcome the debilitating effects of the exposure.
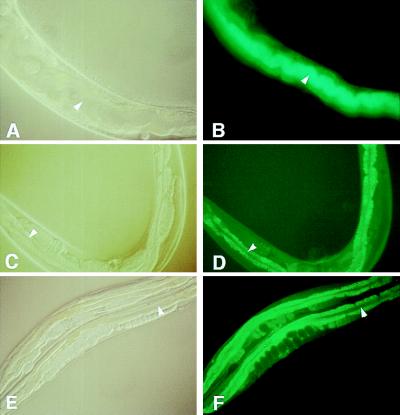
To determine whether PA14 slow killing under the above conditions involves an active infectious process, we constructed E. coli and PA14 strains expressing the Aequorea victoria GFP. As shown in Fig. 4 A and B, after 48 h of feeding on PA14/GFP, intense green fluorescence was observed throughout the lumen of worm intestines, indicating the accumulation of GFP-expressing PA14. In contrast, very little fluorescence was observed in the control E. coli DH5α/GFP strain (Fig. 4 C and D). Importantly, consistent with its attenuated virulence phenotype, the nonpolar gacA mutant expressing GFP (gacA SW7-4/GFP) failed to accumulate in the worm gut to a significant extent at this time point, although some accumulation was observed 72 h after feeding on gacA SW7-4/GFP (Fig. 4 E and F). These data suggest that the killing of C. elegans by PA14 is associated with the establishment and proliferation of pathogenic bacteria within the nematode gut.
Figure 4.
Fluorescence micrographs of worms fed on PA14/GFP (B), DH5α/GFP (D), and gacA SW7-4/GFP (F) for 48 h. The cellular structures of the same worms visualized under Nomarski phase contrast are shown in A, C, and D, respectively. (B) The entire lumen of worms fed on PA14/GFP was filled with GFP-expressing PA14. PA14/GFP began to accumulate after 36 h of feeding. In contrast, the lumen of worms feed on DH5α/GFP (D) and gacA SW7-4/GFP (F) showed no detectable GFP (see arrows), indicating that very few, if any, of the nonpathogenic bacteria were present. The intestinal cells were highly fluorescent in D and E because of autofluorescence. For gacA SW7-4/GFP, GFP expression was detected at 72 h. The micrographs for PA14/GFP are shown at ×400 magnification, whereas the micrographs for DH5α/GFP and gacA SW7-4/GFP are at ×160 magnification.
Because PA14-mediated killing of C. elegans seems to require active replication of PA14 in the worm intestine, we reasoned that decreasing the rate of PA14 replication by feeding on PA14 at lowered temperatures might spare the worms. Therefore, plates containing PA14 lawns grown for 24 h at 37°C were seeded with worms and subsequently incubated at 15 or 25°C; the generation time of PA14 on NG media at 15°C is about 5.2 times longer than at 25°C (not shown). Over 95% of worms were dead after 60 h at 25°C but no mortality was seen in plates incubated at 15°C (not shown). These data suggested that the attenuated killing at 15°C is a consequence of the slower growth rate of PA14.
To rule out the possibility that a slower rate of bacterial consumption at 15°C was the cause of decreased killing, we compared the killing of wild-type and eat-1(ad427) worms by PA14 at 25°C. The eat-1 mutant has a defect in pharyngeal muscle motion and consequently pumps more slowly than wild type, consuming bacteria at a slower rate (40). As shown in Fig. 3B, no difference in killing rate was observed between eat-1 and N2 adults. This result provides further evidence that the rate of killing by PA14 is a function of the establishment and proliferation of bacteria within the gut and is independent of the rate at which bacteria enter the gut.
DISCUSSION
P. aeruginosa is an opportunistic pathogen of vertebrates and a primary pathogen of insects. There are also sporadic reports that P. aeruginosa can elicit disease in a variety of plants. In this paper, we have shown that various P. aeruginosa strains can also kill C. elegans. The C. elegans–P. aeruginosa pathogenesis models that we have developed use a strain of P. aeruginosa, PA14, that we have previously shown to be a pathogen of both plants and mice. To the best of our knowledge, there are no other reported examples in which a single strain of a bacterial pathogen can establish a pathogenic interaction with so many evolutionarily diverse hosts.
Interestingly, we have shown that PA14 most likely kills C. elegans by at least two distinct underlying mechanisms: slow killing, which requires the consumption of live bacteria and is correlated with the proliferation of live bacteria in the worm gut, and fast killing, which does not require live bacteria and is therefore most likely mediated by exported toxins. Although the data in Figs. 3 and 4 suggest that slow killing involves an infectious process, they do not rule out the possibility that PA14 proliferation in the C. elegans gut is a consequence rather than the cause of killing. In the case of fast killing by testing isogenic toxA and plcS mutants of PA14, we showed that two known toxins, the hemolytic phospholipase C and exotoxin A, are not essential for killing worms under the fast-killing conditions. Consistent with this conclusion, we have shown that fast killing is mediated at least in part by phenazines, low molecular-weight diffusible toxins excreted by P. aeruginosa (37). The observation that two P. fluorescens strains were as lethal as PA14 in fast killing but were not pathogenic under slow-killing conditions is consistent with the conclusion that fast and slow killing are mechanistically distinct.
Our observation that some but not all strains of P. aeruginosa kill C. elegans is consistent with previous reports that some unspecified strains of P. aeruginosa are able to support growth and reproduction of C. elegans (18, 41). These observations are also consistent with the fact that different strains of P. aeruginosa have been shown to exhibit varying degrees of virulence in different mammalian (11, 42) and plant (11, 12, 14) infection models.
Importantly, several nonessential P. aeruginosa genes including toxA, gacA, and dsbA that are involved in pathogenic interactions in mice and plants are also required for the maximally effective killing of C. elegans. From an evolutionary perspective, the identification of many common virulence determinants required for pathogenesis in three distinct groups of organisms—plants, invertebrates, and vertebrates—provides firm experimental evidence that the molecular basis of P. aeruginosa pathogenicity may be conserved in three evolutionarily divergent hosts. For example, the requirement of exotoxin A for full virulence in plant, nematode, and mammalian hosts suggests that these evolutionarily distant organisms have conserved not only the target for this ADP-ribosylating toxin but may have also conserved the receptor for the toxin and the transport of the toxin into the host cells.
The development of the C. elegans–P. aeruginosa pathogenesis model enables the application of systematic genetic analysis to study the interaction between a bacterial pathogen and an animal host from the perspectives of both the pathogen and the host. Importantly, because P. aeruginosa mutations that affect fast or slow killing of C. elegans also affect mouse pathogenesis, both fast and slow killing seem to be relevant models of various aspects of mammalian pathogenesis. We have already used the C. elegans–P. aeruginosa model to identify P. aeruginosa PA14 mutants that are impaired in fast (37) or slow (M.-W.T. and F.M.A., unpublished work) killing and have found that many of these mutants are also less pathogenic in a mouse-burn model. In addition, we have isolated C. elegans mutants that are more resistant to both fast and slow killing and C. elegans mutants that are more susceptible to slow killing (M.-W.T. and S.M.-M., unpublished data). Analysis of these latter mutants would help dissect the complex mechanisms involved in the host response to pathogen attack. Finally, our laboratory has also shown that P. aeruginosa strain PA14 is a potent pathogen of several insects, including D. melanogaster, Galleria mellonella (greater wax moth), and Plutella xylostella (diamondback moth) (G. Jander and S.M.-M., unpublished data). Like C. elegans, D. melanogaster holds much promise for the genetic analysis of the innate immune response as demonstrated in several recent publications (3, 43).
Acknowledgments
C. Darby and C. Manoil have also developed a C. elegans–P. aeruginosa pathogenesis model and we thank them for sharing data before publication. We also thank G. Ruvkun and D. Elkes for helpful suggestions regarding growing C. elegans at the initial stage of this work. This work was supported by a grant from Hoechst to Massachusetts General Hospital. M.-W.T. is a Harvard Junior Fellow, and S.M.-M. is a Helen Hay Whitney Fellow.
ABBREVIATIONS
- GFP
green fluorescent protein
- LT50
time required to kill 50%
- NG
nematode growth
- PGS
peptone-glucose-sorbitol
References
- 1.Ghosh S, Baltimore D. Nature (London) 1990;344:678–682. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Preston-Hurlburt P, Janeway C A., Jr Nature (London) 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffmann J A. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 4.Hammond-Kosack K E, Jones J D G. Ann Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 5.Ganz T, Lehrer R I. Curr Opin Immunol. 1994;6:584–589. doi: 10.1016/0952-7915(94)90145-7. [DOI] [PubMed] [Google Scholar]
- 6.Broekaert W F, Terras F R G, Cammue B P A, Osborn R W. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann J A. Curr Opin Immunol. 1995;7:4–10. doi: 10.1016/0952-7915(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 8.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finley B B, Falkow S. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor C B. Plant Cell. 1998;10:873–876. doi: 10.1105/tpc.10.6.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 12.Rahme L G, Tan M-W, Le L, Wong S M, Tompkins R G, Calderwood S B, Ausubel F M. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elrod R P, Braun A C. J Bacteriol. 1942;44:633–645. doi: 10.1128/jb.44.6.633-645.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroth M N, Cho J J, Green S K, Kominos S D. In: Pseudomonas aeruginosa: Ecological Aspects and Patient Colonization. Young V M, editor. New York: Raven; 1977. pp. 1–29. [Google Scholar]
- 15.Lysenko O. J Insect Pathol. 1963;5:78–82. [Google Scholar]
- 16.Elrod R P, Braun A C. Science. 1941;94:520–521. doi: 10.1126/science.94.2448.520. [DOI] [PubMed] [Google Scholar]
- 17.Fink R B. Pseudomonas aeruginosa the Opportunist: Pathogenesis and Disease. Boca Raton, FL: CRC; 1993. [Google Scholar]
- 18.Andrew P A, Nicholas W L. Nematologica. 1976;22:451–461. [Google Scholar]
- 19.Devidas P, Rehberger L A. Plant Soil. 1992;145:115–120. [Google Scholar]
- 20.Holloway B W. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 21.Ishimoto K S, Lory S. Proc Natl Acad Sci USA. 1989;86:1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens E J, Ryan C M, Friedberg J S, Barnhill R L, Yarmush M L, Tompkins R G. J Burn Care Rehabil. 1994;15:232–235. doi: 10.1097/00004630-199405000-00005. [DOI] [PubMed] [Google Scholar]
- 23.Davis K R, Schott E, Ausubel F M. Mol Plant–Microbe Interact. 1991;4:477–488. [Google Scholar]
- 24.O’Toole G A, Kolter R. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 25.Weller D M, Cook R J. Phytopathology. 1983;73:463–469. [Google Scholar]
- 26.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts R C, Burioni R, Helinski D R. J Bacteriol. 1990;172:6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahana J A, Silver P A. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J D, Smith J A, Struhl K, editors. New York: Wiley; 1996. pp. 9.6.13–9.6.19. [Google Scholar]
- 29.King E O, Ward M K, Raney D E. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in Molecular Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 32.Ditta G, Stanfield S, Corbin D, Helinski D R. Proc Natl Acad Sci USA. 1980;27:7347–7451. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard A E, Vasil M L. J Bacteriol. 1986;176:291–298. doi: 10.1128/jb.167.1.291-298.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohman D E, Sadoff J C, Iglewski B H. Infect Immun. 1980;28:899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 36.Bardwell J C A, McGovern K, Beckwith J. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 37.Mahajan-Miklos, S., Tan, M.-W., Rahme, L. G. & Ausubel, F. M. (1999) Cell, in press. [DOI] [PubMed]
- 38.Hodgkin J, Horvitz H R, Brenner S. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward S, Miwa J. Genetics. 1978;88:285–303. doi: 10.1093/genetics/88.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avery L. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grewal P S. Nematologica. 1991;37:72–82. [Google Scholar]
- 42.Preston M J, Fleiszig S M J, Zaidi T S, Goldberg J B, Shortridge V D, Vasil M L, Pier G B. Infect Immun. 1995;63:3497–3501. doi: 10.1128/iai.63.9.3497-3501.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemaitre B, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]